Abstract
Small non-coding RNAs (sncRNAs), such as microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), and transfer RNA (tRNA)-derived small RNAs (tsRNAs), play essential roles in regulating various cellular and developmental processes. Over the past three decades, researchers have identified novel sncRNA species from various organisms. These molecules demonstrate dynamic expression and diverse functions, and they are subject to intricate regulation through RNA modifications in both healthy and diseased states. Notably, certain sncRNAs in gametes, particularly sperm, respond to environmental stimuli and facilitate epigenetic inheritance. Collectively, the in-depth understanding of sncRNA functions and mechanisms has accelerated the development of small RNA-based therapeutics. In this review, we present the recent advances in the field, including new sncRNA species and the regulatory influences of RNA modifications. We also discuss the current limitations and challenges associated with using small RNAs as either biomarkers or therapeutic drugs.
Keywords: Small non-coding RNAs, RNA modifications, Human diseases, RNA therapeutics, Biomarkers
1. Introduction
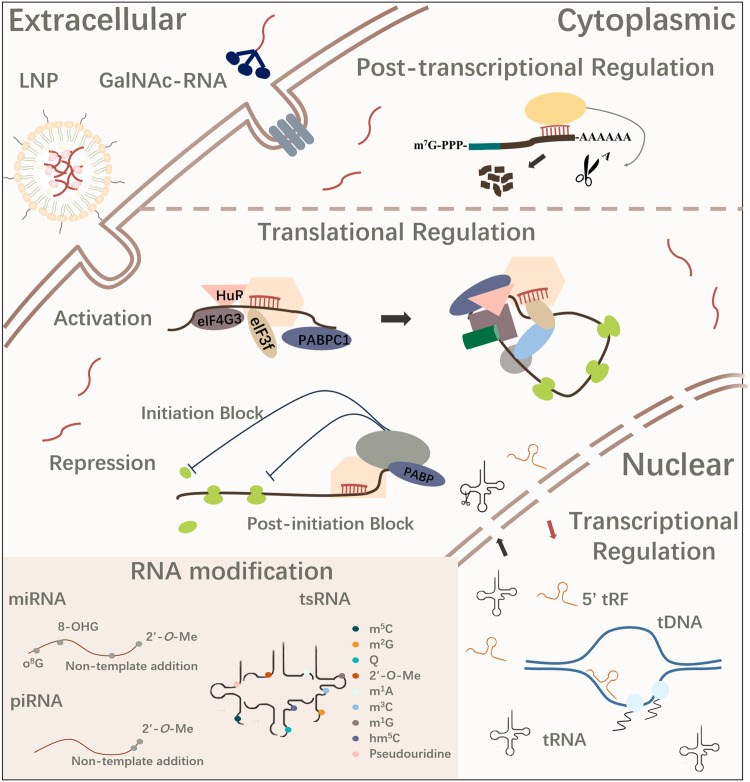
Small non-coding RNAs (sncRNAs) refer to non-coding RNAs (ncRNAs) less than 200-nucleotide (nt) and were first discovered in C. elegans nearly three decades ago [1,2]. Since then, the kingdom of sncRNAs has rapidly expanded with increasing numbers of novel species identified in various organisms. SncRNAs are broadly classified into two categories: the housekeeping ones, including small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) [3], and the regulatory ones, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs). Such regulatory sncRNAs are known to play critical roles in gene expression at post-transcriptional, translational, and epigenetic levels. Recent advances in next-generation sequencing technologies have identified new sncRNA species derived from other RNA molecules, such as transfer RNAs (tRNAs) [4,5] and ribosomal RNAs (rRNAs) [6]. While the functions of such sncRNAs require further elucidation, their involvement in specific biological processes has been suggested. Additionally, studies have demonstrated that RNA modifications play an important role in the biogenesis and functionality of sncRNAs, thus shaping their regulatory roles. These findings increase the potential of sncRNAs in the diagnosis, prognosis, and therapeutics for human diseases. In this review, we summarize the recent advances and breakthroughs within the field, including novel sncRNA species and the regulatory roles of RNA modifications to the biogenesis and functionality of sncRNAs. Moreover, we discuss the potential applications of sncRNAs in clinical practice (Fig. 1).
Fig. 1.
Schematic diagram summarizing the regulatory roles of sncRNAs, modifications and delivery approaches for RNA therapeutics. sncRNAs are key regulators for various biological processes, which function to regulate gene expression at transcriptional, post-transcriptional, and translational levels. Multiple sncRNA species have been identified with advanced detection and annotation methods, among which many are carried with various types of chemical modifications, adding another layer of complexity to the sncRNA kingdom. sncRNAs have strong potentials in therapeutics, and a few sncRNA-based therapeutics have been approved by FDA since 2018.
2. Sequential and functional diversity of sncRNAs
The most intensely researched classes of endogenous sncRNAs are miRNAs, endogenous small interfering RNAs (endo-siRNAs), and piRNAs. The mechanisms that distinguish these three classes from one another are primarily based on the differences in their biogenesis and protein partners. To create miRNAs, RNase III enzymes Drosha and Dicer cleave hairpin-shaped precursors whereas endo-siRNAs are generated from double-stranded RNA (dsRNA) through Dicer [7,8]. On the other hand, the precursors of piRNAs are single-stranded. Their production is independent of Drosha and Dicer [9,10]. miRNAs (∼22-nt) and endo-siRNAs (∼21-nt) form complexes with AGO-family proteins such as AGO2, which mediate post-transcriptional regulation by inducing RNA cleavage [11] and translation repression [8,12]. piRNAs (24–32-nt), as their name indicates, are mainly associated with PIWI proteins, a germline-specific AGO subfamily. piRNAs primarily repress transposable elements (TE) at both post-transcriptional and epigenetic levels [13]. Furthermore, piRNAs also repress protein-coding genes in animal germlines via siRNA and miRNA-like mechanisms [14], [15], [16]. Newly emerging roles and functional mechanisms of these well-researched sncRNAs are still continually discovered. For instance, recent studies highlight that piRNAs and PIWI proteins promote the activation of translation in mice and flies, and the underlying molecular mechanisms are remarkably conserved between the two species. In mouse spermatids and fly embryos, PIWI proteins interact with translation initiation factor 3 (eIF3) subunits and other RNA-binding proteins, forming a translation-activating complex [17,18]. In mouse spermatids, the selectivity of this complex depends on a specific base pairing between piRNAs and the 3′ UTR of their target mRNAs [17]. Additionally, a new class of piRNAs was recently identified in human, monkey, and hamster oocytes, which are associated with rodent-lacking PIWIL3 and termed oocyte short piRNAs (os-piRNAs) [19], [20], [21].
