Abstract
Cyanovirin-N (CV-N) is a cyanobacterial protein with potent neutralizing activity against human immunodeficiency virus (HIV). CV-N has been shown to bind HIV type 1 (HIV-1) gp120 with high affinity; moreover, it blocks the envelope glycoprotein-mediated membrane fusion reaction associated with HIV-1 entry. However, the inhibitory mechanism(s) remains unclear. In this study, we show that CV-N blocked binding of gp120 to cell-associated CD4. Consistent with this, pretreatment of gp120 with CV-N inhibited soluble CD4 (sCD4)-dependent binding of gp120 to cell-associated CCR5. To investigate possible effects of CV-N at post-CD4 binding steps, we used an assay that measures sCD4 activation of the HIV-1 envelope glycoprotein for fusion with CCR5-expressing cells. CV-N displayed equivalently potent inhibitory effects when added before or after sCD4 activation, suggesting that CV-N also has blocking action at the level of gp120 interaction with coreceptor. This effect was shown not to be due to CV-N-induced coreceptor down-modulation after the CD4 binding step. The multiple activities against the HIV-1 envelope glycoprotein prompted us to examine other enveloped viruses. CV-N potently blocked infection by feline immunodeficiency virus, which utilizes the chemokine receptor CXCR4 as an entry receptor but is CD4 independent. CV-N also inhibited fusion and/or infection by human herpesvirus 6 and measles virus but not by vaccinia virus. Thus, CV-N has broad-spectrum antiviral activity, both for multiple steps in the HIV entry mechanism and for diverse enveloped viruses. This broad specificity has implications for potential clinical utility of CV-N.
Cyanovirin-N (CV-N) is a protein from the cyanobacterium Nostoc ellipsosporum, first isolated in an effort to discover agents from natural extracts that block infection by human immunodeficiency virus (HIV) (5). The protein displays potent activity against diverse isolates of HIV type 1 (HIV-1), as well as HIV-2 and simian immunodeficiency virus (5, 12), but does not inhibit human herpesvirus 1 (HHV-1), cytomegalovirus, and adenovirus type 5 (5). In tissue culture cells, CV-N is nontoxic at concentrations much higher than those required for anti-HIV activity. Recombinant CV-N has been produced in Escherichia coli, and the recombinant protein is indistinguishable from its native counterpart (19). High-resolution structures have been obtained by both solution nuclear magnetic resonance spectroscopy (4) and X-ray crystallography (34). The biological and biochemical features of CV-N have led to the suggestion that it may have clinical utility against HIV, particularly as a topical agent to prevent sexual transmission (5, 12).
The mechanism(s) underlying the HIV-1-inhibitory activity of CV-N remain unclear. CV-N binds with high affinity to gp120, the external subunit of the HIV envelope glycoprotein (Env) (5); evidence suggests the anti-HIV-1 effects of CV-N are mediated through this interaction (5, 12). Moreover, CV-N inhibits the Env-mediated membrane fusion reaction associated with HIV entry into target cells (12). The molecular basis for this block to fusion and entry are poorly understood. There are reports in the literature arguing both for (12) and against (18) the notion that CV-N blocks binding of gp120 to CD4; moreover, it has been proposed that CV-N acts primarily at a post-CD4 step in the entry process, although this has not been directly examined (18).
In this study, we focus on the effects of CV-N at specific molecular stages of the HIV-1 Env-mediated fusion pathway. Fusion is a multistep process involving binding of gp120 to CD4, conformational changes in gp120 induced by CD4 binding, interaction of CD4-activated gp120 with a target cell coreceptor (a suitable chemokine receptor [CCR5, CXCR4, etc.]), and finally fusion between the viral and target cell membrane, mediated by the transmembrane gp41 subunit of Env (reviewed in references 3 and 33). Here, we show directly that CV-N blocks binding of HIV-gp120 to CD4; we also provide evidence for inhibition of the interaction of CD4-activated Env with coreceptor. Additionally, we show that CV-N potently inhibits other enveloped viruses that do not use the same target cell receptors for entry. The antiviral activity of CV-N is thus complex, displaying much broader specificity than previously indicated. The implications of these findings for potential clinical uses of CV-N are discussed.
MATERIALS AND METHODS
Cells.
Human HeLa and murine NIH 3T3 cells were obtained from the American Type Culture Collection. PM1 human T lymphocytes (17) were obtained from M. Norcross, Food and Drug Administration, Bethesda, Md. HEK293-CCR5 transfectant cells (9) were obtained from P. Murphy, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). All of the above cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 50 μg of gentamicin/ml. HEK293-CCR5 cells were maintained in the presence of Geneticin (2 mg/ml; Gibco BRL, Bethesda, Md.). The feline T-cell line MCH5-4 (16) and the brain-derived glial cell line G355-5 were kindly provided by Don Blair, Frederick Cancer Research and Development Center, NIH. Both cell lines were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, minimal essential medium (MEM)-vitamins, nonessential amino acids, sodium pyruvate, 200 μM l-glutamine, 5.5 × 10−5 M β-mercaptoethanol, and 50 μg of gentamicin/ml. The MCH5-4 cells were supplemented with recombinant human interleukin 2 (50 U/ml; a generous gift of Hoffmann-La Roche, Nutley, N.J.) and concanavalin A (2.5 μg/ml; Boehringer Mannheim, Indianapolis, Ind.).
Viruses.
All vaccinia virus recombinants were derived from the WR wild-type strain. Expression of foreign genes was driven by vaccinia virus promoters unless otherwise specified. The recombinant virus vCB-3 (8) encodes full-length human CD4; vCB-32 encodes SF162 Env (7); vT7-HMV encodes measles virus (MV) hemagglutinin and vT7-FMV encodes MV fusion protein, both linked to the bacteriophage T7 promoter (21); vTF7-3 (13) and vP11T7gene1 (1) both encode bacteriophage T7 RNA polymerase, and vCB21R-LacZ contains the E. coli lacZ gene linked to the bacteriophage T7 promoter (2). Two infectious molecular clones of feline immunodeficiency virus (FIV), namely, FIV-34TF10, originally isolated from a cat in Petaluma, Calif. (27), and FIV-PPR, an isolate obtained from a San Diego cat (22), were used in this study. HHV-6 GS is a prototype strain of subgroup A, initially isolated from the peripheral blood of immunocompromised patients with lymphoproliferative disorders (24).
