Abstract
Kidney disease is a leading cause of death worldwide. Currently, the diagnosis of kidney diseases and the grading of their severity are mainly based on clinical features, which do not reveal the underlying molecular pathways. More recent surge of ∼omics studies has greatly catalyzed disease research. The advent of artificial intelligence (AI) has opened the avenue for the efficient integration and interpretation of big datasets for discovering clinically actionable knowledge. This review discusses how AI and multi-omics can be applied and integrated, to offer opportunities to develop novel diagnostic and therapeutic means in kidney diseases. The combination of new technology and novel analysis pipelines can lead to breakthroughs in expanding our understanding of disease pathogenesis, shedding new light on biomarkers and disease classification, as well as providing possibilities of precise treatment.
Keywords: Artificial intelligence, Kidney diseases, Multi-omics, Network, Pathway, Precision medicine
1. Introduction
Kidney diseases (KDs), including acute kidney injury (AKI), acute kidney disease (AKD), and chronic kidney disease (CKD), are major health problems globally. KDs have high rates of death and morbidity, and induce significant economic losses. It has been estimated that globally, around 850 million people are suffering from KDs of different causes. By 2040, CKD will increase from the 16th to the 5th ranked leading cause of death around the world, which is one of the largest projected increases of any major cause of death [1], [2], [3]. KDs are especially challenging to diagnosis and treatment, because they have diverse pathophysiologies and are always asymptomatic. When KD is not detected and treated promptly, medical care requires specialized resources and imposes high costs for both patients and medical systems [4]. In the USA, medicare costs for CKD and end stage kidney disease (ESKD) were over $114 billion in 2016. In China, from the 2016 Annual Data Report of China Kidney Disease Network (CK-NET), the total medical expenditure on patients with CKD was about $4 billion USD, accounting for 6.50% of the overall Chinese medical expenditure [5]. Worldwide, it was reported that the median cost to treat a patient with CKD was significantly higher than that to treat a patient without CKD.
Currently, the diagnosis of KDs and the grading of their severity are based on clinical features, such as measurement of markers (e.g., creatinine), proteinuria, imaging examination, such as ultrasound, and biopsy findings. However, this approach does not always reveal the underlying molecular pathways, because different diseases might have similar phenotypes, symptoms, clinical parameters, or renal biopsy findings. In addition, patients with the same diagnosis might show high heterogeneity in terms of disease progression and response to the same regimen. Thus, the current classification systems remain unsatisfactory, which hampers our ability to predict long-term prognosis and apply targeted therapies. Unsurprisingly, there is a dearth of positive findings in clinical trials regarding kidney diseases.
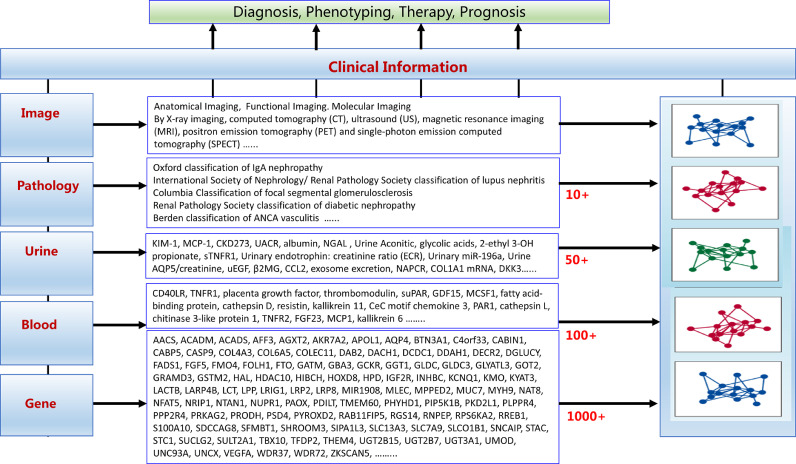
However, there has been a surge of ∼omics studies in the past decade, generating large amounts of genetic, transcriptomic, proteomic, and metabolomic data. Such studies have greatly expanded our understanding of disease pathogenesis, shedding new light on biomarkers and disease classification, as well as providing possibilities for precise treatment (Fig. 1) [6], [7], [8], [9]. In the age of ∼omics, blood, urine, and kidney biopsy samples can be taken to generate transcriptomic, proteomic, and metabolomic datasets; and digital images can be taken to produce high-dimensional, descriptive, and quantitative data for computer analysis. As biomedical research transitions into data-rich science, the era of “big data” has emerged [10]. The integration of such multi-layered datasets with longitudinal assessments of patient outcomes has the capacity to reveal different aspects of disease pathogenesis, progression, and cell-specific responses, which will be useful to guiding the design of targeted therapies.
Fig. 1.
Omics studies have contributed greatly to the diagnosis, phenotyping, therapy, and prognosis of kidney disease in the past ten years. Multiple biomarkers, e.g., genes, in blood and urine have been newly identified, and updated grading or classification systems in pathology and imaging have been proposed.
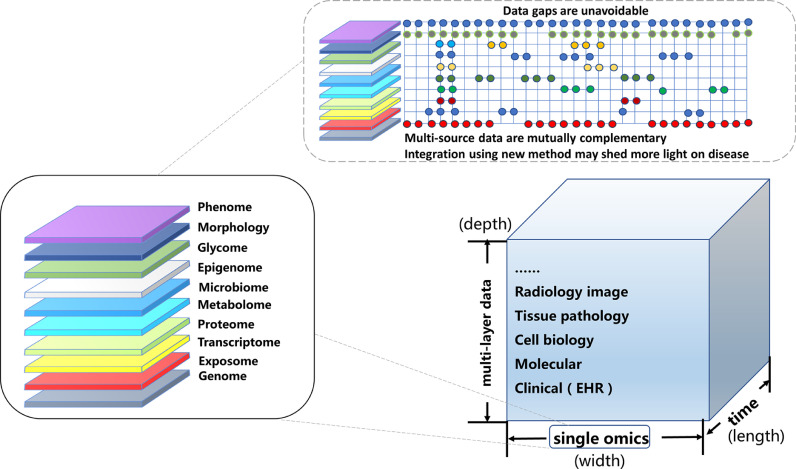
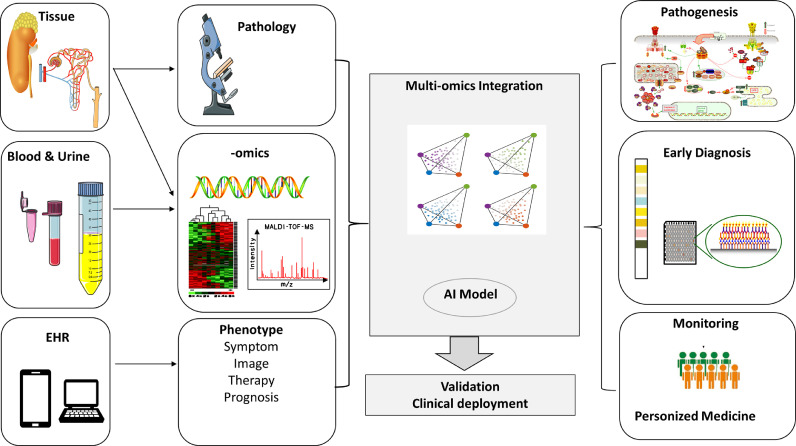
Currently, it is imperative that kidney -omics compendia (genome sequencing, transcriptomics, proteomics, metabolomics, and microbiomics) are integrated with clinical datasets, such as electronic health records (EHRs), digital pathology repositories, and medical images (Fig. 2). Multi-source big data are thus suggested to be the major driver of precision medicine. However, without proper analysis methods, data alone cannot be transformed into clinically actionable knowledge. Thanks to the advances in computing science, artificial intelligence (AI) has been developed for robust data analysis [11], [12], [13], [14], [15], [16], [17].
Fig. 2.
Multi-layered -omics should be integrated into a whole “data ocean”. Single omics such as environmental data, epigenomics, genomics, transcriptomics, proteomics, metabolomics, microbiomics, and phenomics, represent just one dimension of data: The "width of data". Big data are multi-layered, representing the "depth of data". However, data may change or correlate with disease progression and time: The "length of data". Currently, omics data frequently have inherent defects and do not fully match, thus we should incorporate robust data analysis using computing science, such as artificial intelligence.
This review discusses how AI and multi-omics techniques can be applied and integrated to develop novel diagnostic and therapeutic strategies for KDs. In general, the use of AI in nephrology is still in a research stage. Several other recent reviews with related foci might also be of interest [10], [11], [12], [14], [15], [16].
2. Omics study
As introduced, recent -omics approaches, including genomics, transcriptomics, epigenomics, microbiomics, metabolomics, and proteomics, offer the potential to refine current disease classification paradigms and identify novel sub-phenotypes. The addition of “omics” (http://omics.org/) to a molecular item indicates an unbiased systemic assessment of a set of molecules. Omics studies systematically capture and assess a molecular dimension at the genome, epigenome, transcriptome, proteome, metabolome, or microbiome level. For example, transcriptomics measure all transcripts at the same time in a sample (databases for kidney such as Renal gene Expression Database, http://rged.wall-eva.net; Nephroseq, http://nephroseq.org; Kidney interactive Transcriptomics, http://humphreyslab.com/SingleCell), and proteomics measure all proteins (i.e. Humana Kidney and Urine Proteome Project, http://www.hkupp.org). All of these “omics” approaches inquire into human health and disease in an unbiased manner. The prosperity of this field is attributed to technological advances, which have made the analysis of biological molecules cost-efficient and high-throughput. Although comprehensive at each layer, each single “-ome” can be regarded as checking only one dimension at the molecular level. Some -omics studies, such as transcriptomics, metabolomics, and proteomics, might be different at the bulk or single cell level, and offer limited correlations, reflecting reactive associations rather than causative ones. Combined and/or integrated analyses of multiple “ome”-wide profiles were designated as “multi-omics”. This strategy is used to determine causative changes or treatment targets, because incidental association is reduced and the cause and effect can be tested.