In addition to the increasing understanding of well-known sncRNA species, the sncRNA realm is expanding owing to advanced detection and annotation methods. NGS (Next-Generation Sequencing) is probably the most widely used method for the detection and annotation of sncRNAs. Many sncRNA species that were initially overlooked during earlier NGS data analysis are mappable to longer and often structured RNA species [22]. Apart from tsRNAs and rsRNAs, fragments originating from yRNAs (a class of soluble ribonucleoproteins (Ro RNPS)-associated non-coding RNAs. The “y” prefix is used to emphasize their primarily cytoplasmic localization and to distinguish them from nuclear-localized snRNAs such as U6) [23], [24], [25], vault RNAs (vtRNAs) [26], snRNAs [27], snoRNAs [28,29], long non-coding RNAs (lncRNAs) [30], and mRNAs [31,32] have also been recently reported. Although most small RNA sequencing approaches aimed to profile well-studied sncRNAs (such as miRNAs), improvements in library preparation, in particular removal of RNA modifications interfering with adaptor ligation and/or reverse transcription (RT), have paved the way for new advancements in the sncRNA landscape [33]. One example is the recently developed panoramic RNA display by overcoming RNA modification aborted sequencing (PANDORA-seq) [34] which utilizes a combination of enzymes to remove RNA methylation and 3′-phosphate (3′-P) of sncRNAs. Bioinformatics analysis is an integral part of small RNA sequencing. It is no surprise that newly built specialized computer tools and pipelines have improved the speed and achieved better identification and annotation of different types of sncRNAs, in particular the non-canonical ones [35,36]. Nevertheless, accurately quantifying and comparing those novel sncRNAs between various conditions, as well as identifying them in low-input scenarios like early embryos or extracellular biofluids, remain significant challenges.
Comprehending the functions of sncRNAs necessitates understanding their biogenesis, tissue abundance, and biological importance. A portion of sncRNAs is derived from tRNAs and rRNAs, demonstrating that tsRNAs and rsRNAs could be functional sncRNAs [37], [38], [39], [40]. Recent studies have likewise shown tsRNAs and rsRNAs are abundant in multiple tissues [34], implying their biological significance. The cleavage of mature tRNAs or tRNA precursors can occur on several sites, contributing to the generation of diverse tsRNAs. Both the tRNA structure and the RNA modifications on them, such as DNMT2 and NSUN2-mediated 5-methylcytosine (m5C) modification [41], [42], [43], [44], [45], [46], contribute to tRNA cleavage site preference. Similar to tsRNAs, the biogenesis of rsRNAs is also under strict regulation. Various preferred cleavage sites in rRNAs are often observed, depending on the rRNA types. Hence, different rRNAs can generate rsRNAs with favored lengths, leading to highly context-specific profiles of rsRNAs [47], [48], [49]. In particular, the tsRNA and rsRNA population in mammalian sperm has become a leading topic in the field [50,51]. In 2016, two separate groups found that the 30–40 nt small RNA population in mature sperm, primarily composed of tsRNAs and rsRNAs, was sensitive to environmental stimuli. More importantly, these sncRNAs could be carried into oocytes during fertilization and presumably play a part in the intergenerational inheritance of acquired traits [34,52,53]. The majority of tsRNAs in mammalian sperm is derived from 5′ half of mature tRNAs, termed 5′-tRNA halves (5′ tRNAF or 5′ tRFl) [54]. On the other hand, the majority of rsRNAs are derived from 28S rRNA [51]. Subsequent studies using different lifestyle models, as well as environmental stress and toxicants, have further validated the involvement of sperm-borne tsRNAs in epigenetic inheritance [53,[55], [56], [57], [58], [59]]. Despite the proposal that epididymis and epididymosomes might be potential sources [60], the biogenesis of sperm-borne sncRNAs remains a myth. tsRNAs have a varied range of functions, including Argonaute (Ago) family-dependent gene silencing [61], [62], [63], translational inhibition [64], [65], [66], and retrotransposon repression [67]. A recent study further indicates some 5′-tRNA halves are vital for early embryonic development in zebrafish, where they may enhance tRNA transcription by forming stable, transcriptionally inhibitory hybrids with the template strand of DNA, competing with existing tRNA [68]. This suggests that RNA-derived fragments may function by mimicking their precursors based on the sequence and structure similarity [67,69]. Although tsRNAs [70] and rsRNAs [47] are now thoroughly profiled, the molecular mechanisms underlying their roles in different biological processes remain largely unknown, including epigenetic inheritance. Moreover, knowledge about additional types of RNA-derived fragments, such as snRNA- and snoRNA-derived sncRNAs, is limited. Some of these RNA-derived fragments were mistakenly identified as miRNAs [71] or piRNAs [72], but their unique biogenesis, cellular abundance, and functionality remain unknown. Although intriguing, it will be a challenge to comprehensively capture those sncRNAs in various biological situations and decipher their complex information in future studies [33].
3. RNA modifications and structures: the additional layers of complexity in the sncRNA kingdom
The complexity of the sncRNA field isn't solely due to diversity in sequence and genomic sources. Various factors have been discovered over the past decade, influencing sncRNA functionality. Among these factors, several well-characterized RNA modifications can regulate sncRNA function (Table S1), particularly in mammalian germ cell development and epigenetic inheritance.