Proteins and antibodies.
Purified recombinant CV-N (expressed in E. coli) (19) was used in this study. Purified gp120 protein (expressed in a stable, hygromycin-resistant Drosophila Schneider 2 cell line and affinity purified over an F105 column from the JR-FL strain of HIV-1) (32) was a gift from R. Wyatt, Dana Farber Cancer Institute, Boston, Mass.; the purified protein is active both for binding to CD4 and for CD4-induced binding to CCR5. Purified two-domain soluble CD4 protein (sCD4; amino acids 1 to 183) was donated by S. Johnson, Pharmacia Upjohn, Kalamazoo, Mich. Rabbit polyclonal serum 2143, raised against recombinant Env IIIB gp140, was gift of P. Earl, NIAID. T4-4 rabbit polyclonal sera (catalog no. 806) raised against recombinant human sCD4 was obtained from the NIAID AIDS Research and Reference Reagent Program. Several murine monoclonal antibodies (MAbs) were used. Phycoerythrin (PE)-conjugated anti-human CCR5 MAb 2D7 (isotype immunoglobulin G2a [IgG2a], κ) and PE-conjugated anti-human CXCR4 MAb 12G5 (isotype IgG2a, κ) were purchased from Pharmingen International, San Diego, Calif. MAbs against HHV-6 glycoproteins gp102 and gp116 were obtained from Virotech International, Inc., Rockville, Md.
Binding of gp120 to cell-associated CD4.
NIH 3T3 cells were infected with recombinant vaccinia virus vCB-3 (encoding full-length human CD4) at a multiplicity of infection of 10, using MEM plus Earle's balanced salts supplemented with 2.5% FBS (EMEM-2.5); control cells were infected with wild-type vaccinia virus WR. After overnight incubation at 31°C to allow surface expression of CD4, the cells were washed once with EMEM-2.5 and resuspended at the concentration of 4 × 106/ml. JR-FL gp120 (40 ng) was preincubated with different concentrations of CV-N in total 100 μl of EMEM-2.5 for 15 min at room temperature. The protein mix was then added to the cells (2 × 106 in a total volume of 500 μl) and incubated at 37°C for 1 h with constant rotation. The cells were then washed once with prechilled EMEM-2.5 and once with prechilled phosphate-buffered saline, pH 7.4 (PBS). The cell pellet was resuspended in lysis buffer containing 10 mM Tris-HCl (pH 8.0), 0.1% Triton X-100, 10 mM MgSO4, 2 mM CaCl2, and freshly added 1% aprotinin, 1 mM [4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride], 1 mM dithiothreitol, and 50 U of micrococcal nuclease/ml. Lysis was achieved by three repeats of freezing and thawing cycles. The lysate was further incubated with 25% volume of 5× sodium dodecyl sulfate sample buffer for 30 min at room temperature with intermittent vortexing. A fraction of the total cell lysate was then directly analyzed by polyacrylamide gel electrophoresis on a 10% gel, followed by Western transfer, probing for gp120 by using 1:7,000 dilution of the 2143 antiserum, and finally chemiluminescent detection of gp120 with horseradish peroxidase-conjugated goat anti-rabbit IgG (Boehringer Mannheim) and SuperSignal chemiluminescent substrates (Pierce Chemical Co., Rockford, Ill.). To further show specificity of gp120 binding, one aliquot of gp120 was preincubated with excess sCD4 (1 μM) before addition to the CD4-expressing cells.
Anti-CD4 antibody binding to cell-associated CD4.
NIH 3T3 cells infected with WR or vCB-3 were resuspended at 5 × 106 cells/ml in EMEM-2.5. Aliquots (100 μl) of cells were treated with 2 μl of T4-4 antiserum in the presence or absence of 100 nM CV-N and incubated 1 h at room temperature with constant rotation. Cell lysates were prepared as described above. Samples were resolved by polyacrylamide gel electrophoresis, and the anti-CD4 IgG heavy chain was detected by Western blotting with horseradish peroxidase-conjugated goat anti-rabbit IgG and SuperSignal chemiluminiscent substrates.
Binding of gp120 to coreceptor.
JR-FL gp120 (40 ng) was preincubated with different concentrations of CV-N for 15 min at room temperature and then activated by incubation with 600 nM of sCD4 in a total 100-μl volume of EMEM-2.5, first on ice for 30 min and then at room temperature for 30 min. The protein mix was added to 500 μl of HEK293-CCR5 cells (2 × 106 cells) and rotated for 1.25 h at room temperature. The cells were washed once with pre-chilled EMEM-2.5 and once with prechilled PBS. Preparation of cell lysate and detection of gp120 with 2143 antiserum by Western blotting are as described above.
Flow cytometric analysis.
Effects of CV-N on surface expression of HIV coreceptors were determined by flow cytometric analysis of HEK293-CCR5 transfectant cells and HeLa cells (endogenously expressing CXCR4). The cells (106) were first suspended in 100 μl of PBS without or with 250 nM of CV-N and incubated for 1 h at 37°C. Excess CV-N was removed by pelleting the cells and discarding the supernatant. The cell pellets were resuspended in 80 μl of PBS plus 20 μl of the indicated PE-conjugated MAb and incubated for 20 min at room temperature. After two washes in PBS, the cells were resuspended in 500 μl of PBS, followed by flow cytometric analysis using a FACScan analyzer (Becton Dickinson). Dead cells were excluded by appropriate gating. Approximately 6,000 events were accumulated for each sample.
HIV Env-mediated cell fusion.