As suggested, the concept of -omics integration can be divided into two kinds: post-analysis and the combination approach [6]. The first protocol integrates -omics data by analyzing each dataset separately and then validating the results with another omics dataset independently. There are two methods to achieve this: top-down and bottom-up. The top-down data reduction method uses genomic and transcriptomic data to suggest phenotypic responses, and to find related molecular pathways. This can be validated by further metabolomics and proteomics. However, changes in genes, proteins, and metabolites might not correlate with each other directly. In the bottom-up approach, metabolomics and proteomics are used to assess the upstream events responsible for their changes. However, the low molecule coverage limits comprehensive interpretation. In the combination method, -omics data are combined together before data interoperation and visualization. The idea is to find similarities between different -omics data using mathematical methods, such as canonical correlation analysis (CCA), knowledge-based approaches, and orthogonal two-way projection on latent structures (O2PLS) [18], [19].
3. Artificial intelligence
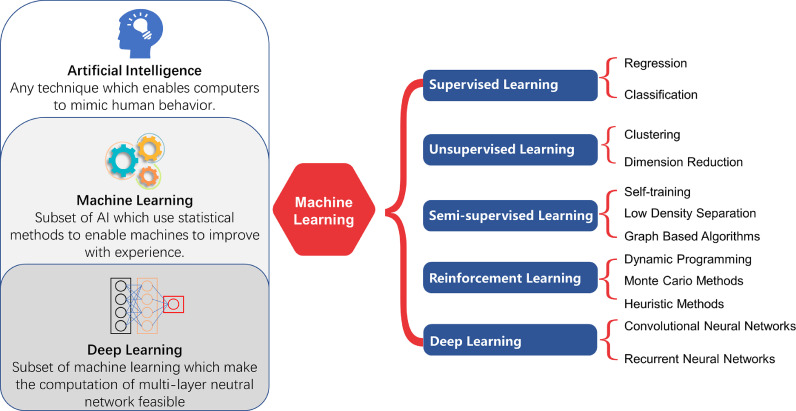
Artificial intelligence (AI) is the science of studying computations, making it possible to perceive, reason, and act. Machine learning (ML) is a branch of AI, representing a collection of computationally intensive statistical learning techniques, to learn and improve from learning. Current AI is mostly based on ML. ML has two major branches: supervised and unsupervised. Unsupervised ML is adopted in conditions where no available outcome and ground truth annotations are present. For example, using EHR data, an unsupervised method could predict patients subgroups with an increased risk of developing outcomes such as CKD and ESKD. In contrast, currently, we more rely on supervised ML. In this scenario, a computer learns to predict the label only after being trained on a large number of training samples with ‘ground truth’ labels. For example, we can recognize specific histological features in a biopsy using supervised ML, thanks to the rapid expansion of whole slide imaging by digital slide scanners (generating whole-slide images, WSIs). The most important breakthrough in the last decade in the field of ML, may be ‘deep learning’ (DL) to train multilayered (‘deep’) neural networks (DNNs) (Fig. 3). It is anticipated that convolutional neural networks (CNNs), a specific subtype of DNNs, will create new opportunities to analyze entire histopathological slides at high resolutions. In a typical development, available WSIs are divided into training, validation, and test sets. The CNN is trained with the training set, while the process is monitored by its performance on the validation set. The test set is used to produce unbiased performance data after an optimal CNN has been achieved.
Fig. 3.
Classification of artificial intelligence algorithms and relationships between artificial intelligence, machine learning, and deep learning. Artificial intelligence (AI) is a set of algorithms that enable computations making it possible to perceive, reason, and act. Machine learning (ML) is a branch of artificial intelligence in which algorithms have the ability to learn and improve from experience, without being explicitly programmed for a specific task. A popular way of classifying machine learning algorithms is by how much human supervision they require when training. Deep learning is a supervised or unsupervised machine learning algorithm based on neural networks, often specialized in image recognition, which has multiple layers of nonlinear processing units for feature extraction and transformation.
4. Application of multi-omics in kidney disease
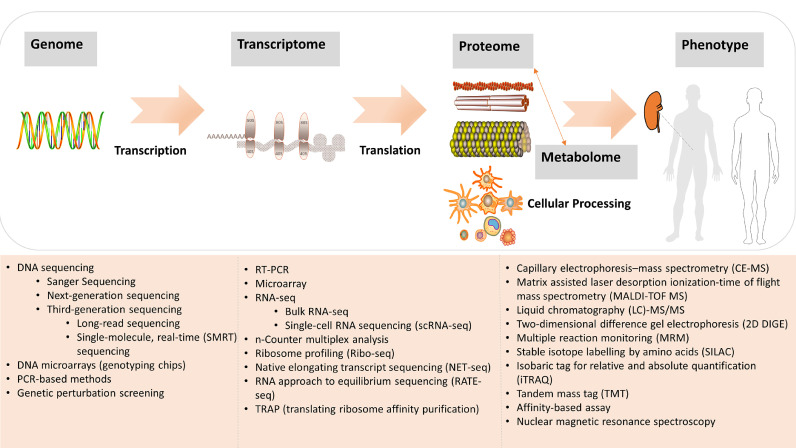
It is important to bear in mind that the “central dogma”, representing the basic flow of information in bio-systems, from DNA (genome) to RNA (transcriptome) to proteins (proteome) and metabolites (metabolome) (Fig. 4). Significant technological advances have enabled us to analyze 3 billion DNA base pairs of the human genome at reduced cost. The advent of genome-wide associations studies (GWASs) and whole exome sequencing (WES) have greatly benefitted our understanding of pathophysiology, risk prediction, and treatment of KDs. A GWAS has the ability show that a disease is associated with some single-nucleotide polymorphisms (SNPs). The identification of genetic factors associated with KDs has the potential to shed critical insights into disease mechanisms. For example, several loci are associated with CKD across different ethnicities, including UMOD, SHROOM3, MPPED2, BCAS3, and UNCX. UMOD was the first locus associated with CKD, with an OR of about 1.3 per copy of the risk allele, which remained unchanged after baseline estimated glomerular filtration rate (eGFR) adjustment [20], [21], [22], [23], [24], [25]. And this risk variant directly increases the expression of uromodulin in a dose-dependent manner, causing salt-sensitive hypertension and kidney damage in mice and humans. Observational studies in the general population showed that urinary uromodulin levels are positively associated with eGFR, markers of tubular transport, and kidney length and volume. It thus suggested that the levels of uromodulin can be considered as biomarkers of kidney tubule function. Low urinary uromodulin is a marker of poorer tubular health in at-risk individuals (aged and early CKD), with decreased levels of uromodulin reflecting decreased production (i.e., decreased renal functional reserve). As different KDs may show differences in prevalence and severity among ancestral groups, for population specific genes, an excess of African Americans with focal segmental glomerulosclerosis (FSGS) and pathogen-triggered HIV-associated nephropathy (HIVAN) prompted the discovery of genetic factor of APOL1 by GWASs [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43]. It was observed that APOL1-associated kidney disease followed a recessive pattern of inheritance. APOL1 genotype also influences the clinical course in kidney transplant recipients, hypertension-attributed ESKD, severe forms of lupus-associated kidney disease and subtypes of membranous nephropathy. Despite many reports of different cellular phenotypes, such as cell death, actin cytoskeletal structure, or mitochondrial dysfunction, the fundamental differences in behavior between the APOL1 alleles at a molecular level have remained elusive. Anyhow, environmental triggers may be important. The overwhelming interaction between HIV infection and the APOL1 risk variants may be the most powerful evidence, with an odds ratio (OR) of 29–89. And it was estimated that, in the pre- HAART (Highly Active Anti-Retroviral Therapy) era, 50% of individuals with the high-risk genotype developed HIVAN. One potential explanation is that both the high-risk APOL1 genotype and high APOL1 expression (driven in some cases by virus or the innate immune response to virus) may be required for disease to occur, as APOL1 can play as a multipurpose viral restriction factor. Several GWASs in European and East Asian populations also have identified almost 20 risk variants for IgA nephropathy (IgAN), suggesting it is shaped by selective forces from host-pathogen interactions [44], [45]. However, it seems that the genetic burden of membranous nephropathy and steroid-sensitive nephrotic syndrome is determined by significantly fewer variants with much larger effects [46]. Apart from case–control GWAS, the Chronic Kidney Disease Genetics (CKDGen) Consortium has focused on identifying genetic loci associated with quantitative traits, such as the glomerular filtration rate (GFR), albuminuria, blood urea nitrogen (BUN), and serum urate levels [47]. In a more recent meta-GWAS in 42 longitudinal studies from the CKDGen Consortium and UK Biobank, Gorski et al. identified that seven variants located in the UMOD, PRKAG2, WDR72, OR2S2, GATM, and LARP4B genes were associated with rapid kidney function decline [48]. Although GWAS has provided significant insights into pathogenic mechanisms, most loci identified still need to be followed-up to find the real causal variants and specific mechanisms. The majority of SNPs identified in GWASs are noncoding, nevertheless they implicate candidate genes or gene regions tagged by the SNPs. Further studies are needed to reveal the exact genetic changes driving the association and to prove causal inferences. However, in genomics, whether for single-gene Mendelian disorders or multi-gene complex diseases, in principle, the germ-line changes in the DNA sequence precede disease onset, indicating that the temporal relationship between genetic variation and disease onset is unidirectional.
Fig. 4.
Overview of the "central dogma" and related methods in -omics data generation. The upper panel depicts the central dogma and regulations of different omics layers. The lower panel presents multiple analytical platforms which can be used to interrogate samples and systems at a molecular level. In general, nucleotide-based experiments, such as genomics (genome-wide association studies), and transcriptomics studies, use PCR, microarrays and sequencing technologies. Proteomics, and metabolomics approaches share common technologies, such as mass spectrometry and nuclear magnetic resonance (NMR) spectroscopy.