To date, more than 150 chemical modifications, termed ‘epitranscriptomics’, have been identified in different RNA species including sncRNAs [73]. Advances in NGS methods for detecting RNA modifications demonstrate that RNA modifications play key roles in RNA metabolism and functions. Alterations in RNA modifications may lead to abnormalities in biological processes and cause diseases such as cancers [74,75]. In particular, sncRNAs and their precursors have multiple modifications, such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), ADAR-mediated adenosine-to-inosine (A-to-I) editing, non-templated uridine addition and pseudouridylation (Ψ). The m6A may enhance the recognition and processing of primary miRNAs (pri-miRNAs) by DGCR8, promoting the maturation of miRNAs [76,77]. Conversely, miRNAs may affect m6A abundance on mRNAs by modulating the activity of METTL3, an m6A writer [78]. A-to-I editing, the most studied RNA editing form, produces an RNA sequence different from its template DNA. A-to-I editing is catalyzed by the Adenosine deaminase (AD) domain-containing proteins ADAR1 and ADAR2 in humans (ADAR and ADARB1 in mice) [79]. ADAR-mediated A-to-I editing is quite prevalent in miRNAs and their precursors [80] and is known to affect miRNA processing [81,82] as well as miRNA-mRNA interaction [83,84]. However, A-to-I editing, although essential for multiple biological events including embryonic development [82], appears to be dispensable for mammalian male fertility [85]. Non-templated addition of nucleotides (NTA), mainly consisting of 3′ mono- and poly-adenylation or uridylation, is generated by terminal nucleotidyltransferases (TENTs). NTA occurs on multiple RNA species, including miRNAs and snoRNAs [86], and is involved in the regulation of miRNA processing [87], [88], [89], interaction with target mRNAs [90], and stability [91,92]. Importantly, NTA is essential for miRNA functionality during various processes such as embryonic development [93,94]. In mouse male germ cells, the loss of TUT4 and TUT7, which are the primary uridylyl transferases, resulted in a significant loss of 3′ uridylation of piRNAs [95]. During spermatogenesis, 3′ uridylation, with mono-uridylation as a major form, enhances the targeting efficacy of piRNAs bound to MIWI [96]. Intriguingly, 3′ uridylation appears to compete with the signature 2′-O-methylation modification at the 3′ end of piRNAs. Hen1 (Henmt1 in mammals) deposits 2′-O-methylation on the 3′ terminal of piRNAs [97], [98], [99], [100], [101]. Loss of 2′-O-methylation is often linked with increased 3′ uridylation and enhanced degradation [102], [103], [104], [105]. However, mono-uridylation of MIWI-bound piRNAs can co-exist with 2′-O-methylation [96], which suggests that HENMT1 may compete with TUT4/7, leading to most mono-uridylated piRNAs immediately methylated by HENMT1 to prevent further extension of the 3′ tailing. tRNAs and tsRNAs might be the sncRNA species that are most heavily decorated with RNA modifications. Up to 39 types of modifications have been identified in human tRNAs and 50 tRNA modification enzymes have been deposited in MODOMICS [73,106,107]. These modifications provide a regulatory mechanism for tRNA functions and stability, including tRNA cleavage or generation of tsRNAs. For example, m5C modification, catalyzed by DNMT2 or NSUN2, promotes tRNA stability by inhibiting the binding of endonuclease angiogenin (ANG), thereby protecting the tRNAs from being cleaved into tsRNAs [44,45]. Loss of DNMT2 leads to dysregulation of tsRNAs and rsRNAs in spermatozoa, and in turn, impairs the transmission of diet-induced metabolic disorders [44]. N1-methyladenine (m1A), N3-methylcytidine (m3C), and N1-methylguanine (m1G) modifications on tRNAs have similar ANG-inhibiting roles and thus repress the generation of tsRNA [108], [109], [110], [111]. TET2, which converts the m5C on tRNAs to 5-hydroxymethylcytosine (hm5C), enhances the generation of 5′ tRFs (i.e. tRFs containing the 5′ sequence of tRNAs) but surprisingly also represses the generation of some 3′ tRFs (i.e. tRFs containing the 3′ sequence of tRNAs). tsRNAs generated from modified tRNAs can inherit those chemical modifications, although it remains elusive whether certain tsRNAs might also be targets of some tRNA modifiers. Multiple RNA modifications, such as m5C, N2-methylguanosine (m2G), m6A, and pseudouridylation, have been detected in tsRNAs in different biological samples, including mature sperm in mammals [44]. Pseudouridylation at position 8 (Ψ8) of tRFs, potentially mediated by Pseudouridine synthase 7 (PUS7) and additional factors [112], was shown to repress translation initiation by sequestering polyadenylate-binding protein 1 (PABPC1), a key translation factor [66]. This suggests that the chemical modifications in tsRNAs may have unique biological relevance beyond being inherited remnants from tRNAs.
4. The clinical application of small RNAs in human diseases
4.1. sncRNAs as biomarkers in diagnosis and prognosis
sncRNAs have great potential to be novel biomarkers for various diseases, including cancers [113], neurological [114,115], metabolic [116], cardiovascular [117], reproductive [118,119], and infectious diseases [120,121]. sncRNAs in biological fluids, which are often encapsulated in exosomes or bound with carrier proteins [122], [123], [124], can be stable and abundant enough for use as clinical biomarkers. Clinical usage of these extracellular sncRNAs is an especially appealing concept in liquid biopsy. Studies have profiled the sncRNA population in tissues and biological fluids (such as blood and seminal plasma) and identified particular sets of sncRNAs of which the abundance is strongly correlated with diagnosis, prognosis, and treatment in human diseases. While current studies primarily focus on miRNAs in liquid biopsy as biomarkers, as technology improves, other sncRNA species may become applicable as biomarkers in clinics. For example, multiple piRNAs have been suggested as diagnostic or prognosis biomarkers in cancers [125]. The quality of in vitro cultured embryos may also be significantly associated with a panel of miRNAs, tsRNAs, and rsRNAs in sperm. Thus, this specific set of sncRNAs in sperm may be considered biomarkers for successful in vitro fertilization (IVF) [126]. Furthermore, sncRNAs, including miRNAs and piRNAs, have also been detected in the used culture media of IVF embryos, indicating that in vitro cultured embryos secret sncRNAs-containing extracellular vesicles [127]. As a result, the panels of sncRNAs from the spent culture media may have an indicative potential for embryo quality, such as ploidy and pregnancy outcomes [128], [129], [130], [131], [132], [133], [134].
The major challenge in clinical usage is that technical differences in sample handling procedures (e.g. contaminations and degradation) and detection methods (e.g. qRT-PCR, microarray, or NGS) [135] can readily affect the sncRNA profile. Therefore, the sncRNA signatures reported by different studies tend to be quite different and even contradictory. Cross-validation is crucial to determine the clinical utility of sncRNA signatures, and standardization of handling and detection procedures is much needed. Once cross-validated, these sncRNA signatures can be effective molecular targets for early diagnosis and effective treatment. In addition, the sncRNA profiles of spermatozoa, which are sensitive to environmental disturbances such as cigarettes [136,137], alcohol [138,139], endocrine disruptors [140,141], and air pollution [142], can reflect parental exposures. Furthermore, the sncRNA profiles of seminal plasma and follicular fluid may respond to parental exposures. Although not affecting sperm morphology, the sperm RNA code may still transmit adverse information to the next generations in response to environmental exposures. Therefore, the sncRNA profiles of sperm, seminal plasma, and follicular fluid may have the potential to serve as indicators for certain exposures in parents, which would be highly valuable in understanding disease predisposition in the Developmental Origins of Health and Disease (DOHaD) field.
4.2. Small RNA-based therapeutics
There have been significant advancements in the field indicating that sncRNAs have strong therapeutic potential. RNA-based therapy is an active and promising area of pharmaceutical development, which expands the range of druggable targets from disease-causing proteins to RNAs. Currently, there are 11 RNA-based therapeutics that have been approved by the FDA and/or the European Medicines Agency (EMA), and a few more are in late-stage clinical development (beyond phase II trial) [143], [144], [145]. Among the approved RNA-based drugs, 4 are double‐stranded siRNA that exploit the endogenous RNAi pathway to silence target genes. The first RNAi drug that was approved by the FDA was Patisiran (Onpattro) [146,147], which was followed by Givosiran in 2019 [148], Lumasiran in 2020 [149] and Inclisiran in 2021 [150] (Table S2). These approvals marked the beginning of a new generation of RNA therapeutics.