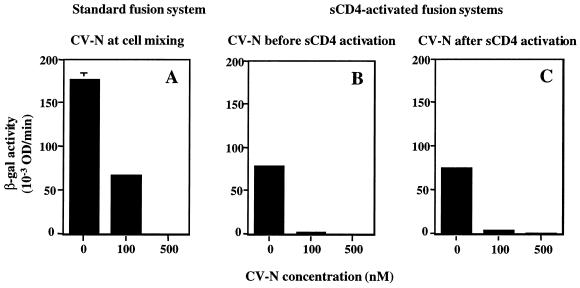
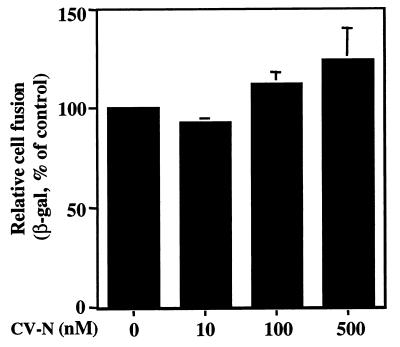
The effect of CV-N on HIV-1 Env-mediated cell fusion was analyzed with either the previously described standard reporter gene activation assay (20) or a modification of that assay involving sCD4 activation of Env for fusion with CD4-negative cells expressing a suitable coreceptor (25). Effector cells were prepared by infecting HeLa cells in suspension with the recombinant vaccinia virus vCB-32 (encoding the HIV-1 Env SF162) and vP11T7gene1 (encoding the bacteriophage T7 RNA polymerase gene driven by a vaccinia virus promoter). For the standard assay (Fig. 4A), target cells were prepared by infecting HEK293-CCR5 cells with two recombinant vaccinia viruses, vCB21R-LacZ (encoding lacZ linked to the T7 promoter) and vCB-3 (encoding human CD4). For the sCD4-activated assay (Fig. 4B and C), target cells were infected with vCB21R-LacZ alone. Following overnight incubation at 31°C to allow protein expression, effector and target cells were each washed and resuspended. For Fig. 4A, effector cells (100 μl, 2 × 106 cells/ml) were added to duplicate wells of 96-well plates and preincubated for 15 min at room temperature with 10 μl of PBS containing different concentrations of CV-N. Target cells (100 μl, 2 × 106 cells/ml) were then mixed with these effector cells. For Fig. 4B, effector cells were first incubated with CV-N for 15 min at room temperature, and then 200 nM (final concentration) sCD4 was added to these cells before mixing with the target cells. For Fig. 4C, effector cells were first incubated for 30 min at 37°C with 200 nM (final concentration) sCD4, followed by incubation with 200 nM CV-N and then mixing with target cells. The cell mixtures were incubated for 2.5 h at 37°C to allow fusion. The cells were then lysed with Nonidet P-40, and β-galactosidase (β-Gal) activity was measured at 570 nm in the presence of chlorophenol-red-β-d-galactopyranoside.
FIG. 4.
Inhibition of SF162 Env function by CV-N when added before or after sCD4 activation. HIV Env-mediated cell fusion was assayed. (A) CV-N was tested in a standard fusion assay using target cells expressing CD4 and CCR5. CV-N was added to effector cells before addition of target cells. (B and C) CV-N was tested in sCD4-activated fusion assay using target cells expressing CCR5 but not CD4. CV-N was added to effector cells before (B) or after (C) addition of sCD4 (200 nM). The indicated CV-N concentrations represent those in the final fusion mixtures. The background β-Gal activity value (0.53), obtained with CD4-negative target cells in the absence of CD4, was subtracted from each value to give the data shown. Error bars indicate standard deviation of the mean values obtained from duplicate samples. OD, optical density.
MV glycoprotein-mediated cell fusion.
NIH 3T3 cells were infected in suspension with the recombinant vaccinia viruses encoding the MV hemagglutinin and fusion glycoproteins and also with vTF7-3. PM1 cells (endogenously expressing CD46) were infected with vCB21R-LacZ. Following overnight incubation at 31°C to allow protein expression, effector and target cells were washed and resuspended. Effector cells (100 μl, 106 cells/ml) were added to duplicate wells of 96-well plates and preincubated for 30 min at 37°C with 50 μl of PBS containing different concentrations of CV-N. Target cells (50 μl, 2 × 106 cells/ml) were then mixed with these effector cells, and the plates were incubated for 2.5 h at 37°C. The cell lysates were assayed for β-Gal activity as described above.
HHV-6-mediated cell fusion and infection.
Adaptation of the cell fusion assay for HHV-6 has been reported elsewhere (26). Briefly, peripheral blood mononuclear cells (PBMCs), isolated from healthy donors, were activated with phytohemagglutinin M and 48 h later were infected with HHV-6 strain GS at a multiplicity of infection of 0.5. After the appearance of large, refractile cells (3 to 7 days postinfection), the cells were infected with vCB21R-LacZ. HeLa cells, which are permissive for HHV-6-mediated cell fusion (26), were infected with vTF7-3. PBMCs (100 μl, 106 cells/ml) were added to duplicate wells of 96-well plates and preincubated for 30 min at 37°C with 50 μl of PBS containing different concentrations of CV-N. HeLa cells (50 μl, 2 × 106 cells/ml) were then mixed with these effector cells. The cell mixtures were incubated for 2.5 h at 37°C to allow fusion. The cell lysates were assayed for β-Gal activity as described above.
For infectivity assays, activated PBMCs (106) were suspended in 350 μl of RPMI 1640 plus 10% FBS, in duplicate, and mixed with 50 μl of PBS containing different concentrations of CV-N at room temperature. Viral stocks (100-μl aliquots of HHV-6 strain GS) were added at a multiplicity of infection of 0.1 and incubated for 1.5 h at 37°C. The cells were washed once, resuspended in 450 μl of RPMI 1640 plus 10% FBS, and incubated with 50 μl of different dilutions of CV-N in PBS for 3 days at 37°C. The cells were then washed with PBS, resuspended in 50 μl of PBS, and dried onto microscopic slides. For immunodetection, the cells were fixed in acetone for 5 min at −20°C, followed by incubation for 20 min at room temperature with a mixture of MAbs to HHV-6 gp102 (final concentration of 125 μg/ml) and HHV-6 gp116 (final concentration of 25 μg/ml) diluted in PBS. The cells were washed in PBS, followed by incubation with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (10 μg/ml, final concentration; Boehringer Mannheim). The cells were washed as before, mounted with 50% glycerol in PBS, and photomicrographed using Bio-Rad MRC 1024 confocal laser scanning microscope.