One way to address the impact of variants on disease is by assessing the expression quantitative trait loci (eQTLs) [49], [50]. eQTLs are chromosomal loci where variants explain some of the variations in RNA or in protein expressions for the candidate gene. The representative available databases for kidney include NephQTL (https://nephqtl.org) and the Human Kidney eQTL Atlas (https://susztaklab.com/eqtl). By this strategy, lysosomal beta A mannosidase (MANBA), disabled-2 (DAB2), and Dachshund homolog 1 (DACH1) have been suggested as potential targets in CKD [51], [52]. In one early study integrating eQTL in renal transcriptomics, Martini S et al. selected 18 SNPs associated with eGFR in a meta-analysis of genome-wide association data by the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) and CKDGen consortia as candidates, and found that renal transcript levels for 18 genes in proximity to the SNPs significantly correlated with GFR (in -cis). They further constructed a co-expression network of 97 pathways linked by those genes [53]. Of these pathways, 56 pathways have been previously reported to be associated with CKD and 41 novel pathways. 78 pathways and 95% of the connections among those pathways were verified in an independent North American biopsy cohort. Of importance, the study revealed a signaling dichotomy in CKD, between activated inflammatory and immune processes and suppressed metabolic processes [54]. Several of the target factors and networks identified by this process, including the transcription factor NRF2, and the JAK-STAT and retinoid signaling pathways, have been clinically investigated or are currently underway in clinical trials. And most pathways are shared among different glomerular diseases, highlighting common pathways that mediate kidney injury. In a more recent report, integrating large-scale GWAS on kidney function with mRNA expression identified the kidney and liver as the primary organs for some traits of kidney function [55]. The proximal tubule is the critical cell type for eGFR, urate, and monogenic electrolyte or metabolic disease-related genes. Podocytes show enrichment of genes implicated in glomerular disease [55]. Notably, rare variants of some of these genes also cause Mendelian kidney diseases, providing important clues for further mechanistic studies. In the post-GWAS era, to take genetic data into clinical application, it is suggested that polygenic risk scores (PRSs) can bridge this gap by aggregating results from risk allele numbers. For example, Gorski et al. computed a PRS and found that individuals with a high-risk PRS (8–14 risk alleles) showed a 1.2 to 1.3- fold increased risk of AKI compared with those with a low-risk score (0–5 risk alleles) [48].
The transcriptome comprises all RNA molecules in cells. Structural similarities between DNA and RNA mean that analysis methods for genomics and transcriptomics are similar. Transcriptome analysis checks for biologically dynamic alterations, which are unstable. Thus, appropriate sampling strategies are crucial. An analysis of longitudinal blood transcriptomes showed that type 1 diabetes (T1D) was characterized by early changes in gene expression before T1D and islet autoimmunity [56]. By a genome-wide transcriptome study of tubules from biopsy samples [57], it was suggested that inflammation and metabolism as the top dysregulated pathways. Those involved genes encode key enzymes of fatty acid oxidation, including CPT1A, CPT2, ACOX1, ACOX2 and their transcriptional regulators, PPARA and PPARGC1A. Further assays from mouse models with fibrosis showed a reduction in transcript and protein levels of fatty acid oxidation enzymes associated with elevated lipids and triglyceride levels in tubular epithelial cells. The phenomenon can be observed prior to the occurrence of fibrosis, indicating likely pathologically causal and novel interventional targets [58]. Urinary epidermal growth factor (EGF) and monocyte chemoattractant protein-1 (MCP-1) were independently associated with disease progression as noninvasive indicators, which was supported by tissue transcriptomics and proteomics approach [59], [60]. Tubulointerstitial injury was correlated with increased TGF-β and TNF-α mRNA and decreased EGF mRNA. It was reported that for a 10U decrease in the glomerular filtration rate, there was an estimated increase of 5% and 10% in TGF-β and TNF-α mRNA, respectively, whereas EGF mRNA decreased by an estimated 15% [61]. And angiotensin-converting enzyme inhibitor enalapril was effective in decreasing albumin and increasing EGF excretion [62]. Thus measurement of urinary EGF may provide a new valuable index of renal function. A “bulk” transcriptome measurement cannot remove or solve the problem of cell heterogeneity, and might be influenced by cell composition or time changes. Such data describe only averaged gene expression across the great heterogeneity of kidney cell types. Single-cell RNA sequencing (scRNA-seq) was developed to reveal transcriptome differences in heterogeneous samples, defining the transcriptome at cell resolution [63]. Using single-cell RNA sequencing, mesenchymal cells were identified as the major contributor and NKD inhibitor of WNT signaling pathway 2 (NKD2) as a myofibroblast-specific target in renal fibrosis [64]. Cadherin 11 (CDH11), SPARC related modular calcium binding 2 (SMOC2), and pigment epithelium-derived factor (PEDF) were identified as promising non-invasive biomarkers of kidney fibrosis [65]. In humans, scRNA-seq data have been used to find novel segment-specific proinflammatory responses in kidney allograft rejection [66], [67], lupus nephritis [68], diabetic nephropathy [69], and IgA nephropathy [10], [70], [71].
Proteins are the major transcriptional products. They conduct most of the functional work in cells, playing roles as structural proteins, transcription factors, receptors, antibodies, hormones, transporters, and enzymes. The relative abundance of proteins, distinct proteins from alternative splice variants, posttranslational modifications, specific protein-protein interactions (PPIs), or expression in specific cellular components, all contribute to the complexity of the proteome. Immunoglobulins and cytokines can be detected or quantified using assays such as enzyme-linked immunosorbent assay (ELISA), immunofluorescent staining, enzyme multiplied immunoassay technique (EMIT), or mass spectrometry (MS). The identification of the M-type phospholipase A(2) receptor (PLA2R) in membranous nephropathy illustrated the potential of proteomic investigation to inform pathobiology, promoting disease diagnosis, and guiding treatment [72], [73], [74]. Importantly, by adding genomic findings in an unbiased manner, PLA2R1 gene variants associated with membranous nephropathy in diverse populations were discovered [75], [76], [77], [78], [79], supporting a causal effect. In a trans-ethnic meta-analysis of 993 plasma proteins among 2,882 participants, it was observed that testican-2 might be a clinically relevant physiological marker in disease progression [80]. Urinary glycine might be a protective biomarker for IgAN [81] and higher levels of C-glycosyltryptophan might predict adverse kidney outcomes and overall mortality [82]. The gut microbiota are also important determinants of the serum metabolome, i.e. uremic toxins and secondary bile acids (SBAs) [83]. Wang et al. suggested that ESKD-enriched Eggerthella lenta and Fusobacterium nucleatum increased uremic toxins production and promoted the development of CKD, and a probiotic Bifidobacterium animalis reduced the levels of toxins and the disease severity in rats [84]. To date, however, there is still a lack of data from large cohorts with serial measurements of the proteome and metabolome over time, together with longitudinal outcomes. Furthermore, the spatial heterogeneity of the proteome and metabolome only provided limited information about intra-organ biology.
To reduce the risk of false positives in -omics studies, multiple correction testing is required. Moreover, AI and pathway analyses might be helpful in organizing and decoding -omics datasets. In these datasets, the amount of variables might be collinear, which can be accounted for by random forest regression. The least absolute shrinkage and selection operator or elastic net are suitable methods to create a parsimonious risk model. Support-vector machines or clustering can be used to categorize variables in supervised or unsupervised ML. These approaches can be adopted to select the “top hits” with an appropriate number to meet the research goal. Pathway analysis includes several methods, aiming to organize many datasets into functional biological links. Many programs can be used for pathway analysis, such as Gene Set Enrichment Analysis (GSEA); Database for Annotation, Visualization, and Integrated Discovery (DAVID); Search Tool for the Retrieval of Interacting Genes/Proteins (STRING); and Ingenuity Pathway Analysis. Biological networks aim to analyze different biological substances such as metabolites, genes, and proteins as an interacting system. There are several types of biological networks including protein–protein interaction, signaling networks, gene regulatory networks (GRN), neuronal networks, and metabolomics networks. Many bio-informatic tools have been developed to create, visualize, and analyze these networks. Recently WGCNA [85], webCEMiTool (https://cemitool.sysbio.tools/) [86] and DiffCorr [87] have been applied to compare networks across health and disease stages. Other user friendly tools are also available, such as Networkanalyst (https://www.networkanalyst.ca/) [88], RegNetwork (http://www.regnetworkweb.org/home.jsp) [89], OmicsNet (http://www.omicsnet.ca) [90], Mibiomics (https://shiny-bird.univ-nantes.fr/app/Mibiomics) [91], Paintomics (http://www.paintomics.org/) [92], or Metaboanalyst (https://www.metaboanalyst.ca/) [93]. One of most popular visualization and integration tool is Cytoscape (https://cytoscape.org/) [94].
These integration protocols can be adopted in Mendelian randomization (MR) studies to test whether candidate markers lie in causal pathways. For example, in a “multi-omics” study using integrated analysis of the genome, transcriptome, and DNA methylome from 430 human kidneys, Eales et al observed associations between 1,038 genes and 479 GWAS association signals related to blood pressure [95]. By utilizing kidney SNPs as genetic instruments in MR analyses, causal effects of blood pressure on clinical kidney outcomes (urinary albumin-to-creatinine ratio and CKD) were identified. It provided evidence indicating the kidney as the tissue mediator of the genetic effects on blood pressure.
What's more, innovative integration approaches have also been adopted to identify optimal CKD model systems with which to test potential therapeutic targets. For example, a cross-species comparison of glomerular transcriptional networks was previously checked between human and murine diabetic kidney disease (DKD). By a network-matching algorithm, it was observed that STAT3 was one of the central nodes [96], suggesting the involvement of Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway in DKD. It was further reinforced by functional assay, i.e., podocyte-specific JAK2 overexpression worsened DKD in mice [97], and treatment with a specific JAK1/2 inhibitor for 2 weeks could partially reverse the major phenotypic changes of DKD. This effect was not DKD specific, which can be also observed in focal segmental glomerular sclerosis (FSGS) and IgA nephropathy [98], [99].
5. Exemplars of novel approaches to combine AI with ∼omics in kidney disease
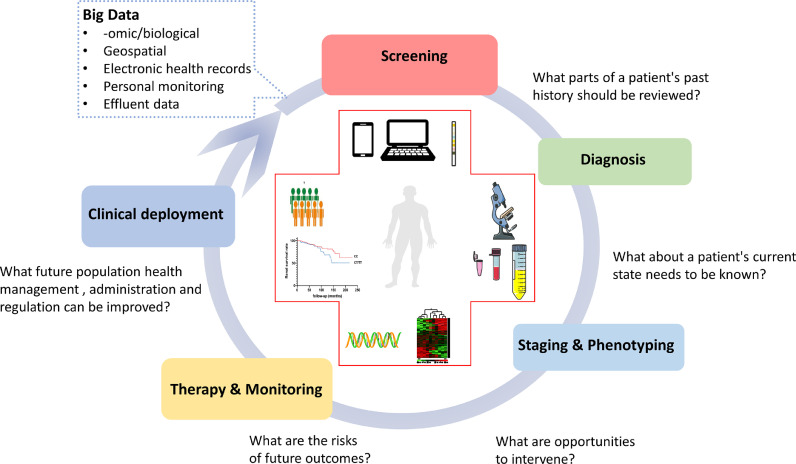
Computers have been used for different tasks for several years, especially for repetitive and cumbersome work, as well as to enhance reproducibility. Technological AI-based approaches could provide a good solution for precision medicine by delivering perfectly reproducible results. AI is also suited to precise and exhaustive extraction of the multi-dimensional data, including -omics (mRNA, microRNA, protein, and others). In clinical medicine, AI has shown promising roles across a wide spectrum of applications, including diagnostics, therapeutics, public health management, administration, and regulation. The vast quantity and accessibility of EHRs or wearable devices will influence decision-making for physicians, patients, healthcare providers, and healthcare systems (Fig. 5) [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116].