Compared to small molecule drugs, The hydrophilic nature and larger size of unmodified RNAs make it difficult for them to efficiently move across cell membranes. In addition, naked RNAs are prone to degradation by RNases and unlikely delivered into specific tissues. Therefore, designing RNA-based drugs must consider key properties like Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET), similar to other fields of pharmaceutical development. To improve the ADMET properties, a variety of delivery approaches have been developed for RNA-based drugs [143,151,152]. Patisiran, which is administrated intravenously, is encapsulated in a lipid nanoparticle (LNP) that protects the siRNA during plasma circulation. The LNP absorbs apolipoprotein E (ApoE), the ligand of low-density lipoprotein receptor (LDLR), which leads to a selective accumulation of patisiran in hepatocytes that express a high level of LDLR [153,154]. The other three RNAi drugs adopt alternative delivery approaches. Instead of being incorporated into nanoparticulate carriers, the therapeutic siRNA is directly conjugated to trimeric N‐acetyl galactosamine (GalNAc) which is a tissue-targeting ligand that binds to asialoglycoprotein receptor (ASGPR) primarily expressed on hepatocytes, allowing effective and specific transport of the drug into the liver [[148], [149], [150],155].
Aside from siRNAs, microRNA-based drugs, including microRNA mimics and antagomiR (i.e. microRNA inhibitors), show promise as therapeutic agents [144,156,157]. While no miRNA-based candidate drug has been approved by the FDA, a handful have reached phase II clinical trials (Table S2). As one miRNA can target multiple mRNAs often involved in one pathway, miRNA-based therapeutics, compared to siRNA-based therapeutics, may elicit a broader therapeutic effect. In contrast to gene silencers such as siRNAs and microRNAs, small activating RNAs (saRNAs) are double-stranded RNAs that activate target genes, making them a valuable addition to RNA therapeutics [158,159]. The first and currently the only saRNA drug entering clinical trials is MTL-CEBPA which upregulates C/EBP-α, a master transcriptional factor that regulates hepatic and myeloid functions. MTL-CEBPA is being developed to treat advanced hepatocellular carcinoma (HCC). [160,161].
5. Conclusion and future perspective
Over the past decades, studies have largely expanded our knowledge of the sncRNA kingdom, revealing the diversity of sncRNA classes, the complexity of their regulation, and the abundance of their functions. Yet, it is very likely that our current knowledge represents just the tip of the iceberg. Among the newer classes of sncRNAs, only tsRNAs appear to be actively being studied, and our understanding of these novel sncRNAs is still in its early stages, with little known about their biogenesis, cellular abundance, or context-dependent biological relevance. More importantly, while the characterization of identified sncRNA species may begin soon, another pressing question remains: whether there are more sncRNA species to be discovered. Multiple NGS methods and bioinformatic tools have been developed for the discovery of sncRNAs, yet profiling and quantification remain particularly challenging for sncRNA species that are structured, heavily modified, and/or highly conserved in sequences, such as tRNAs and tsRNAs. As the profiling of sncRNAs using NGS methods can be enormously interfered with by RNA modifications, mapping of RNA modifications and uncovering new sncRNA classes are also challenges. Some base-resolution NGS methods, as well as accompanied bioinformatics analyses, have been developed for a few prevalent RNA modifications, such as m6A, m5C, and pseudouridylation [33,162], but most appear to only profile one modification at one time. Multiple modifications may co-exist on one sncRNA molecule. Therefore, it is tempting to speculate that crosstalk exists between different modifications, and combinations of modifications may elicit a more profound effect compared to individual ones. This awaits the development of new approaches that can simultaneously profile multiple RNA modifications. To explore the sncRNA kingdom further, combining NGS methods with mass spectrometry, such as MS ladder complementation sequencing (MLC-seq) [163], as well as direct sequencing, may provide alternative solutions [33,164]. Additionally, profiling of novel sncRNAs and their modifications in rare samples, such as preimplantation embryos, remains mostly unachievable, presenting obstacles to further understanding in areas such as epigenetic inheritance.
Despite the tremendous breakthrough achieved in the clinical applications of small RNAs in the past decade, the field still faces major challenges when it comes to specificity, delivery, and tolerability, all of which need to be seriously considered during the design of RNA-based therapeutics [144]. Though challenging, with continued advancements in technology and our understanding of the sncRNA kingdom, small RNA-based therapeutics will no doubt have the potential to revolutionize the treatment modality for a wide spectrum of untreatable diseases.
It is important to note that this review only covers a few aspects of the vast and thriving sncRNA field. There are many other active directions in sncRNA research, such as sncRNA-protein interaction [165,166], counteracting or synergistic effects among sncRNAs and other RNA species [167], transport of sncRNAs within or across cells, i.e., the subcellular compartmentalization [168] and extracellular existence of sncRNAs [123,124,169], all of which are integral to our current exploration of this intriguing field. Collectively, we strongly believe that research in the sncRNA field will continue to be active and future studies will increase our understanding of sncRNAs, as well as their pioneering applications.
Declaration of competing interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
We are grateful to members of the M.- F.L. laboratory and the L.-T.G laboratory for assisting with manuscript preparation. This work was supported by grants from National Natural Science Foundation of China (32170815, 91940305, 91640201 and 32300459), the National Key Research & Developmental Program of China (2021YFC2700200 and 2022YFC2702602), and China Postdoctoral Science Foundation (2021M703212).
Biographies

Lan-Tao Gou is Principle Investigator at the Center for Excellence in Molecular Cell Science, CAS. Dr. Gou received his Ph.D. from Shanghai Institute of Biochemistry and Cell Biology in 2013, and continued to work at the institute as Assistant Investigator. He received his postdoctoral training in Dr. Xiang-Dong Fu's lab at University of California, San Diego from 2015 to 2020 and was awarded the NIH Career Transition Award (K99) from 2018 to 2020. Dr. Gou joined the Center for Excellence in Molecular Cell Science, CAS in September 2020 as Principle Investigator. His lab focus on RNA regulation and chromatin remodeling in broad reproduction.

Mo-Fang Liu is Principle Investigator at the Center for Excellence in Molecular Cell Science, CAS. Dr. Liu's current research is focused on the function and mechanism of PIWI/piRNAs in mammal spermatogenesis and human male infertility as well as the function and mechanism of miRNAs in human cancers, especially in inflammation-associated tumorigenesis and cancer cell metabolism. Her researches reveal the physiological and pathological functions of small molecule noncoding RNA, which further promote the development of diagnosis and treatment of male infertility and tumors. Her contribution has been recognized by awards including the National Science Fund for Distinguished Young Scholars and the National Leading Talent of Technological Innovation of Ten-Thousands Talents Program.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fmre.2023.03.003.