Vaccinia virus-mediated low-pH fusion.
Based on the reported observation that brief exposure of vaccinia virus-infected cells to pH 5.5 results in cell-cell fusion (10, 14), we used a newly developed low-pH-induced vaccinia virus-based reporter gene assay (H. Greenstone and E. Berger, unpublished data). Briefly, HeLa cells infected with vCB21R-LacZ were mixed with HeLa cells infected with vTF7-3. The cell mixtures were incubated with different concentrations of CV-N for 15 min at room temperature, exposed briefly to pH 5.5, washed with neutral pH buffer, resuspended in EMEM-2.5 (neutral pH), and then incubated at 37°C for 2.5 h in presence of the same concentrations of CV-N. The cells were lysed with Nonidet P-40, and the β-Gal activity was measured as described above.
FIV infection.
MCH5-4 cells (106) were preincubated, in duplicate, for 30 min with 1 and 10 nM (final concentrations) CV-N in Hanks' balanced salt solution prior to the addition of FIV-PPR. The virus was removed after 1 h, and the cells were cultured for 5 days in presence of appropriate concentrations of CV-N. The negative control cells were incubated with the CV-N diluent. Viral load was quantitated using a reverse transcriptase assay as previously described (16). For pretreatment experiments, FIV-34TF10 and G355-5 cells were separately preincubated with 10 nM CV-N for 30 min. Then either CV-N-treated virus was added to G355-5 cells (106) or untreated virus was added to the CV-N-treated G355-5 cells. After 1 h, the cells were washed and then maintained in media without CV-N for 5 days. All samples were performed in duplicate and assayed for virus as above.
RESULTS
CV-N blocks the gp120-CD4 binding interaction.
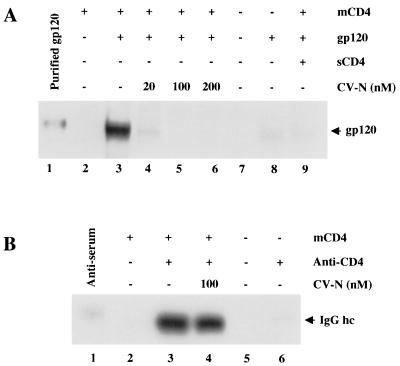
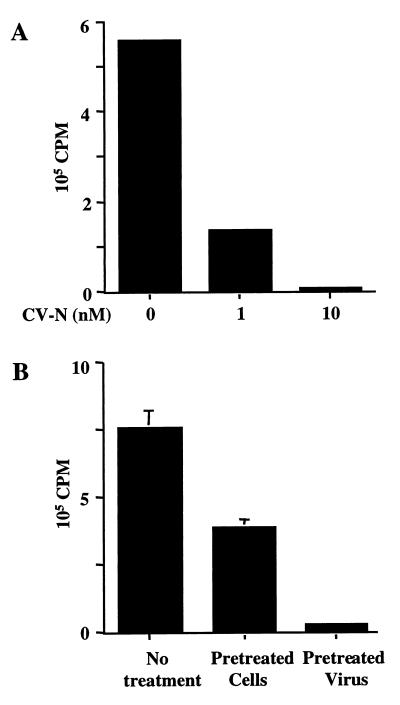
The first interaction of HIV-1 with specific target cell receptors involves the gp120 subunit of Env binding to cellular CD4. In an effort to understand the mechanism of CV-N inhibition of HIV-1 Env-mediated fusion and infection, we tested the effect of CV-N on binding of soluble gp120 to cell-associated CD4 (Fig. 1A). The gp120 protein used in this experiment was from the JR-FL isolate (primary R5, macrophagetropic, non-syncytium inducing). NIH 3T3 cells expressing vaccinia virus-encoded CD4 were incubated with JR-FL gp120, centrifuged, and washed; cell-associated gp120 was detected by Western immunoblot analysis. Since no CCR5 coreceptor is present on the target cell in this system, analysis of the gp120-CD4 interaction is simplified. As shown in Fig. 1A, JR-FL gp120 bound to CD4-expressing cells (lane 3). The specificity of binding was verified by the low background binding observed with CD4-negative cells (lane 8) and by the inhibition caused by excess sCD4 (lane 9). When gp120 was preincubated with CV-N, binding to CD4 was potently inhibited in a dose-dependent fashion (lanes 4 to 6).
FIG. 1.
Effect of CV-N on binding of gp120 and anti-CD4 antibodies to cell-associated CD4. (A) Binding of mock-treated (lane 3) or CV-N-treated (lanes 4 to 6) JR-FL gp120 to CD4-expressing (vCB-3-infected) cells was determined by Western blotting of whole cell lysates using polyclonal anti-gp120 antibody for detection. For lane 9, gp120 was preincubated with excess of sCD4. As negative control, CD4-negative (WR-infected) cells were incubated without (lane 7) or with (lane 8) gp120. The indicated CV-N concentrations represent those in the final incubation mixtures. Purified JR-FL gp120 (lane 1) was run as positive standard for immunodetection. mCD4, membrane-associated CD4. (B) Effect of CV-N on binding of anti-CD4 polyclonal antiserum to CD4-expressing (lanes 2 to 4) or CD4-negative cells (lanes 5 and 6) was determined by Western blotting of whole cell lysates using horseradish peroxidase-conjugated goat anti-rabbit IgG. The indicated CV-N concentrations represent those in the final incubation mixture. In lane 1, diluted antiserum was run as a positive standard. Migration of IgG heavy chain (hc) is indicated.
As an additional test of the specificity of CV-N inhibition of the gp120-CD4 interaction, we examined whether CV-N could block specific binding of unrelated proteins to CD4. As shown in Fig. 1B, polyclonal anti-CD4 antibodies bound to the same CD4-expressing cells used in Fig. 1A but not to the CD4-negative cells (compare lanes 3 and 6). CV-N had no inhibitory effect on this specific antibody binding (lane 4). Thus, the effects of CV-N on protein binding to CD4 showed specificity for gp120. Moreover, these results demonstrate that CV-N does not cause down-modulation of surface CD4; this conclusion was also verified by flow cytometry experiments indicating no effect of CV-N on binding of several anti-CD4 MAbs (BL4 and 13B8.2 [Immunotech Coulter] and Leu3A [Becton Dickinson]) to CD4 endogenously expressed on SupT1 cells (not shown).