Fig. 5.
Use of artificial intelligence can help to address clinically relevant questions. In the medicine, AI is poised to play major roles across a spectrum of application domains, including diagnostics, therapeutics, population health management, administration, and regulation. The vast quantity and accessibility of EHRs and ∼omics will influence future decision-making on multiple fronts, including for patients, physicians, healthcare systems, healthcare providers, and regulatory bodies.
AI has been used in nephrology, for example, to offer solutions for repetitive work requiring significant attention [16], to improve estimation of the glomerular filtration rate [117], to facilitate the early diagnosis of AKI [118] or CKD [119], to personalize anemia management in ESKD [120], and to refine drug dosing [121].
Thus, in the field of nephrology, AI may be leveraged to analyze big data for: (I) clinicians: to make more accurate diagnoses and interpret pipelines; (II) for health systems: to improve workflow and reduce medical errors; (III) for patients: to promote health education and help disease prevention. In this review, we discuss some of the spotlights in the field related to aiding clinical decision-making. Good overviews of AI in other applications can be found in previous expert perspectives [6], [7], [10], [15], [16], [122], [123], [124].
5.1. Digital pathology
The significance of a kidney biopsy is to make a diagnosis, determine the pathology classification and disease stage, assist treatment selection, and predict prognosis. Using light microscopy, immunofluorescence microscopy, and electron microscopy, as well as various staining techniques, recognition of distinctive patterns are reported and interpreted by a pathologist. Meanwhile, classification and grading systems are always needed to assess the severity of pathological changes. Although pathologists are skilled in qualitative pattern recognition and quantification, the pathological scores remain semi-quantitative, which might contribute to a low diagnostic consensus rate among different pathologists. In addition, the scoring task in research with large samples can be tedious. Thus, it is necessary to develop tools to facilitate fast, objective, and consistent assessment of renal pathology, which is a prerequisite for fruitful clinical trials, better prediction, or response evaluation. The development of whole-slide scanners to generate WSIs has made the digitization of histopathology fast and high-throughput. The resulting progress in digital pathology has facilitated the implementation of AI in computer-aided diagnostics [125].
In general, the use of AI in kidney pathology is still in its infancy and mostly at the research stage [124]. Most reported studies have focused on segmentation tasks [126], [127]. This can efficiently segment kidney histopathology in periodic acid–Schiff (PAS) stained sections, and it was reported that some outputs correlate well with specific scoring systems [128], [129]. Several automatic segmentation [129] and classification [130] methods have been developed using AI, mostly using a supervised deep-learning approach. High accuracy of the computational method can be obtained compared with the results produced by expert nephropathologists. To provide a continuous risk model instead of a discrete category, AI has shown supportive value for clinical diagnostics. Although PAS staining is always readily available in different kidney pathology laboratories, the full workup of kidney pathology includes further staining, such as methenamine silver and periodic acid–methenamine silver staining (Jones), trichrome staining, Masson trichrome staining, and hematoxylin-eosin (HE) staining. For a definitive diagnosis, in addition to bright field microscopy, immunofluorescence (IF) and electron microscopy (EM) are essential. For both clinical use and research, AI might increase efficiency by replacing rigid tasks. Developed AI algorithms would yield objective and reproducible data from WSIs [131]. Applied in a clinical setting, this might increase accuracy and reproducibility. In addition, it might have significance in pathology education, or in areas where nephrology pathologists are unavailable. This might aid tailored treatment for individual patients if accurate quantitative descriptors are available. In research, the potential of AI to analyze large numbers of WSIs in a standardized manner could help to develop novel biomarkers. To harness these opportunities, the establishment of large, well-curated multi-center data sets is required, as well as the training and involvement of clinicians. However, currently, large cohorts are not available in kidney pathology, and all the reported studies just analyzed tens of WSIs. Even in these small cohorts, annotation takes immense effort by pathologists. Although the renal pathology field has emphasized the precise definition of pathological findings and universally accepted definitions, gold standards for most annotations are lacking. The involvement of deep learning in pathology has emerged very recently, and compared with established diagnostic algorithms, the lack of multi-center validation and efficacy studies is not surprising. As introduced above, because molecular diagnoses are derived from objective data from measurements, they are likely to be more accurate than histology. It was suggested that an combination of molecular algorithms could improve the estimate stability [132] either by human experts or using an automated system.
5.2. Prediction and sub-phenotyping
Different approaches have been developed for sub-phenotyping, which usually take clinical characteristics as the first step. In past years, several clinical risk scores have been developed to identify patients with high risk [133]. Access to large datasets in EHRs (Table 1), and multi-omics data have contributed to recent ML approaches to identify sub-phenotypes in patients at risk of AKI [134], CKD, and ESKD.
Table 1.
Examples of current application of machine learning in CKD (taking IgA nephropathy as example) and AKI.
| Year | Disease | Type of data | Sample size | Purpose of AI | Output Data (Label) | Results | Algorithms | Notes | PMID |
|---|---|---|---|---|---|---|---|---|---|
| 1998 | IgAN | Phenomics (6 input variables) | 54 patients with IgAN | Method investigation | Outcome was assigned as ‘stable’ if serum creatinine was < 150 µmol/l after 7 years and ‘non-stable’ if serum creatinine was ≥ 150 µmol/l. | The ANN assigned the correct outcome to 47/54 (87.0%) patients: sensitivity 19/22(86.4%), specificity 28/32(87.5%). The mean score for nephrologists was 37.5/54 (69.4%, range 35–40), mean sensitivity 72% and mean specificity 66%. | artificial neural network (ANN) | One of the main purposes of the study was to demonstrate the potential application of artificial neural networks to clinical nephrology. Studies of ANN to predict 10-, 15- and 20-year outcome would be of value and would require data from more than one centre to provide sufficient training data. | 9481717 |
| 2016 | IgAN | Phenomics (6 input variables) | 1040 patients with IgAN | Risk prediction | predict ESKD status and the time to ESKD (defined as three categories: ≤ 3 years, between > 3 and 8 years and over 8 years) | The ANNs demonstrated high performance for both the prediction of ESKD (with an AUC of 89.9, 93.3 and 100% in the Italian, Norwegian and Japanese IgAN population, respectively) and its timing (f-measure of 90.7% in the cohort from Italy and 70.8% in the one from Norway). | artificial neural network (ANN) | The first Clinical decision support system for end-stage kidney disease risk estimation in IgAN patients | 26047632 |
| 2018 | IgAN | Phenomics (35 input variables) | 262 patients with IgAN | Risk prediction | predict the ESKD status | This RF model with obove CDSS (http://www.igan.net/) 6 predictors (gender, age, hypertension, 24-h urine protein levels and histological grading) achieved an F-measure of 0.8 and the an AUC of 92.57%. | random forest model | In addition to the predictors in the CDSS, Oxford-MEST scores, C3 staining and eGFR conveyed additional information for ESRD prediction in Chinese IgAN patients using a RF model. | 30537719 |
| 2019 | IgAN | Phenomics (36 input variables) | 2,047 patients with IgAN | Risk prediction | combined event of end-stage kidney disease (ESKD) or 50% reduction in estimated glomerular filtration rate within 5 years after diagnostic kidney biopsy | a C statistic of 0.89 for the derivation cohort and 0.84 for the validation cohort while using the 10 most important variables |
eXtreme Gradient Boosting (XGBoost) | a simplified scoring scale model (SSM) included 3 variables: urine protein excretion, global sclerosis, and tubular atrophy/interstitial fibrosis was derived |
31031086 |
| 2019 | IgAN | Phenomics (14 input variables) | 1,370 patients with IgAN | Risk prediction | predict the status of ESRD | Logistic regression [area under the curve (AUC) =96.1%] outperformed the other models [random forest (AUC =95.5%), SVM (AUC =95.8%), decision tree (AUC =84.3%), ANN (AUC =92.2%) and KNN (AUC =94.6%)]. | random forest model | The formula was employed: P = eX/(1+eX) where X= –1.354–(1.887*M) – (2.970*T) + (0.22*24 h urine protein) + (1.792*treatment), e is the base of the natural logarithm. For M and T score, 0 and 1 were defined as absent and present. For treatment, 1 and 2 were defined as no glucocorticoid use and glucocorticoid use. A P value of 0.645 was selected as a cut-off point and P value >0.645 should be considered positive ESRD status and P<0.645 should be considered negative. | 31317004 |
| 2021 | IgAN | Phenomics (7 input variables) | 948 patients with IgAN | Risk prediction | a two-step procedure of a classifier model that predicts ESKD, and a regression model that predicts development of ESKD over time |
The classifier model showed a performance value of 0.82 (area under the receiver operating characteristic curve) in patients with a follow-up of five years, which improved to 0.89 at the ten year follow-up. The regression model showed a mean absolute error of 1.78 years and a root mean square error of 2.15 years. |
artificial neural network | A clinical decision support system (CDSS) (https://igan.poliba.it) may predict ESKD in patients with IgAN with a median follow-up of 5 and 10 years. | 32889014 |
| 2021 | IgAN | Phenomics (7 input variables) | 80 patients with IgAN | Risk prediction | analyze regression techniques, estimating the deterioration of renal function, as expressed by the difference in creatinine concentration, compared to the baseline | The performance of the tested models was obtained: Random Forest Classifier showed an accuracy of 0.8–1.0, Multi-Layer Perceptron an Area Under Curve of 0.8842–0.9035 and an accuracy of 0.7527–1.0) and regressors with a low estimation error (Decision Tree Regressor showed MAE 0.2059, RMSE 0.2645). | Gaussian Naive Bayes Classifier; Support Vector Machine; Random Forest Classifier; K-nearest Neighbor Classifier; amd Multi-Layer Perceptron (an artificial neural network, ANN) | The choice of input data and the choice of the right model have a direct impact on the performance. The limitation of our study is that it uses a retrospective design and a small number of samples. | 33920611 |
| 2015 | IgAN | Phenomics (8 input variables) | 1,174 patients with IgAN | Risk prediction |
identify significant predictors of ESKD and time-to-ESKD |
ANNs was superior performance compared to the other models. The ANN for ESKD prediction has accuracy greater than 90% as well as precision, recall, and f-measure for the class of patients not reaching ESKD, while precision, recall, and f-measure for the class of patients reaching ESKD are slightly lower. |