Contributor Information
Lan-Tao Gou, Email: goulantao@sibcb.ac.cn.
Mo-Fang Liu, Email: mfliu@sibcb.ac.cn.
Appendix. Supplementary materials
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8(3):209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 4.Thompson D.M., Lu C., Green P.J., et al. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14(10):2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y.S., Shibata Y., Malhotra A., et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei H., Zhou B., Zhang F., et al. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS One. 2013;8(2):e56842. doi: 10.1371/journal.pone.0056842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 9.Gunawardane L.S., Saito K., Nishida K.M., et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 10.Czech B., Munafò M., Ciabrelli F., et al. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T., Totoki Y., Toyoda A., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 12.Su R., Fan L.-H., Cao C., et al. Global profiling of RNA-binding protein target sites by LACE-seq. Nat. Cell Biol. 2021;23(6):664–675. doi: 10.1038/s41556-021-00696-9. [DOI] [PubMed] [Google Scholar]
- 13.Y.W. Iwasaki, M.C. Siomi, H. Siomi, PIWI-Interacting RNA: Its Biogenesis and Functions, 84(1) (2015) 405–433. [DOI] [PubMed]
- 14.Gou L.-T., Dai P., Yang J.-H., et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24(6):680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P., Kang J.-Y., Gou L.-T., et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res. 2015;25(2):193–207. doi: 10.1038/cr.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. Watanabe, E.-c. Cheng, M. Zhong, et al., Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline, 25(3) (2015) 368–380. [DOI] [PMC free article] [PubMed]
- 17.Dai P., Wang X., Gou L.T., et al. A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell. 2019;179(7) doi: 10.1016/j.cell.2019.11.022. 1566–1581 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramat A., Garcia-Silva M.R., Jahan C., et al. The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 2020;30(5):421–435. doi: 10.1038/s41422-020-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q., Li R., Lyu Q., et al. Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat. Commun. 2019;10(1):3389. doi: 10.1038/s41467-019-11312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roovers Elke F., Rosenkranz D., Mahdipour M., et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10(12):2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- 21.Ishino K., Hasuwa H., Yoshimura J., et al. Hamster PIWI proteins bind to piRNAs with stage-specific size variations during oocyte maturation. Nucleic. Acids. Res. 2021;49(5):2700–2720. doi: 10.1093/nar/gkab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuck A.C., Tollervey D. RNA in pieces. Trends Genet. 2011;27(10):422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Cambier L., de Couto G., Ibrahim A., et al. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol. Med. 2017;9(3):337–352. doi: 10.15252/emmm.201606924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billmeier M., Green D., Hall A.E., et al. Mechanistic insights into non-coding Y RNA processing. RNA Biol. 2022;19(1):468–480. doi: 10.1080/15476286.2022.2057725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magaña S.M., Peterson T.E., Evans J.E., et al. Pediatric brain tumor cell lines exhibit miRNA-depleted, Y RNA-enriched extracellular vesicles. J. Neurooncol. 2022;156(2):269–279. doi: 10.1007/s11060-021-03914-4. [DOI] [PubMed] [Google Scholar]
- 26.Hussain S., Sajini A.A., Blanco S., et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.-J., Heard E. Small RNAs derived from structural non-coding RNAs. Methods. 2013;63(1):76–84. doi: 10.1016/j.ymeth.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Ender C., Krek A., Friedländer M.R., et al. A human snoRNA with MicroRNA-like functions. Mol. Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Taft R.J., Glazov E.A., Lassmann T., et al. Small RNAs derived from snoRNAs. RNA. 2009;15(7):1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Röther S., Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93(11):1905–1915. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 31.Pircher A., Bakowska-Zywicka K., Schneider L., et al. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol. Cell. 2014;54(1):147–155. doi: 10.1016/j.molcel.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giraldez M.D., Spengler R.M., Etheridge A., et al. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019;38(11) doi: 10.15252/embj.2019101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J., Zhou T., Chen Q. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022;24(4):415–423. doi: 10.1038/s41556-022-00880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J., Zhang Y., Tan D., et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 2021;23(4):424–436. doi: 10.1038/s41556-021-00652-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi J., Ko E.A., Sanders K.M., et al. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA- and tRNA-derived small RNAs. Genom. Proteom. Bioinform. 2018;16(2):144–151. doi: 10.1016/j.gpb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J.H., Chen W.X., Mei S.Q., et al. TsRFun: a comprehensive platform for decoding human tsRNA expression, functions and prognostic value by high-throughput small RNA-Seq and CLIP-Seq data. Nucleic. Acids. Res. 2022;50(D1):D421–D431. doi: 10.1093/nar/gkab1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. García-López, L. Alonso, D.B. Cárdenas, et al., Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization, 21(5) (2015) 946–962. [DOI] [PMC free article] [PubMed]
- 38.Chak L.L., Mohammed J., Lai E.C., et al. A deeply conserved, noncanonical miRNA hosted by ribosomal DNA. RNA. 2015;21(3):375–384. doi: 10.1261/rna.049098.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda S., Kawamura T., Loher P., et al. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic. Acids. Res. 2017;45(15):9108–9120. doi: 10.1093/nar/gkx537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larriba E., Rial E., del Mazo J. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genomics [Electronic Resource] 2018;19(1):634. doi: 10.1186/s12864-018-5020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Haussecker, Y. Huang, A. Lau, et al., Human tRNA-derived small RNAs in the global regulation of RNA silencing, 16(4) (2010) 673–695. [DOI] [PMC free article] [PubMed]
- 42.C. Cole, A. Sobala, C. Lu, et al., Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs, 15(12) (2009) 2147–2160. [DOI] [PMC free article] [PubMed]
- 43.Yamasaki S., Ivanov P., Hu G.F., et al. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Zhang X., Shi J., et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018;20(5):535–540. doi: 10.1038/s41556-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuorto F., Liebers R., Musch T., et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19(9):900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 46.He C., Bozler J., Janssen K.A., et al. TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol. 2021;28(1):62–70. doi: 10.1038/s41594-020-00526-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan L., Grigoriev A. Computational meta-analysis of ribosomal RNA fragments: potential targets and interaction mechanisms. Nucleic. Acids. Res. 2021;49(7):4085–4103. doi: 10.1093/nar/gkab190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherlin T., Magee R., Jing Y., et al. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol. 2020;18(1):38. doi: 10.1186/s12915-020-0763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Lambert, A. Benmoussa, P. Provost, Small Non-Coding RNAs Derived from Eukaryotic Ribosomal RNA, 5(1) (2019) 16. [DOI] [PMC free article] [PubMed]