CV-N blocks sCD4-induced binding of gp120 to the CCR5 coreceptor.
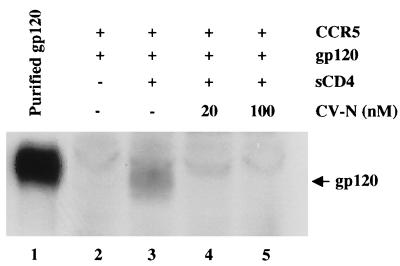
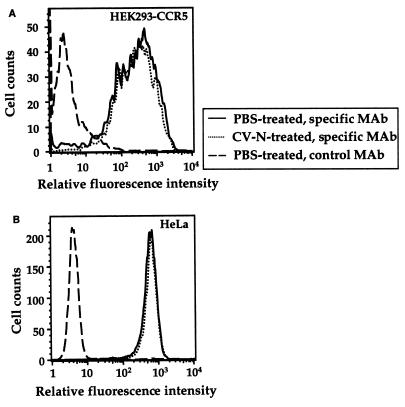
To elucidate whether CV-N can inhibit gp120 binding at other target cell sites involved in HIV-1 fusion and infection, we tested binding of gp120 from the JR-FL R5 isolate to HEK293-CCR5 cells that express CCR5 but not CD4. It has been shown previously that gp120 binds to CCR5 only upon activation by CD4 (28, 32). Therefore, purified gp120 was preincubated with sCD4 and then added to HEK293-CCR5 cells. Following incubation for 1 h at 37°C, the cells were centrifuged and washed; cell-associated gp120 was detected by Western immunodetection analysis. The sCD4-activated gp120 bound to the CCR5-expressing cells (Fig. 2, lane 3). The specificity of binding was confirmed by its strict dependence on sCD4 activation of gp120 (compare lanes 2 and 3). Preincubation of gp120 with 20 or 100 nM CV-N prior to sCD4 activation strongly blocked its binding to CCR5-expressing cells (lanes 4 and 5). We investigated the possible effect of CV-N on surface expression of the two major HIV coreceptors, CCR5 and CXCR4. As shown in Fig. 3A, flow cytometric analysis using the 2D7 anti-CCR5 MAb with HEK293-CCR5 cells showed no effect of 250 nM CV-N on CCR5 surface expression. Parallel experiments using the anti-CXCR4 MAb 12G5 demonstrated that CV-N had no effect on endogenous CXCR4 expression on HeLa cells (Fig. 3B). Taken together, the results in this and the previous section indicate that the CV-N blocking effects are manifested at distinct interactions between gp120 and its essential target cell receptors.
FIG. 2.
Effect of CV-N on CD4-dependent gp120 binding to target cell coreceptor. Binding of mock-treated (lanes 2 and 3) or CV-N-treated (lanes 4 and 5) JR-FL gp120 to CCR5-expressing cells, upon activation by sCD4, was determined by Western blotting of whole cell lysates using polyclonal anti-gp120 antibody for detection. Lane 1 shows purified gp120 used as positive control for immunodetection. The indicated CV-N concentrations represent those in the final incubation mixtures. The faint band above gp120 in lanes 2 to 5 is a cellular protein that nonspecifically reacts with the antiserum.
FIG. 3.
Effect of CV-N on cell surface expression of HIV coreceptors. Cells were incubated with PBS alone or PBS plus 250 nM CV-N prior to staining with the indicated MAbs, followed by flow cytometric analysis. (A) For detection of CCR5, HEK293-CCR5 cells were stained with the specific MAb (2D7, PE conjugated); as a negative control, the same cells were stained with an irrelevant isotype-matched MAb (12G5, PE conjugated). (B) For detection of CXCR4, HeLa cells were stained with the specific MAb (12G5, PE conjugated); control cells were stained with the isotype-matched 2D7 MAb, conjugated to PE.
CV-N blocks Env interaction with coreceptor before or after sCD4 activation.
The experiments described above do not distinguish whether the CV-N blocking of gp120-coreceptor binding is a direct effect or simply the consequence of CV-N blocking of sCD4 binding to gp120 and resulting activation for coreceptor binding. To address this issue, we used an experimental variation of the standard cell fusion system that enables analysis of discrete steps in the sequential cascade by which Env associates with CD4 followed by coreceptor (25). In this system, sCD4 activates Env-expressing cells to fuse with cells bearing coreceptor but lacking CD4. We used this sCD4-activated fusion system to characterize further the mechanisms by which CV-N can block Env functional interaction with target cell receptors (Fig. 4). The effector cells used in these experiments expressed the R5 HIV-1 Env from the SF162 strain; the target cells expressed CCR5 plus CD4 (standard fusion system [Fig. 4A]) or CCR5 alone (sCD4-activated system [Fig. 4B and C]). The results using the sCD4-activated system indicate similar fusion inhibition patterns irrespective of whether CV-N was present during the preincubation with sCD4 (Fig. 4B) or after the sCD4 activation (Fig. 4C). These findings suggest that CV-N can block not only the binding of gp120 to CD4 but also the CD4-induced binding of gp120 to coreceptor. Figure 4 also shows that the potency of CV-N in both modes in the sCD4-activated system was somewhat greater than that obtained in the standard system (Fig. 4A), consistent with similar potency difference obtained with fusion-blocking antibodies (25).
CV-N does not block vaccinia virus.
We performed a control experiment to rule out the possibility that the apparent CV-N inhibition of HIV-1 Env-mediated fusion is simply an artifact reflecting impairment of the vaccinia virus-based reporter gene read-out. We adapted the cell fusion assay to measure fusion mediated by vaccinia virus, an activity known to be induced by low-pH treatment of infected cells (10, 14). Figure 5 shows that CV-N had no inhibitory effect on vaccinia virus-mediated cell fusion, thereby indicating that the vaccinia virus-based reporter gene activation system was unaffected by CV-N. Moreover, the failure of CV-N to block vaccinia virus-mediated fusion provides added specificity to the previously described anti-HIV effects. However, the experiments in the following sections reveal that the blocking effects of CV-N are not restricted to HIV.