artificial neural networks (ANNs), neuro fuzzy systems (NFSs), support vector machines (SVMs), and decision trees (DTs) |
Four different data-driven models (artificial neural networks, neuro fuzzy systems, support vector machines, and decision trees) have been trained. Difference is due to the peculiarities of the dataset which includes a lower number of clinical records. | 26453758 |
| 2021 | IgAN | Phenomics (36 input variables) | 2,047 patients with IgAN | Risk prediction | predict the prognosis of IgAN patients by taking the time-to-event information | The XGBoost-Surv achieved overwhelmed performance over the cox regression (0.655), lasso-cox (0.795), RSF (0.773) and SVM-Surv (0.781) on the external validation set. | EXtreme Gradient Boosting for survival (XSBoost-Surv) | Shapley Additive exPlanations (SHAP) was utilized to interpret the prediction result of our model. The Tubular atrophy/Interstitial fibrosis (%) and global sclerosis (%) were the strongest contributors to the model's final decision. | 33936448 |
| 2020 | IgAN | Phenomics (40 input variables) | 4,047 patients with IgAN | Subphenotyping | different benefit from immunosuppression (IS) therapy measured by ESKD or 30%-50% reduction in estimated glomerular filtration rate | Three identified subgroups obtained a significant IS benefits. In patients with serum creatinine ≤1.437 mg/dl, the benefits of IS were observed in those with proteinuria > 1.525 g/24h, especially in those with proteinuria >2.480 g/24h. In patients with serum creatinine > 1.437 mg/dl, those with high proteinuria and crescents benefitted from IS. | model-based recursive partitioning | This study shed some light on individualised therapy in IgAN. | 32062356 |
| 2018 | AKI | Phenomics (EHR data from the the University of Chicago) | 121,158 patients | Risk prediction | develop an acute kidney injury risk prediction model using electronic health record data for longitudinal use in hospitalized patients | The AUC (95% CI) was 0.90 for predicting stage 2 acute kidney injury within 24 hours and 0.87 within 48 hours. The AUC was 0.96 for receipt of renal replacement therapy (n = 821) in the next 48 hours. Accuracy was similar across hospital settings and admitting serum | Gradient Boosting Machine algorithm | The algorithm provides a nearly 2-day lead time in advance of more traditional SCr-based AKI definitions. This window of time between evidence of increased AKI risk and clinical AKI being present is the ideal period for a clinical intervention. |
29596073 |
| AKI | creatinine groupings. At a probability threshold of greater than or equal to 0.022, the algorithm had a sensitivity of 84% and a specificity of 85% for stage 2 acute kidney injury and predicted the development of stage 2 a median of 41 hours prior to the development of stage 2 acute kidney injury. | ||||||||
| 2018 | AKI | Phenomics (institutional EHR data) | 1211 patients who underwent living donor liver transplantation (LDLT) or deceased donor liver transplantation (DDLT) | Risk prediction | predict AKI after liver transplantation | Gradient boosting machine showed the largest test AUROC (0.90) and highest accuracy (84%). AUROC of the multivariable logistic prediction model for the test dataset was 0.61. Decision tree and random forest techniques showed moderate performance (AUROC 0.86 and 0.85, respectively). | Scikit-learn, XGboost, and Keras. | Variants important in the gradient boosting machine were SvO2 (mixed venous oxygen saturation); Hb(hemoglobin); MEDL (model for end-stage liver disease); EBL(estimated blood loss); BMI(body-mass index); and ABP(arterial blood pressure) . | 30413107 |
| 2018 | AKI | Phenomics (42 input variables) | 2010 patients who underwent open heart surgery and thoracic aortic surgery | Risk prediction | predict acute kidney injury after cardiac surgery | During the first postoperative week, AKI occurred in 770 patients (38.3%). The best performance regarding AUC was achieved by the gradient boosting machine to predict the AKI of all stages (0.78) or stage 2 or 3 AKI. The AUC of logistic regression analysis was 0.69. Decision tree, random forest, and support vector machine showed similar performance to logistic regression. | decision tree, random forest, extreme gradient boosting, support vector machine, neural network classifier, and deep learning | Important covariates in the final model included angiotensin receptor blocker,body-mass index, coronary artery bypass graft, calcium channel blocker, chronic kidney disease, creatinine, history of cerebrovascular accident, ejection fraction, preoperative ratio of early transmitral flow velocity to early diastolic velocity of the mitral annulus, fresh frozen plasma, hematocrit, hypertension, intraoperative mean mixed venous oxygen saturation, three vessel coronary disease, preoperative packed red blood cells. | |
| 2018 | AKI | Phenomics (45 input variables) | 970,869 patients undergoing PCIs | Risk prediction | AKI risk prediction after percutaneous coronary intervention (PCI) from the National Cardiovascular Data Registry (NCDR) CathPCI registry | A total of 69,826 (7.4%) patients developed AKI. The XGBoost model using permutation selection, had the highest AUC (0.725), best Brier score (0.0630), highest resolution (0.0043), and best reliability (0.0004 × 10−2). | XGBoost | The improvement in performance of the proposed machine learning model over the current AKI model was a result of employing all available variables to modeling, using permutation test for variable selection and implementing XGBoost to model the relationship between variables and outcome. The best machine learning model only entailed the use of 2 more variables than the current AKI model, posing minimal additional burden on data extraction and processing. | 30481186 |
| 2019 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | 23,950 patients | Risk prediction | develop and validate a data driven multivariable clinical predictive model for early detection of AKI among a large cohort of adult critical care patients | It demonstrated that using machine learning models (multivariate logistic regression, random forest and artificial neural networks) with demographics and physiologic features can predict AKI onset as defined by the current clinical guideline with a competitive AUC (mean AUC 0.783 by our all-feature, logistic-regression model). | multivariate logistic regression, random forest and artificial neural networks | The comprehensive demographics and physiologic features can accurately predict max serum creatinine level during Day 2 and Day 3 with a root mean square error of 0.224 mg/dL. | 30700291 |
| 2019 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | 6,682 patients | Risk prediction | prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care | The machine learning XGBoost model outperformed the traditional logistic regression model in differentiating between the volume-responsive (VR) and volume-unresponsive (VU) groups (AU-ROC, 0.860 vs. 0.728). | extreme gradient boosting (XGBoost), logistic regression | Using advanced machine learning techniques, it was observed that some important clinical factors associated with VR-AKI such as age, urinary creatinine concentration, maximum BUN concentration, and albumin. These results have some implications and require further consideration. | 30961662 |
| 2019 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | 58,976 patients | Risk prediction | prediction of mortality risk of AKI patients who are stratified according to their AKI stages | Comparing the performance of four predictors, GBDT acquires better results. Comparing the Stages, Stage-III shows a higher mortality predictive performance. | Logistic Regression (LR), L2 norm regularized Logistic Regression (Ridge), Random Forest (RF), and Gradient Boosting Decision Tree (GBDT) |
The technique of stratifying patients with AKI into different stages obtained a better performance compared to the mixed cohort. The technique of constructing a mortality predictor based on different cohorts has the potential to consider different AKI causes. It might be useful to incorporate more information (e.g., imaging and genomic biomarkers) to further improve the performance of dynamic clinical mortality risk prediction models. | 31437966 |
| 2019 | AKI | Phenomics (EHR data with 3,599 clinically relevant features) | 703,782 adult patients across 172 inpatient and 1,062 outpatient sites | Risk prediction | develop a deep learning approach for the continuous risk prediction of future deterioration in patients from diverse clinical environments | It predicts 55.8% of all inpatient episodes of acute kidney injury, and 90.2% of all acute kidney injuries that required subsequent administration of dialysis, with a lead time of up to 48 h and a ratio of 2 false alerts for every true alert. This corresponds to an area under the receiver operating characteristic curve of 92.1%, and an area under the precision–recall curve of 29.7%. | Recurrent neural networks | Recurrent neural networks achieve the highest performance for both PR AUC and ROC AUC. Feed-forward models (deep MLP, shallow MLP, Logistic Regression, Random Forest, Gradient Boosted Trees) do not have the capacity to aggregate the information about a patient over time, which necessitates manual collection and engineering of patient historical features. Gradient Boosted Trees (GBTs) benefited from heavy overweighting of observations with positive-labels while equivalent oversampling for random forest and neural-networkbased models did not bring a similar improvement. |
31367026 |
| 2019 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | 19044 patients with AKI in the ICU | Risk prediction | consturct a mortality prediction model using the random forest (RF) algorithm for acute kidney injury (AKI) patients in the intensive care unit (ICU) | The observed in-hospital mortality of AKI patients is 13.6% in the final cohort. The results of model performance comparison show that the Brier score of the RF model is 0.085 and AUROC of the RF model is 0.866. The accuracy of the RF model is 0.728. The F1 score of the RF model is 0.459. The calibration plots show that the RF model slightly overestimates mortality in patients with low risk of death and underestimates mortality in patients with high risk of death. | Random forest model, Artificial neural network model, Support vector machine model, Customized SAPS II model | The top five important predictor variables that have influence on the in-hospital mortality of AKI patients in the ICU produced by the RF model are urine output, systolic blood pressure, age, serum bicarbonate level and heart rate. | 30914181 |
| 2020 | AKI | Phenomics (13 input variables) | 212 patients received thoracoabdominal aortic aneurysm repair (TAAAR) | Risk prediction | acute renal failure (ARF) | Five-fold cross-validation showed that among the four classification models, random forest was the most precise model for predicting ARF, with an average area under the curve (AUC) of 0.89 ± 0.08. | logistic regression, linear kernel SVM, Gaussian kernel SVM, and RF | Machine learning models can precisely predict ARF and paraplegia during early stages after surgery. This program allows cardiac surgeons to address complications earlier and may help improve the clinical outcomes of TAAAR. | 31765025 |
| 2020 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | a total of 37,486 ICU stays including 7,657 AKI cases from 6,933 unique patients, and 29,829 controls from 24,366 unique patients | Subphenotyping | discover AKI sub-phenotypes using structured and unstructured electronic health record (EHR) data of patients before AKI diagnosis | Three distinct sub-phenotypes are obtained by using cluster method based on patients representations. | memory networks (MN), Logistic Regression (LR), Random Forest (RF), and Gradient Boosting Decision Tree (GBDT) | After Age adjustment, the following features are significantly different across the three subtypes: Glucose, Albumin, Creatinine, White Blood Count (WBC), Urine, and estimated Glomerular Filtration Rate (eGFR). | 31911172 |