- 50.Peng H., Shi J., Zhang Y., et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22(11):1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu C., Yu L., Wu B., et al. A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation. J. Mol. Cell Biol. 2017;9(3):256–259. doi: 10.1093/jmcb/mjx016. [DOI] [PubMed] [Google Scholar]
- 52.Chen Q., Yan M., Cao Z., et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 53.Sharma U., Conine C.C., Shea J.M., et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hua M., Liu W., Chen Y., et al. Identification of small non-coding RNAs as sperm quality biomarkers for in vitro fertilization. Cell Discov. 2019;5 doi: 10.1038/s41421-019-0087-9. 20-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nätt D., Kugelberg U., Casas E., et al. Human sperm displays rapid responses to diet. PLoS Biol. 2019;17(12) doi: 10.1371/journal.pbio.3000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarker G., Sun W., Rosenkranz D., et al. Maternal overnutrition programs hedonic and metabolic phenotypes across generations through sperm tsRNAs. Proc. Natl. Acad. Sci. U. S. A. 2019;116(21):10547–10556. doi: 10.1073/pnas.1820810116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida K., Maekawa T., Ly N.H., et al. ATF7-dependent epigenetic changes are required for the intergenerational effect of a paternal low-protein diet. Mol. Cell. 2020;78(3) doi: 10.1016/j.molcel.2020.02.028. 445–458.e6. [DOI] [PubMed] [Google Scholar]
- 58.Gong P., Bailbé D., Bianchi L., et al. Paternal high-protein diet programs offspring insulin sensitivity in a sex-specific manner. Biomolecules. 2021;11(5):751. doi: 10.3390/biom11050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaz C., Kermack A.J., Burton M., et al. Short-term diet intervention alters the small non-coding RNA (sncRNA) landscape of human sperm. Biorxiv. 2021 2021.07.08.451257. [Google Scholar]
- 60.Sharma U., Sun F., Conine C.C., et al. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell. 2018;46(4) doi: 10.1016/j.devcel.2018.06.023. 481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haussecker D., Huang Y., Lau A., et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar P., Anaya J., Mudunuri S.B., et al. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Fazio A., Schlackow M., Pong S.K., et al. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic. Acids. Res. 2022;50(3):1734–1752. doi: 10.1093/nar/gkac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keam S.P., Sobala A., Ten Have S., et al. tRNA-Derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 2017;16(2):413–420. doi: 10.1021/acs.jproteome.6b00267. [DOI] [PubMed] [Google Scholar]
- 65.Lyons S.M., Gudanis D., Coyne S.M., et al. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017;8(1) doi: 10.1038/s41467-017-01278-w. 1127-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guzzi N., Cieśla M., Ngoc P.C.T., et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173(5) doi: 10.1016/j.cell.2018.03.008. 1204–1216.e26. [DOI] [PubMed] [Google Scholar]
- 67.Schorn A.J., Gutbrod M.J., LeBlanc C., et al. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170(1) doi: 10.1016/j.cell.2017.06.013. 61–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L., Xu W., Liu K., et al. 5′ Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 2021;7(47) doi: 10.1126/sciadv.abh0494. eabh0494-eabh0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L., Xu W., Liu K., et al. Social Science Electronic Publishing; 2020. Trna-Derived 5′tiRNAs are Essential for Embryonic Development by Facilitating tRNA Gene Transcription. [Google Scholar]
- 70.Zuo Y., Zhu L., Guo Z., et al. TsRBase: a comprehensive database for expression and function of tsRNAs in multiple species. Nucleic. Acids. Res. 2021;49(D1):D1038–D1045. doi: 10.1093/nar/gkaa888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ender C., Krek A., Friedländer M.R., et al. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 72.Zhong F., Zhou N., Wu K., et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic. Acids. Res. 2015;43(21):10474–10491. doi: 10.1093/nar/gkv954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boccaletto P., Stefaniak F., Ray A., et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic. Acids. Res. 2021;50(D1):D231–D235. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han J., Wang J.Z., Yang X., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18(1):110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H., Deng Q., Lv Z., et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer. 2019;18(1):181. doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Alarcón C.R., Goodarzi H., Lee H., et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alarcón C.R., Lee H., Goodarzi H., et al. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen T., Hao Y.-J., Zhang Y., et al. m6A RNA methylation is regulated by MicroRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 79.Tan M.H., Li Q., Shanmugam R., et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550(7675):249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawahara Y., Megraw M., Kreider E., et al. Frequency and fate of microRNA editing in human brain. Nucleic. Acids. Res. 2008;36(16):5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang W., Chendrimada T.P., Wang Q., et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13(1):13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ota H., Sakurai M., Gupta R., et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153(3):575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawahara Y., Zinshteyn B., Sethupathy P., et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315(5815):1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shoshan E., Mobley A.K., Braeuer R.R., et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015;17(3):311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Snyder E.M., Licht K., Braun R.E. Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1. Biol. Reprod. 2017;96(1):244–253. doi: 10.1095/biolreprod.116.145151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu S., Kim V.N. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat. Rev. Mol. Cell Biol. 2020;21(9):542–556. doi: 10.1038/s41580-020-0246-8. [DOI] [PubMed] [Google Scholar]
- 87.Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16(10):1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim B., Ha M., Loeff L., et al. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J. 2015;34(13):1801–1815. doi: 10.15252/embj.201590931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim H., Kim J., Yu S., et al. A mechanism for microRNA arm switching regulated by uridylation. Mol. Cell. 2020;78(6) doi: 10.1016/j.molcel.2020.04.030. 1224–1236.e5. [DOI] [PubMed] [Google Scholar]