FIG. 5.
Effect of CV-N on vaccinia virus-mediated low-pH-induced cell fusion assay. The vaccinia virus-based low-pH-induced cell fusion assay was used. The background β-Gal activity value (27), obtained at neutral pH in the absence of CV-N, was subtracted from each value to give the data shown. The value of β-Gal activity (660) obtained at pH 5.5 in absence of CV-N was defined as 100%. The indicated CV-N concentrations represent those in the final incubation mixtures. Error bars indicate standard deviation of the mean values obtained from duplicate samples.
CV-N blocks infectivity of FIV, a CD4-independent, nonprimate immunodeficiency virus.
Since CV-N inhibits HIV at more than one step in the receptor interaction cascade, we questioned whether the effects of this agent might extend beyond HIV-1 (and its close relatives HIV-2 and simian immunodeficiency virus). We investigated the effect of CV-N on FIV, a nonprimate immunodeficiency virus that infects feline cells independent of CD4 (23) but strictly dependent on CXCR4 (6, 23, 30, 31). As shown in Fig. 6A, CV-N at concentrations as low as 10 nM strongly blocked FIV infection of CXCR4-expressing MCH5-5 cells. In view of the previous experiment (Fig. 3B) showing that CXCR4 surface expression is unaffected by CV-N, we conclude that the CV-N effects on FIV are not due to down-modulation of this coreceptor. Figure 6B demonstrates that the CV-N effects were much more pronounced with pretreatment of the FIV virions compared to pretreatment of the host cells, even though in both cases the CV-N was maintained in the virus-cell mixture during the 1-h preincubation. This result suggests that the inhibition resulted predominantly from the CV-N–virion interaction; similar conclusions have been reached for CV-N inhibition of HIV-1 (5, 12). In the FIV pretreatment experiments, modest inhibitory effects were consistently observed upon pretreatment of the host cells (Fig. 6B).
FIG. 6.
CV-N's effect on FIV infection. The viral load of FIV-infected cells cultured for 5 days was determined by a reverse transcriptase assay. All samples were done in duplicate. (A) Dose-dependent inhibition of FIV-PPR on the feline T-cell line MCH5-4. The infected cells were cultured in the presence of the indicated concentrations of CV-N. (B) Separate preincubation of either FIV-34TF10 or G355-5 feline glial cells with 10 nM CV-N. Infected cells were cultured in absence of CV-N. Error bars indicate standard deviation of the mean values obtained from duplicate samples.
Antiviral activity of CV-N extends beyond immunodeficiency viruses.
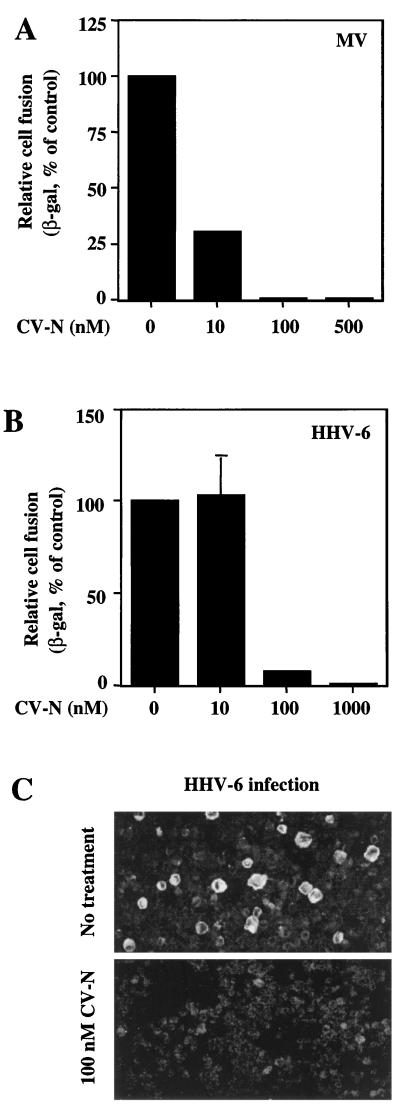
We further investigated the breadth of CV-N antiviral activity by studying other nonretroviral enveloped viruses, the entry of which does not involve either CD4 or chemokine receptors. MV employs CD46 as the receptor for entry and infection (11); this process is mediated by the specific interaction between the viral hemagglutinin and CD46 and displays no requirement for CD4. We previously used the reporter gene cell fusion assay to study the MV hemagglutinin and fusion glycoproteins (21). Figure 7A indicates that CV-N inhibited MV fusion, with a potency equivalent or greater than that observed for HIV-1 (Fig. 4A; also data in reference 12).
FIG. 7.
Inhibitory effects of CV-N on MV and HHV-6. For the experiments shown in panels A and B, the vaccinia virus-based reporter gene cell fusion assays were used. The indicated CV-N concentrations represent those in the final fusion mixtures. (A) Effect of CV-N on MV envelope glycoprotein-mediated fusion. The background β-Gal activity value (0.6), obtained with the effector cells expressing the hemagglutinin protein alone, was subtracted from each value obtained with effector cells expressing both hemagglutinin and fusion glycoproteins. The β-Gal activity (72.4) obtained from MV envelope glycoproteins-mediated fusion in absence of CV-N was defined as 100%. (B) Effect of CV-N on HHV-6-mediated cell fusion. As a background control, the β-Gal activities from effector cells (PBMCs) and target cells (HeLa) incubated alone were combined, and this value (2.48) was subtracted from each point to give data shown. The β-Gal activity (6.24) obtained from fusion in absence of CV-N was defined as 100%. In panels A and B, β-Gal values represent the means of duplicate samples; error bars indicate standard deviation of the mean values obtained from duplicate samples. (C) Effect of CV-N on HHV-6 infection. Infection of PBMCs was performed in the absence (top) or continuous presence (bottom) of 100 nM CV-N. After 3 days, the cells were assayed by immunofluorescence microscopy for the presence of the HHV-6 glycoproteins gp102 and gp116. The large, brightly stained cells in the top panel are typical of HHV-6 infection.