| 2020 | AKI | Phenomics and some biomarkers (9 input variables) | 101 adult patients with ≥20% total body surface area (TBSA) burns or non-burn trauma-related injuries requiring surgery | Risk prediction | early eecognition of burn- and trauma-related AKI | The AI/ML algorithm helped predict AKI 61.8 (32.5) hours faster than the Kidney Disease and Improving Global Disease Outcomes (KDIGO) criteria for burn and non-burned trauma patients. | logistic regression (LR), k-nearest neighbor (k-NN), random forest (RF), support vector machine (SVM), and our multi-layer perceptron (MLP) deep neural network (DNN) | NGAL was analytically superior to traditional AKI biomarkers such as creatinine and UOP. With ML, the AKI predictive capability of NGAL was further enhanced when combined with NT-proBNP or creatinine. | 31937795 |
| 2020 | AKI | Phenomics (17 input variables) | 22,542 patients in the Medical Information Mart for Intensive Care dataset | Risk prediction | prediction of acute kidney injury | Random forest was the best classifier using structural temporal pattern detection. The accuracy of the classifier with local and global trend features was significantly higher than that while using symbolic temporal pattern detection and the last recorded value (81.3% vs 70.6% vs 58.1%; P<.001). | Random Forest, Extreme Gradient Boosting Tree, Kernel-based Bayesian Network, Support Vector Machine (SVM), Logistic Regression, Naïve Bayes, K-Nearest Neighbor, and Artificial Neural Network (ANN) | It highlights the importance of using all information in time series data rather than using a single value. | 32181753 |
| 2020 | AKI | Phenomics (13 input variables) | 1,173 hepatectomy patients, 77 (6.6%) of whom had AKI and 1,096 (93.4%) who did not | Risk prediction | predict the likelihood of acute kidney injury after liver cancer resection | The AUC values for the four algorithms were: Gbdt (0.772), gbm (0.725), forest (0.662) and DecisionTree (0.628). | random forest (forest), LightGBM (gbm), decision tree (tr), gradient boosting (Gbdt) | Age, cholesterol, tumor size, surgery duration and PLT influence the likelihood and development of postoperative acute kidney injury. | 32140301 |
| 2020 | AKI | Phenomics (the EHRs of 15 hospitals that are part of Aurora Health Care system) | 36,614 patients with 44,691 total hospital stays | Risk prediction | continually predicts AKI to occur any time during the rest of the hospital stay | The continual prediction model obtained statistically significantly better AUC than the one-time prediction model (0.724 vs. 0.653; p < 0.05; two-tailed paired t-test). | Weka machine learning software | Top ten medication and comorbidity features whose prescription or diagnosis during a hospital stay were associated with a change in the model's prediction from non-AKI to AKI. Those include Cisplatin, Aminoglycosides, Hypercalcemia, Diuretics, Acyclovir, ACE inhibitors or NSAIDS or Diuretics, ARB or ACE inhibitors or NSAIDS or Diuretics, Respiratory failure, Rhabdomyolysis, K sparing. | 32001013 |
| 2020 | AKI | Phenomics (32 input variables) | 1571 adult patients who started CRRT for acute kidney injury | Risk prediction | predict mortality in patients undergoing continuous renal replacement therapy | For the ICU mortality, the random forest model showed the highest AUC (0.784), and the artificial neural network and extreme gradient boost models demonstrated the next best results (0.776). The AUC of the random forest model was higher than 0.611, 0.677, and 0.722, as achieved by APACHE II, SOFA, and MOSAIC, respectively. | κ-nearest neighbor (KNN), support vector machine (SVM), multivariate adaptive regression splines (MARS), random forest (RF), extreme gradient boost (XGB), and artificial neural network (ANN) | The models developed using machine learning algorithms better predicted ICU and in-hospital mortalities than conventional scoring systems such as APACHE II and SOFA, and MOSAIC. | 32028984 |
| 2021 | AKI | Phenomics (64 input variables) | 200,859 admissions between 2014 and 2015 at 208 hospitals of the United States | Risk prediction | use XGBoost to construct a predictive mortality model for AKI patients in the ICU | The overall in-hospital mortality of AKI patients was 16.35%. The best performing algorithm in this study was XGBoost with the highest AUROC (0.796, p < 0.01), F1(0.922, p < 0.01) and accuracy (0.860). The precision (0.860) and recall (0.994) of the XGBoost model rank second among the four models. | the XGBoot (eXtreme Gradient Boosting) decision tree model, LR (logistic regression), SVM (support vector machines), and RF (random forest) | It indicates that the minimum creatinine was more useful in predicting AKI mortality than any of the other laboratory measurements or vital signs | 33539390 |
| 2021 | AKI | Phenomics (64 input variables) | 2,009 patients | Risk prediction | develop a predictive model for contrast induced nephropathy (CIN) in patients with coronary artery disease (CAD) with relatively normal renal function (NRF) | With baseline UA ≥450 μmol/L, CK-MB ≥48 U/L, and NT-proBNP ≥850 pg/mL as the cut-off values, patients were devided into two risk groups (low risk and high risk, corresponding to a total score of <10 and ≥10, respectively).The CIN incidence of patients in the low-risk group was 1.0%, while the incidence increased to 14.8% when the total score was ≥10. | elastic net | Three independent risk factors in patients with CAD with relatively NRF for CI-AKI were: baseline UA level, CK-MB level, and NT-proBNP level of CIN. | 34778413 |
| 2021 | AKI | Metabolomics | 214 individuals (122 patients with AKI, 92 patients without AKI as controls) | Risk prediction | develop and evaluate the diagnostic use of metabolomics-based biomarkers in patients with Cardiac surgery-associated acute kidney injury (CSA-AKI) | Gluconic acid, fumaric acid, and pseudouridine were significantly upregulated in patients with AKI. A random forest model constructed with selected clinical parameters and metabolites exhibited excellent discriminative ability (area under curve, 0.939). In the AKI swine model, plasma levels of the 3 discriminating metabolites increased in a time-dependent manner. The predictive model remained robust when tested in a subset of patients with early-stage AKI in the validation cohort (area under curve, 0.943). | random forest (RF), support vector machine (SVM), and logistic regression (LR) |
High-resolution metabolomics is sufficiently powerful for developing novel biomarkers. Plasma levels of 3 metabolites were useful for the early identification of CSA-AKI. | 34719239 |
| 2021 | AKI | Phenomics (NA) | 4,289 hospitalized adult patients with acute kidney injury at admission | Subphenotyping | cluster patients with acute kidney injury at hospital admission into clinically distinct subtypes using an unsupervised machine learning approach and assess the mortality risk among the distinct clusters | Consensus clustering analysis identified four distinct clusters. Cluster 2 patients had lower serum bicarbonate, strong ion difference, and hemoglobin, but higher serum chloride, whereas cluster 3 patients had lower serum chloride but higher serum bicarbonate and strong ion difference. Cluster 4 patients were younger and more likely to be admitted for genitourinary disease and infectious disease but less likely to be admitted for cardiovascular disease. Cluster 4 patients also had more severe acute kidney injury, lower serum sodium, serum chloride, and serum bicarbonate, but higher serum potassium and anion gap. Cluster 2, 3, and 4 patients had significantly higher hospital and one-year mortality than cluster 1 patients (p < 0.001). | unsupervised consensus clustering analysis | It demonstrated using machine learning consensus clustering analysis to characterize a heterogeneous cohort of patients with acute kidney injury on hospital admission into four clinically distinct clusters with different associated mortality risks. | 34698185 |
| 2021 | AKI | Metabolomics | 1st group included 61 patients (non-AKI: 23, mild AKI: 24, severe AKI: 14) and 2nd group included 60 additional patients (non-AKI: 40, mild AKI: 20). | Risk prediction | identify metabolites potentially associated with the progression of AKI | Uinre glycine and ethanolamine were decreased in patients with AKI compared with non-AKI patients at 6-24 h in the two groups. The linear statistical model constructed at each time point by machine learning achieved the best performance at 24 h (median AUC 89%) for the 1st group. When cross-validated between the two groups, the AUC showed the best value of 70% at 12 h. | the least absolute shrinkage and selection operator (LASSO) | Urinary metabolomic profile analyses identified metabolites and time points that show patterns specific to patients who develop AKI. | 34677386 |
| 2021 | AKI | Phenomics (52input variables) | 6093 patients (2442 in training and 3651 in external validation) | Risk prediction | predict acute dialysis initiation in AKI of hospitalized patients with coronavirus disease 2019 (COVID-19) | Of the five machine learning models tested, the nonimputed XGBoost had the best performance, with an AUROC of 0.98 at 1 day, 0.96 at 3 days, 0.94 at 5 days, and 0.93 at 7 days on internal validation. The AUPRC was 0.82 at 1 day, 0.80 at 3 days, 0.79 at 5 days, and 0.78 at 7 days. Using the optimal threshold derived using the Youden J statistic, nonimputed XGBoost had the highest sensitivity (ranging between 0.84 and 0.95) and specificity (0.9–0.96) across time points in the internal validation cohort. | logistic regression, Least Absolute Shrinkage and Selection Operator (LASSO), random forest, and eXtreme GradientBoosting (XGBoost0 | During all time horizons, serum creatinine at admission was one of the major features driving model predictions. Other clinically relevant features included BUN, systolic BP, age, and oxygen saturation. | 34031183 |
| 2021 | AKI | Phenomics (EHR data from the Medical Information Mart for Intensive Care III (MIMIC-III) database) | 12,347 patient | Risk prediction | evaluate the ability of a machine learning algorithm to predict for AKI as defined by KDIGO stage 2 or 3 up to 48 hours | On a hold-out test set, the CNN algorithm attained an AUROC of 0.86 and PPV of 0.24, relative to a cohort AKI prevalence of 7.62%, for long-horizon AKI prediction at a 48-hour window before onset. | XGBoost, and convolutional neural networks (CNNs) | A CNN machine learning-based AKI prediction model outperforms XGBoost and the SOFA scoring system, revealing superior performance in predicting AKI 48 hours before onset, without reliance on serum creatinine (SCr) measurements. | 34013107 |
An increasing amount of studies taking artificial intelligence for predicting, diagnosing and sub-phenotyping. And there are many promising models, but exhibit variable performance. The variability in ML prediction can be attributed, in part, to the specific ML model utilized, variable selection and processing, study and subject characteristics, and the steps associated with model training, validation, testing, and calibration. Apart from traditional models, there are some studies investigating ∼omics in AKI prediction. IgAN: IgA nephropathy; AKI: Acute kidney injury; EHRs: Electronic health records; PCI: Percutaneous coronary intervention; GBM: Gradient boosting machine; DT: Decision tree; RF: Random forest; SVM: Support vector machine; LR: Logistic regression; RNN: Recurrent neural network; ANN: Artificial neural network; ROC: Receiver operating characteristic curve; AUC: Area under curve.