- 90.Yang A., Bofill-De Ros X., Shao T.J., et al. 3′ Uridylation confers miRNAs with non-canonical target repertoires. Mol. Cell. 2019;75(3) doi: 10.1016/j.molcel.2019.05.014. 511–522.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.D'Ambrogio A., Gu W., Udagawa T., et al. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012;2(6):1537–1545. doi: 10.1016/j.celrep.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gutiérrez-Vázquez C., Enright A.J., Rodríguez-Galán A., et al. 3′ Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA. 2017;23(6):882–891. doi: 10.1261/rna.060095.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fernandez-Valverde S.L., Taft R.J., Mattick J.S. Dynamic isomiR regulation in drosophila development. RNA. 2010;16(10):1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee M., Choi Y., Kim K., et al. Adenylation of maternally inherited microRNAs by Wispy. Mol. Cell. 2014;56(5):696–707. doi: 10.1016/j.molcel.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morgan M., Kabayama Y., Much C., et al. A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res. 2019;29(3):221–232. doi: 10.1038/s41422-018-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhao M.-Z., Lin D.-H., Zuo H., et al. piRNA 3′ uridylation facilitates the assembly of MIWI/piRNA complex for efficient target regulation in mouse male germ cells. Cell Res. 2022 doi: 10.1038/s41422-022-00659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kirino Y., Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13(9):1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kirino Y., Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14(4):347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 99.Ohara T., Sakaguchi Y., Suzuki T., et al. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14(4):349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 100.Saito K., Sakaguchi Y., Suzuki T., et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21(13):1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liang H., Jiao Z., Rong W., et al. 3′-Terminal 2′-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic. Acids. Res. 2020;48(13):7027–7040. doi: 10.1093/nar/gkaa504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kamminga L.M., Luteijn M.J., den Broeder M.J., et al. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29(21):3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lim S.L., Qu Z.P., Kortschak R.D., et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLos Genet. 2015;11(10) doi: 10.1371/journal.pgen.1005620. e1005620-e1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gainetdinov I., Colpan C., Cecchini K., et al. Terminal modification, sequence, length, and PIWI-protein identity determine piRNA stability. Mol. Cell. 2021;81(23) doi: 10.1016/j.molcel.2021.09.012. 4826–4842.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pastore B., Hertz H.L., Price I.F., et al. pre-piRNA trimming and 2′-O-methylation protect piRNAs from 3′ tailing and degradation in C. elegans. Cell Rep. 2021;36(9) doi: 10.1016/j.celrep.2021.109640. 109640-109640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.de Crécy-Lagard V., Boccaletto P., Mangleburg C.G., et al. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic. Acids. Res. 2019;47(5):2143–2159. doi: 10.1093/nar/gkz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021;22(6):375–392. doi: 10.1038/s41580-021-00342-0. [DOI] [PubMed] [Google Scholar]
- 108.Liu F., Clark W., Luo G., et al. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016;167(3) doi: 10.1016/j.cell.2016.09.038. 816–828.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cosentino C., Toivonen S., Diaz Villamil E., et al. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic. Acids. Res. 2018;46(19):10302–10318. doi: 10.1093/nar/gky839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Z., Qi M., Shen B., et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic. Acids. Res. 2019;47(5):2533–2545. doi: 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rashad S., Han X., Sato K., et al. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020;17(8):1092–1103. doi: 10.1080/15476286.2020.1779492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guegueniat J., Halabelian L., Zeng H., et al. The human pseudouridine synthase PUS7 recognizes RNA with an extended multi-domain binding surface. Nucleic. Acids. Res. 2021;49(20):11810–11822. doi: 10.1093/nar/gkab934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Z., Zhang J., Diao L., et al. Small non-coding RNAs in human cancer: function, clinical utility, and characterization. Oncogene. 2021;40(9):1570–1577. doi: 10.1038/s41388-020-01630-3. [DOI] [PubMed] [Google Scholar]
- 114.Kern F., Fehlmann T., Violich I., et al. Deep sequencing of sncRNAs reveals hallmarks and regulatory modules of the transcriptome during Parkinson's disease progression. Nat. Aging. 2021;1(3):309–322. doi: 10.1038/s43587-021-00042-6. [DOI] [PubMed] [Google Scholar]
- 115.Watson C.N., Belli A., Di Pietro V. Small non-coding RNAs: new class of biomarkers and potential therapeutic targets in neurodegenerative disease. Front. Genet. 2019;10:364. doi: 10.3389/fgene.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chi T., Lin J., Wang M., et al. Non-coding RNA as biomarkers for type 2 diabetes development and clinical management. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.630032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akat K.M., Moore-McGriff D., Morozov P., et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc. Natl. Acad. Sci. U. S. A. 2014;111(30):11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.M.B.R. Alves, E.C.C. Celeghini, C. Belleannée, From sperm motility to sperm-borne microRNA signatures: new approaches to predict male fertility potential, 8(791) (2020). [DOI] [PMC free article] [PubMed]
- 119.Vashisht A., Gahlay G.K. Using miRNAs as diagnostic biomarkers for male infertility: opportunities and challenges. Mol. Hum. Reprod. 2020;26(4):199–214. doi: 10.1093/molehr/gaaa016. [DOI] [PubMed] [Google Scholar]
- 120.de Araujo L.S., Ribeiro-Alves M., Leal-Calvo T., et al. Reprogramming of small noncoding RNA populations in peripheral blood reveals host biomarkers for latent and active mycobacterium tuberculosis infection. mBio. 2019;10(6) doi: 10.1128/mBio.01037-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tribolet L., Kerr E., Cowled C., et al. MicroRNA biomarkers for infectious diseases: from basic research to biosensing. Front. Microbiol. 2020;11:1197. doi: 10.3389/fmicb.2020.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Izzotti A., Carozzo S., Pulliero A., et al. Extracellular MicroRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am. J. Cancer Res. 2016;6(7):1461–1493. [PMC free article] [PubMed] [Google Scholar]
- 123.Turchinovich A., Weiz L., Langheinz A., et al. Characterization of extracellular circulating microRNA. Nucleic. Acids. Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang K., Zhang S., Weber J., et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic. Acids. Res. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guo B., Li D., Du L., et al. piRNAs: biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020;39(2):567–575. doi: 10.1007/s10555-020-09863-0. [DOI] [PubMed] [Google Scholar]
- 126.Hua M., Liu W., Chen Y., et al. Identification of small non-coding RNAs as sperm quality biomarkers for in vitro fertilization. Cell Discov. 2019;5(1):20. doi: 10.1038/s41421-019-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vyas P., Balakier H., Librach C.L. Ultrastructural identification of CD9 positive extracellular vesicles released from human embryos and transported through the zona pellucida. Syst. Biol. Reprod. Med. 2019;65(4):273–280. doi: 10.1080/19396368.2019.1619858. [DOI] [PubMed] [Google Scholar]
- 128.McCallie B., Schoolcraft W.B., Katz-Jaffe M.G. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil. Steril. 2010;93(7):2374–2382. doi: 10.1016/j.fertnstert.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 129.Rosenbluth E.M., Shelton D.N., Sparks A.E.T., et al. MicroRNA expression in the human blastocyst. Fertil. Steril. 2013;99(3) doi: 10.1016/j.fertnstert.2012.11.001. 855–861.e3. [DOI] [PubMed] [Google Scholar]