Finally we analyzed the effects of CV-N on HHV-6. This virus has recently been found to also use CD46 as its receptor for entry, fusion, and infection (26). We found that CV-N potently inhibited fusion of HHV-6-infected cells with permissive target cells (Fig. 7B), as well as acute HHV-6 infection of PBMCs (Fig. 7C).
DISCUSSION
We demonstrate herein that the cyanobacterial protein CV-N blocks multiple steps in the pathway of HIV-1 interaction with specific target cell receptors, leading to membrane fusion and virus entry. Previous efforts to assess these issues have yielded conflicting results. Mariner et al. (18) reported an enzyme-linked immunosorbent assay in which CV-N failed to block sCD4 binding to components in an HIV-1 lysate captured with polyclonal anti-gp120; also, the same study presented data indicating failure of CV-N to block HIV-1 binding to various CD4-positive T-cell lines. However, it was not verified that the binding activities measured in these experiments truly reflected the specific gp120-CD4 interaction. The effects of CV-N on gp120 binding to CD4 were subsequently addressed by Esser et al. (12). In experiments analyzing binding of different anti-gp120 neutralizing MAbs to virions or to monomeric gp120, no CV-N inhibitory effects were noted for MAb F105 or IgG1b12, both directed against the CD4 binding site. We note that the relationship of this result to the effects of CV-N on gp120-CD4 binding is indirect; moreover, the interpretation is complicated by the extensive conformational changes believed to be required for the high-affinity gp120-CD4 interaction (15, 33). Esser et al. (12) also presented flow cytometry experiments indicating that CV-N blocked HIV-1 virion-mediated occlusion of the Leu3A neutralizing epitope on CD4; moreover, CV-N inhibited the Leu3A-sensitive (CD4-dependent) component of virion binding to CD4-positive cells. The dose responses of these effects paralleled those observed for CV-N inhibition of HIV-1 infectivity. The simplest interpretation was that CV-N interferes with the gp120-CD4 interaction, though it was acknowledged that the presence of coreceptor on the CD4-expressing cell represented a caveat to this conclusion. Furthermore, this interpretation was difficult to reconcile with another experiment in the same report showing that CV-N did not impair sCD4-induced enhancement of exposure of certain gp120 epitopes. In the present study, we sought to resolve this issue by simple direct biochemical analyses. By using a Western blot assay verified to measure specific gp120 binding to cell-associated CD4, we demonstrate directly that CV-N specifically blocks the gp120-CD4 interaction in a potent, dose-dependent fashion. These results were not complicated by possible coreceptor interactions, since no coreceptor was present on the murine cells used in this experiment.
CV-N has also been proposed to affect gp120 function after the CD4 binding step (18), although this has not been experimentally tested. In this report, we directly analyzed the effect of CV-N on the binding of gp120 to coreceptor. In a simple biochemical assay, CV-N strongly blocked binding of sCD4-activated gp120 to cell-associated CCR5, an effect that was shown not to be due to coreceptor down-modulation. To distinguish whether this inhibition simply reflected CV-N blockade of CD4 binding to, and activation of, gp120, versus CV-N action at a subsequent stage, we used a functional assay measuring sCD4-activated, CCR5-dependent cell fusion. The ability of CV-N to block sCD4-activated fusion when added after, as well as before, sCD4 treatment suggests a direct effect on activated gp120 binding to coreceptor. However, we cannot exclude the possibility that the sCD4 binding and activation steps are largely reversible and in dynamic equilibrium; if so, CV-N added after sCD4 might bind to gp120 molecules as the sCD4 dissociates, thereby preventing sCD4 rebinding and activation. Additional experiments are required to address these complex kinetic issues. Acknowledging this caveat, our results suggest that CV-N can exert inhibitory effects through at least two discrete steps in the pathway of HIV-1 Env interaction with the specific target cell receptors. Presumably both activities contribute to the potency of CV-N against HIV-1 fusion/entry. We also note that the inhibitory effects may not be limited to receptor interactions. Indeed, there are indications that CV-N can interact with the gp41 subunit of Env (B. R. O'Keefe, J. M. Muschik, M. J. Currens, and M. R. Boyd, Abstr. Protein Society Meeting, Boston, Mass., p. 139, 1999); additional modes of inhibition can be envisioned (virion aggregation, Env destabilization, etc.).
Our HIV-1 findings prompted us to broaden the analysis of CV-N to additional enveloped viruses. An earlier report (5) demonstrated that the anti-HIV-1 effects of CV-N did not extend to several other viruses tested, specifically adenovirus type 5 (nonenveloped), human cytomegalovirus (enveloped), and HHV-1 (enveloped). We expanded these findings by showing that vaccinia virus-mediated fusion was not susceptible to CV-N inhibition. However, results were quite different for the other enveloped viruses tested. FIV infectivity was potently blocked by CV-N; this virus uses the chemokine receptor CXCR4 as a primary entry receptor (6, 23, 30, 31), thus providing another illustration of CV-N effects distinct from CD4 binding. Similarly susceptible to CV-N inhibition in fusion and/or infectivity analyses were MV and HHV-6, both of which use CD46 as an entry receptor.
We have also considered the possibility that target cell effects might contribute to the antiviral activities of CV-N. The FIV experiments indicated much more pronounced activity with pretreatment of virions compared to host cells, although some inhibitory effects were consistently observed in the latter case. In these experiments, CV-N was present not only in the preincubation stage but also in the subsequent interaction stage between virion and target cells; the inherent difficulty of physically manipulating retrovirus particles precluded the use of protocols involving extensive virion washing before adding to target cells. In additional experiments (not shown), we analyzed HIV-1 Env-mediated cell fusion, which permits direct comparison of the effects of pretreatment and washing of target versus effector cells. CV-N inhibited only modestly when either cell partner was pretreated and washed prior to initiation of the fusion reaction (in contrast to the strong inhibition observed when CV-N was maintained throughout the fusion reaction); moreover, the inhibitory effects were comparable with pretreatment of either the target or effector cells. These results argue against predominant irreversible effects of CV-N on components of either cell partner.