For AKI, Tomasev et al. applied DL techniques to a large US Veterans Affairs (VA) longitudinal dataset of 703,782 adult patients across 172 inpatient and 1,062 outpatient sites to develop a predictive model for AKI [123], [135], [136]. With a lead time of up to 48 h, it was reported that the model could predict 55.8% of all inpatient episodes and 90.2% of all dialysis cases within 90 days of the initial onset. However, for every true alert, their algorithm generated two false positives. Churpek et al. described a predictive model for AKI developed from 495,971 adult patients. The results showed an area under the curve (AUC) as high as 0.92 for AKI and 0.97 for dialysis. However, considering the low prevalence of outcome events (3.4% stage 2 AKI), the positive-predictive value was low [137]. In an assessment of the MIMIC 3 dataset, Xu et al. identified three phenotypes in patients with a high risk of AKI and associated with the subsequent severity stage of AKI [138]. Sandokji et al. reviewed the EHRs of 6328 hospitalized children retrospectively, and identified a time-updated prediction model with ten readily available variables to predict AKI [139]. The retrospective nature of the study limited its ability to make clear conclusions on the causality between predictor variables and the outcome [140]. Future AI algorithms developed to support real-time decisions in AKI, should be based on complete medical information, including EHRs, medical history, physical signs, laboratory data, and medications. AKI risk scores based on such an algorithm would stratify patients and inform clinical decisions to enable personalized therapy [141]. Utilizing an AI approach through perioperative data-driven learning to predict cardiac surgery-associated acute kidney injury, suggested that the top three most influential features were the intraoperative urine output, the preoperative hemoglobin level, and transfused blood units during surgery [142]. This study represented a commendable attempt to optimize postoperative strategies to minimize complications following cardiac surgery.
CKD screening is a major challenge for both community and primary care settings, even in high-income countries. Sabanayagam et al developed a DL algorithm to detect CKD using retinal images, with an AUC of 0.81–0.94 [143]. Kuo et al. predicted kidney function based on ultrasound images with an overall CKD classification accuracy of 85.6%, offering a possibility of noninvasive assessment of kidney function [144]. It was reported that a deep-learning model based on electrocardiogram (ECG) data might improve the detection of life-threatening hyperkalemia in CKD, enabling noninvasive screening [145]. Shang et al. designed an electronic CKD phenotype [146] using a combination of rule-based and ML methods to identify patients on the staging grid of albuminuria by the glomerular filtration rate (“A-by-G” grid). Applying this phenotype to 1.3 million patients indicated that over 80% of CKD are undetected. This is might significant to improve phenotype definitions in studies of CKD.
Accurate ESKD prediction is vitally important in improving clinical outcomes. To predict rapid kidney function decline, using EHR data from 118,584 patients, Inaguma et al. observed that the amount of urine protein, especially an increasing tendency, could serve as a prominent risk factor [147]. Using an unsupervised latent class mixed model [148], Raynaud et al. identified that donor age, eGFR, proteinuria, and histological features including graft scarring, interstitial inflammation and tubulitis, microcirculation inflammation, and circulating anti-HLA donor specific antibodies, could predict long-term kidney allograft outcomes. Using a retrospective cohort of 948 patients, Schena et al. developed an ANN ESKD prediction model in IgA nephropathy [149]. Chen et al. constructed a scoring scale model (SSM) with three variables (tubular atrophy/interstitial fibrosis, global sclerosis, and urine protein excretion). They reported that the 5-year risks for the combined event using the SSM risk scores for 0–4 points were 2.7%, 4.8%, 16.4%, 30.8%, and 72.4%, respectively. To date, several different models have been developed and tested to predict ESKD in IgAN [150], [151], [152], [153], [154] (Table 1); however, results were conflicting, partially because different predictors were involved and the selected predictors were limited. If the algorithm captures biases in the training data, the conclusions would not be strongly supported. In cases of erroneous discoveries, it will be important to have a better understanding of the disease and relationships among predictors, as well as robust validation and mechanistic follow-up studies. Interestingly, Gonzalo-Calvo et al. suggested that adding -omics data, including circulating microribonucleic acids, to machine learning modeling could improve cardiovascular risk prediction in patients with ESKD on hemodialysis [155]. In addition, the inclusion of metabolites might also improve risk prediction in people with diabetes [156]. However, sample sizes in -omics studies need to be increased for reliable read-outs.
A beneficial impact of AI on life quality and survival in dialysis and kidney transplant patients has been proposed. It was suggested that AI-based algorithms were better than nephrologists to predict volumes, Kt/V, hypotension, and cardiovascular events [157]. The applications of AI in dialysis also include: service process, dialysis procedure, anemia management, diet, arteriovenous fistula assessment, peritoneal techniques, infections, and outcome prediction. Applications of AI in transplantation studies include pretransplant organ-matching, graft rejection prediction, and therapy dose modulation.
5.3. AI based system for rare kidney diseases
There are over 8000 kinds of rare diseases (RDs) worldwide. About 80% of RDs are genetic and about 75% are childhood onset. Although individually rare, it is estimated that they could affect 350 million people in total, representing a major challenge for healthcare. Patients with RDs frequently have several obstacles: multi-organ involvement without specific symptoms leading to late diagnosis and misdiagnosis, lack of targeted therapies, and an absence of monitoring systems. Thus, these patients could derive a particular benefit from AI technologies [158], [159], [160]. To integrate and analyze data from different sources (e.g., multi-omics and patient registries), the use of AI can overcome the above challenges. Opportunities will include increased access to various expertise, improved patient participation and monitoring, and better patient empowerment. AI technologies can be used to identify patients from EHR data [161], to provide real-world information about their medical history, and to determine effective drug selection and therapeutic regimens [162], [163]. In genetic counseling, AI has also shown its superiority to improve the accuracy for variant calling, prediction, and classification [164], [165], [166].
5.4. Drug discovery
A variety of AI methods, such as Bayesian, support vector machines, and DNNs have demonstrated their merits in drug development. These methods leverage big datasets obtained from high-throughput screening and allow the prediction of target properties with enhanced accuracy [167], [168], [169]. AI has also been used to text-mine literature using natural language processing (NLP), to search druggable targets using genome wide data [170], [171], [172], and to perform ‘virtual screens’ on vast numbers of compounds. In the field of structure-based drug development or targeted mutagenesis, knowledge about protein structure is imperative. Despite of decades of effort, only about 100,000 proteins revealed structures, representing 17% of human protein sequences. AlphaFold is a newly developed ML approach that incorporates physical and biological knowledge, leveraging multi-sequence alignments into the algorithm design [173], [174], [175], [176], [177]. By applying AlphaFold2 (https://alphafold.ebi.ac.uk/) to 98.5% of human proteins at scale, it disclosed 58% of the residues with a confident prediction. AlphaFold is a co-evolution-dependent method. By contrast, physics-based approaches, such as end-to-end differentiable models, semi-supervised approaches, and generative approaches, are also gradually maturing, and might provide a complementary path to tackle the prediction of protein structure [178]. These AI tools are predicted to accelerate future drug design. Unfortunately, drug development is a lengthy process.
5.5. Precision medicine
Precision medicine is generalized as the delivery of tailored interventions to specific patients, i.e., “the right drug for the right patient at the right time”. The practice largely depends on detailed molecular characteristics generated from -omics investigations. Omics data must be integrated with accurate clinical measurements and detailed pathological assessments. While there are intrinsic limitations to the use of traditional statistical methods to deal with multi-layered big data, AI is a promising technique to enable the use and integration of -omics data. For instance, in inflammatory bowel disease, ML techniques are able to stratify patients into sub-phenotypes based on the integration of immunological findings, endoscopic observation, and histological data [179], [180]. Decision support systems based on AI have been used in patients receiving hemodialysis to guide drug and dialysis dose prescription [181], [182]. In addition, ML models can help to cluster clinically similar patients into molecularly different phenotypes, based on ‘-omics’ data, to select more effective treatment options [183]. These perspectives can be used as a reference to stratify benefits and risks, as well as to develop individualize treatment, by introducing non-invasive and more specific diagnostic classifications.
Several collaborative projects have been launched to achieve the goal of precision medicine in KDs. The Kidney Precision Medicine Project (KPMP) (https://www.kpmp.org/) is an NIH-funded plan that is actively generating molecular and 3D imaging maps of reference kidneys from patients with AKI and CKD. The goal is to promote data sharing and accelerate molecular diagnosis of common KDs, finally aiming to develop more precise advanced therapy [184]. In this context, a repository of clinical and biospecimen data named the Kidney Tissue Atlas has been created. It will be helpful to use molecular data to define novel subtypes, which currently suffers from insufficient classification. Then, the anticipated targeted and effective therapies could be discovered. Other cohorts/consortia initiated include the ERCB (European Renal cDNA Bank), the CKDGen Consortium (https://ckdgen.imbi.uni-freiburg.de), NEPTUNE (Nephrotic Syndrome Study Network) (https://www.neptune-study.org/), Clinical Phenotyping Resource and Biobank Core (C-Probe) (https://kidneycenter.med.umich.edu), and the CRIC cohort (Chronic Renal Insufficiency Cohort) (http://www.cristudy.org).