- 130.Abu-Halima M., Khaizaran Z.A., Ayesh B.M., et al. MicroRNAs in combined spent culture media and sperm are associated with embryo quality and pregnancy outcome. Fertil. Steril. 2020;113(5) doi: 10.1016/j.fertnstert.2019.12.028. 970–980.e2. [DOI] [PubMed] [Google Scholar]
- 131.Capalbo A., Ubaldi F.M., Cimadomo D., et al. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil. Steril. 2016;105(1) doi: 10.1016/j.fertnstert.2015.09.014. 225–235.e3. [DOI] [PubMed] [Google Scholar]
- 132.Rosenbluth E.M., Shelton D.N., Wells L.M., et al. Human embryos secrete microRNAs into culture media—a potential biomarker for implantation. Fertil. Steril. 2014;101(5):1493–1500. doi: 10.1016/j.fertnstert.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 133.Hawke D.C., Watson A.J., Betts D.H. Extracellular vesicles, microRNA and the preimplantation embryo: non-invasive clues of embryo well-being. Reprod. Biomed. Online. 2021;42(1):39–54. doi: 10.1016/j.rbmo.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 134.A. Timofeeva, Y. Drapkina, I. Fedorov, et al., Small Noncoding RNA Signatures for Determining the Developmental Potential of an Embryo at the Morula Stage, 21(24) (2020) 9399. [DOI] [PMC free article] [PubMed]
- 135.Abu-Halima M., Häusler S., Backes C., et al. Micro-ribonucleic acids and extracellular vesicles repertoire in the spent culture media is altered in women undergoing In Vitro Fertilization. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-13683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marczylo E.L., Amoako A.A., Konje J.C., et al. Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics. 2012;7(5):432–439. doi: 10.4161/epi.19794. [DOI] [PubMed] [Google Scholar]
- 137.Hammer B., Kadalayil L., Boateng E., et al. Preconceptional smoking alters spermatozoal miRNAs of murine fathers and affects offspring's body weight. Int. J. Obes. 2005;45(7):1623–1627. doi: 10.1038/s41366-021-00798-2. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.G.R. Rompala, A. Mounier, C.M. Wolfe, et al., Heavy Chronic Intermittent Ethanol Exposure Alters Small Noncoding RNAs in Mouse Sperm and Epididymosomes, 9(32) (2018). [DOI] [PMC free article] [PubMed]
- 139.Bedi Y., Chang R.C., Gibbs R., et al. Alterations in sperm-inherited noncoding RNAs associate with late-term fetal growth restriction induced by preconception paternal alcohol use. Reprod. Toxicol. 2019;87:11–20. doi: 10.1016/j.reprotox.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Beck D., Ben Maamar M., Skinner M.K. Integration of sperm ncRNA-directed DNA methylation and DNA methylation-directed histone retention in epigenetic transgenerational inheritance. Epigenet. Chromatin. 2021;14(1) doi: 10.1186/s13072-020-00378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ben Maamar M., Sadler-Riggleman I., Beck D., et al. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 2018;4(2) doi: 10.1093/eep/dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen M., Xu Y., Wang W., et al. Paternal exposure to PM2.5 programs offspring's energy homeostasis. Environ. Sci. Technol. 2021;55(9):6097–6106. doi: 10.1021/acs.est.0c08161. [DOI] [PubMed] [Google Scholar]
- 143.Hammond S.M., Aartsma-Rus A., Alves S., et al. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med. 2021;13(4) doi: 10.15252/emmm.202013243. e13243-e13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Winkle M., El-Daly S.M., Fabbri M., et al. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20(8):629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang M.M., Bahal R., Rasmussen T.P., et al. The growth of siRNA-based therapeutics: Updated clinical studies. Biochem. Pharmacol. 2021;189 doi: 10.1016/j.bcp.2021.114432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hoy S.M. Patisiran: first global approval. Drugs. 2018;78(15):1625–1631. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 147.Kristen A.V., Ajroud-Driss S., Conceição I., et al. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 2019;9(1):5–23. doi: 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- 148.Scott L.J. Givosiran: first approval. Drugs. 2020;80(3):335–339. doi: 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- 149.Scott L.J., Keam S.J. Lumasiran: first approval. Drugs. 2021;81(2):277–282. doi: 10.1007/s40265-020-01463-0. [DOI] [PubMed] [Google Scholar]
- 150.Lamb Y.N. Inclisiran: first approval. Drugs. 2021;81(3):389–395. doi: 10.1007/s40265-021-01473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19(10):673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Paunovska K., Loughrey D., Dahlman J.E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 2022;23(5):265–280. doi: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Akinc A., Querbes W., De S., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18(7):1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kanasty R., Dorkin J.R., Vegas A., et al. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 155.Matsuda S., Keiser K., Nair J.K., et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 2015;10(5):1181–1187. doi: 10.1021/cb501028c. [DOI] [PubMed] [Google Scholar]
- 156.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 157.Bajan S., Hutvagner G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells. 2020;9(1):137. doi: 10.3390/cells9010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li L.-C., Okino S.T., Zhao H., et al. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103(46):17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Modarresi F., Faghihi M.A., Lopez-Toledano M.A., et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30(5):453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Voutila J., Reebye V., Roberts T.C., et al. Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol. Ther. 2017;25(12):2705–2714. doi: 10.1016/j.ymthe.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sarker D., Plummer R., Meyer T., et al. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-α, in patients with advanced liver cancer: a first-in-human, multicenter, open-label, Phase I trial. Clin. Cancer Res. 2020;26(15):3936–3946. doi: 10.1158/1078-0432.CCR-20-0414. [DOI] [PubMed] [Google Scholar]
- 162.Frye M., Jaffrey S.R., Pan T., et al. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 2016;17(6):365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 163.Yuan X., Su Y., Zhang X., et al. MLC Seq: <em>De novo</em>sequencing of full-length tRNA isoforms by mass ladder complementation. Biorxiv. 2021 2021.05.22.445286. [Google Scholar]
- 164.Begik O., Lucas M.C., Pryszcz L.P., et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 2021;39(10):1278–1291. doi: 10.1038/s41587-021-00915-6. [DOI] [PubMed] [Google Scholar]
- 165.Corley M., Burns M.C., Yeo G.W. How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell. 2020;78(1):9–29. doi: 10.1016/j.molcel.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kelaini S., Chan C., Cornelius V.A., et al. RNA-binding proteins hold key roles in function, dysfunction, and disease. Exp. Biol. Med. 2021;10(5):366. doi: 10.3390/biology10050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yamamura S., Imai-Sumida M., Tanaka Y., et al. Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci. 2018;75(3):467–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sang L., Yang L., Ge Q., et al. Subcellular distribution, localization, and function of noncoding RNAs. Wiley Interdiscip. Rev. RNA. 2022:e1729. doi: 10.1002/wrna.1729. [DOI] [PubMed] [Google Scholar]
- 169.Zheleznyakova G.Y., Piket E., Needhamsen M., et al. Small noncoding RNA profiling across cellular and biofluid compartments and their implications for multiple sclerosis immunopathology. Proc. Natl. Acad. Sci. U.S.A. 2021;118(17) doi: 10.1073/pnas.2011574118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.