In view of the complexity of CV-N action, what might be the explanation(s) for the inhibitory effects of CV-N against multiple viruses? Esser et al. (12) found that of all the anti-gp120 neutralizing MAbs tested, CV-N blocked the binding of only one, 2G12. This MAb is known to be directed against a conserved conformational epitope that is highly dependent on glycosylation (29), leading to the suggestion that CV-N and 2G12 might act similarly (though not identically). Perhaps the broad effects of CV-N on diverse enveloped viruses simply reflect the protein's affinity for carbohydrate moieties that are extensively represented on many viral envelope glycoproteins. However, it must be noted that the evidence for CV-N interaction with carbohydrate moieties on gp120 is only indirect (12); additional studies are required to determine if specific CV-N–carbohydrate interactions are essential for the antiviral activities. We also note that the carbohydrate model does not easily explain the distinctions between the potent effects seen with HIV, FIV, MV, and HHV-6, versus the negligible effects shown here for vaccinia virus and previously reported for other enveloped viruses (human cytomegalovirus and HHV-1); each of these viruses have glycosylated envelope proteins.
The present results may have important implications for potential clinical applications of CV-N. The protein is minimally toxic to tissue culture cells (5, 12); moreover, antiviral concentrations in blood have been achieved and sustained in vivo in rodents, with minimal toxicity (National Cancer Institute, Developmental Therapeutics Program, unpublished data). Initial studies (NIAID, unpublished data) showing benign effects of CV-N in a rabbit vaginal toxicity model are also encouraging. Additional studies are required to assess whether the broad inhibitory activity of CV-N revealed in this report bode well or poorly for the clinical utility of CV-N.
ACKNOWLEDGMENTS
We thank R. Wyatt for providing purified JR-FL gp120, P. Kennedy for assistance in HHV-6 fusion assay, J. Yewdell for helping with confocal microscopy, H. Greenstone for guidance on low-pH fusion, and K. Salzwedel for helpful comments on the manuscript.
This work was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Broder C C, Berger E A. Cell-type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Bewley C A, Gustafson K R, Boyd M R, Covell D G, Bax A, Clore G M, Gronenborn A M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- 5.Boyd M R, Gustafson K R, McMohan J B, Shoemaker R H, O'Keefe B R, Mori T, Gulakowski R J, Wu L, Rivera M I, Laurencot C M, Currens M J, Cardellina I J H, Buckheit J R W, Nara P L, Pannell L K, Sowder I R C, Henderson L E. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential application to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brelot A, Heveker N, Adema K, Hosie M J, Willet B, Alizon M. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J Virol. 1999;73:2576–2586. doi: 10.1128/jvi.73.4.2576-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broder C C, Dimitrov D S, Blumenthal R, Berger E A. The block to HIV-1 envelope glycoprotein-mediated membrane fusion in animal cells expressing human CD4 can be overcome by a human cell component(s) Virology. 1993;193:483–491. doi: 10.1006/viro.1993.1151. [DOI] [PubMed] [Google Scholar]
- 9.Combadiere C, Salzwedel K, Smith E D, Tiffany H L, Berger E A, Murphy P M. Identification of CX3CR1. A chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J Biol Chem. 1998;273:23799–23804. doi: 10.1074/jbc.273.37.23799. [DOI] [PubMed] [Google Scholar]
- 10.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorig R E, Marcil A, Chopra A, Richardson C D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 12.Esser M T, Mori T, Mondor I, Sattentau Q J, Dey B, Berger E A, Boyd M R, Lifson J D. Cyanovirin-N binds to gp120 to interfere with CD4-dependent human immunodeficiency virus type 1 virion binding, fusion, and infectivity but does not affect the CD4 binding site on gp120 or soluble CD4-induced conformational changes in gp120. J Virol. 1999;73:4360–4371. doi: 10.1128/jvi.73.5.4360-4371.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 15.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerner D L, Grant C K, de Parsaval A, Elder J H. FIV infection of IL-2 dependent and independent feline lymphocyte lines: host cell range distinctions and specific cytokine upregulation. Vet Immunol Immunopathol. 1998;65:277–297. doi: 10.1016/S0165-2427(98)00162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lusso P, Cocchi F, Balotta C, Markham P D, Louie A, Farci P, Pal R, Gallo R C, Reitz M S., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariner J M, McMohan J B, O'Keefe B R, Nagashima K, Boyd M R. The HIV-inactivating protein, cyanovirin-N, does not block gp120-mediated virus-to-cell binding. Biochem Biophys Res Commun. 1998;248:841–845. doi: 10.1006/bbrc.1998.9060. [DOI] [PubMed] [Google Scholar]
- 19.Mori T, Gustafson K R, Pannell L K, Shoemaker R H, Wu L, McMohan J B, Boyd M R. Recombinant production of cyanovirin-N, a potent human immunodeficiency virus-inactivating protein derived from a cultured cyanobacterium. Protein Expression Purif. 1998;12:151–158. doi: 10.1006/prep.1997.0838. [DOI] [PubMed] [Google Scholar]
- 20.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nussbaum O, Broder C C, Moss B, Stern L B, Rozenblatt S, Berger E A. Functional and structural interactions between measles virus hemagglutinin and CD46. J Virol. 1995;69:3341–3349. doi: 10.1128/jvi.69.6.3341-3349.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips T R, Talbott R L, Lamont C, Muir S, Lovelace K, Elder J H. Comparision of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poeschla E M, Looney D J. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–6866. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 25.Salzwedel K, Smith E, Dey B, Berger E. Sequential CD4/coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoro F, Kennedy P E, Locatelli G, Malnati M S, Berger E A, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 27.Talbott R L, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 29.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willet B J, Hosie M J, Neil J C, Turner J D, Hoxie J A. Common mechanism of infection by lentiviruses. Nature. 1997;385:587. doi: 10.1038/385587a0. [DOI] [PubMed] [Google Scholar]
- 31.Willet B J, Picard L, Hoxie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 33.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 34.Yang F, Bewley C A, Louis J M, Gustafson K R, Boyd M R, Gronenborn A M, Clore G M, Wlodawer A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J Mol Biol. 1999;288:403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]