6. Current limitations and future directions
As discussed in this review, new opportunities are opening up for nephrologists because of the development of -omics technologies and AI methods. These new technologies will contribute to a better understanding of the molecular pathophysiology of kidney diseases, their mechanistic classification, and their targeted therapy (Fig. 6). In this scenario, multi-source data integration plays an important role. However, we need to recognize that the meaningful results of computational analysis can be only derived from valid data. Big data, particularly data from real-world clinical practice, are usually biased and might be mis-interpreted. Most reported studies also suffer from the biases resulting from the use of retrospective cohorts. The structures of clinical data are complex, and need to be linked with different data resources. Data storage might result in errors and missing information is almost inevitable. In addition, there is a lack of standardized reporting systems that are easy to process and identify. For -omics studies, the quality of research, the transparency and reproducibility of assay data, generalization issues resulting from results being produced on different platforms or laboratories, are all critical points. Using standardized references, assay protocols, and procedures for quality control will improve reproducibility among studies. In addition, large -omics studies have the potential to find the most noteworthy changes, but are less useful to observe uncommon, yet likely defining, events. Thus, it is necessary to complement strategies based on a priori knowledge and mechanistic models. Considerations of ethical issues are also required when merging data access and sharing. Data disparities among different populations, inappropriate use of datasets, inaccurate or inappropriate disclosures, limitations in deidentification techniques, and little incentive for data sharing in the current healthcare systems, may lead to privacy breaches, minority discriminatory and biased treatment. Technologies of AI will undoubtedly alter the traditional physician-patient relationship, but it may also introduce safety challenges once system performance fluctuates. In addition, for all medical devices and regimens, there is tension between providing ethical medical care and generating profit. One key element to implementing AI in healthcare is development of regulatory standards for assessment of safety and efficacy. One of the organizations leading guidance in this area is the International Medical Device Regulators Forum (IMDRF), whose members currently include Australia, Brazil, Canada, China, Europe, Japan, Russia, Singapore, South Korea, and the United States [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116]. The application of multi-omics, and artificial intelligence in nephrology is still at an early discovery stage. We need to create and execute a roadmap to validate those basic discoveries and further translate into clinical care, creating a new era of personalized medicine to improve health and kidney outcomes.
Fig. 6.
Interrogation of multi-layered omics data using by artificial intelligence to address disease molecular subtypes, to diagnosis and predict diseases early, and to develop precise targeted therapy are the current challenges in kidney diseases. Information from an individual can be de-identified and coded at the individual level, and then captured across multiple data domains, including omics and/or pathology data from tissue, urine or blood samples as well as electronic health records (EHR). Such approaches have the potential to aid risk stratification, provide insights into novel mechanisms associated with kidney disease, and aid the identification of clinically relevant subpopulations or subgroups who may benefit from targeted therapy.
Author contribution statement
ZXJ: Collected data, wrote the manuscript, revised the manuscript, and gave final approval of the version to be published. ZXH: Edited the manuscript. ZXH: Edited the manuscript.
Declaration of competing interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
Support was provided by the National Science Foundation of China (82022010, 82131430172, 81970613); the Beijing Natural Science Foundation (Z190023); Academy of Medical Sciences –Newton Advanced Fellowship (NAFR13\1033); the Fok Ying Tung Education Foundation (171030); the Beijing Nova Program Interdisciplinary Cooperation Project (Z191100001119004); and the Chinese Academy of Medical Sciences Research Unit (2019RU023). The funders had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript. We apologize to those whose work or original publication could not be cited because of space limitations.
Biography

Xu-jie Zhou obtained his PhD degree from Peking University, China. He started his career at Peking University First Hospital since 2011, and is currently the associate professor. His major research interests focus on basic and translational research in kidney diseases. Zhou has published over 90 peer-reviewed papers.
References
- 1.Foreman K.J., Marquez N., Dolgert A., et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: Reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet. 2018;392(10159):2052–2090. doi: 10.1016/S0140-6736(18)31694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luyckx V.A., Al-Aly Z., Bello A.K., et al. Sustainable Development Goals relevant to kidney health: An update on progress. Nat. Rev. Nephrol. 2021;17(1):15–32. doi: 10.1038/s41581-020-00363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalantar-Zadeh K., Jafar T.H., Nitsch D., et al. Chronic kidney disease. Lancet. 2021 doi: 10.1016/S0140-6736(21)00519-5. [DOI] [PubMed] [Google Scholar]
- 4.Luo S., Grams M.E. Epidemiology research to foster improvement in chronic kidney disease care. Kidney Int. 2020;97(3):477–486. doi: 10.1016/j.kint.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Zhao M.H., Zuo L., et al. China Kidney Disease Network (CK-NET) 2016 Annual Data Report. Kidney Int. Suppl. 2020;10(2):e97–e185. doi: 10.1016/j.kisu.2020.09.001. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindenmeyer M.T., Alakwaa F., Rose M., et al. Perspectives in systems nephrology. Cell Tissue Res. 2021 doi: 10.1007/s00441-021-03470-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy S., Mariani L.H., Kretzler M. Integrated multi-omics approaches to improve classification of chronic kidney disease. Nat. Rev. Nephrol. 2020;16(11):657–668. doi: 10.1038/s41581-020-0286-5. [DOI] [PubMed] [Google Scholar]
- 8.Dubin R.F., Rhee E.P. Proteomics and Metabolomics in Kidney Disease, including Insights into Etiology, Treatment, and Prevention. Clin J Am Soc Nephrol. 2020;15(3):404–411. doi: 10.2215/CJN.07420619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes M.B. Classification systems in renal pathology: Promises and problems. Surg Pathol Clin. 2014;7(3):427–441. doi: 10.1016/j.path.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Saez-Rodriguez J., Rinschen M.M., Floege J., et al. Big science and big data in nephrology. Kidney Int. 2019;95(6):1326–1337. doi: 10.1016/j.kint.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 11.Niel O., Bastard P. Artificial intelligence in nephrology: Core concepts, clinical applications, and perspectives. Am. J. Kidney Dis. 2019;74(6):803–810. doi: 10.1053/j.ajkd.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Chan L., Vaid A., Nadkarni G.N. Applications of machine learning methods in kidney disease: Hope or hype? Curr. Opin. Nephrol. Hypertens. 2020;29(3):319–326. doi: 10.1097/MNH.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thongprayoon C., Kaewput W., Kovvuru K., et al. Promises of big data and artificial intelligence in nephrology and transplantation. J Clin Med. 2020;9(4) doi: 10.3390/jcm9041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermsen M., Smeets B., Hilbrands L., et al. Artificial intelligence: Is there a potential role in nephropathology? Nephrol. Dial. Transplant. 2020 doi: 10.1093/ndt/gfaa181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sealfon R., Mariani L.H., Kretzler M., et al. Machine learning, the kidney, and genotype-phenotype analysis. Kidney Int. 2020;97(6):1141–1149. doi: 10.1016/j.kint.2020.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker J.U., Mayerich D., Padmanabhan M., et al. Artificial intelligence and machine learning in nephropathology. Kidney Int. 2020;98(1):65–75. doi: 10.1016/j.kint.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemley K.V. Machine learning comes to nephrology. J. Am. Soc. Nephrol. 2019;30(10):1780–1781. doi: 10.1681/ASN.2019070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedersen H.K., Forslund S.K., Gudmundsdottir V., et al. A computational framework to integrate high-throughput ‘-omics’ datasets for the identification of potential mechanistic links. Nat. Protoc. 2018;13(12):2781–2800. doi: 10.1038/s41596-018-0064-z. [DOI] [PubMed] [Google Scholar]
- 19.Chong J., Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017;7(4) doi: 10.3390/metabo7040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devuyst O., Pattaro C. The UMOD locus: Insights into the pathogenesis and prognosis of kidney disease. J. Am. Soc. Nephrol. 2018;29(3):713–726. doi: 10.1681/ASN.2017070716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köttgen A., Glazer N.L., Dehghan A., et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009;41(6):712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trudu M., Janas S., Lanzani C., et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 2013;19(12):1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devuyst O., Olinger E., Weber S., et al. Autosomal dominant tubulointerstitial kidney disease. Nat. Rev. Dis. Primers. 2019;5(1):60. doi: 10.1038/s41572-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 24.Devuyst O., Olinger E., Rampoldi L. Uromodulin: From physiology to rare and complex kidney disorders. Nat. Rev. Nephrol. 2017;13(9):525–544. doi: 10.1038/nrneph.2017.101. [DOI] [PubMed] [Google Scholar]
- 25.Shlipak M.G., Day E.C. Biomarkers for incident CKD: A new framework for interpreting the literature. Nat. Rev. Nephrol. 2013;9(8):478–483. doi: 10.1038/nrneph.2013.108. [DOI] [PubMed] [Google Scholar]
- 26.Friedman D.J., Pollak M.R. APOL1 and kidney disease: From genetics to biology. Annu. Rev. Physiol. 2020;82:323–342. doi: 10.1146/annurev-physiol-021119-034345. [DOI] [PubMed] [Google Scholar]
- 27.Zhou X.J., Klionsky D.J., Zhang H. Podocytes and autophagy: A potential therapeutic target in lupus nephritis. Autophagy. 2019;15(5):908–912. doi: 10.1080/15548627.2019.1580512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freedman B.I., Sedor J.R. Intensive blood-pressure control in hypertensive chronic kidney disease. N. Engl. J. Med. 2010;363(26):2565–2566. doi: 10.1056/NEJMc1011419. 2565. [DOI] [PubMed] [Google Scholar]
- 29.Parsa A., Kao W.H., Xie D., et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams W.W., Pollak M.R. Health disparities in kidney disease–emerging data from the human genome. N. Engl. J. Med. 2013;369(23):2260–2261. doi: 10.1056/NEJMe1312797. [DOI] [PubMed] [Google Scholar]
- 31.Cohen S.D., Kopp J.B., Kimmel P.L. Kidney diseases associated with human immunodeficiency virus infection. N. Engl. J. Med. 2017;377(24):2363–2374. doi: 10.1056/NEJMra1508467. [DOI] [PubMed] [Google Scholar]
- 32.Nadkarni G.N., Gignoux C.R., Sorokin E.P., et al. Worldwide frequencies of APOL1 renal risk variants. N. Engl. J. Med. 2018;379(26):2571–2572. doi: 10.1056/NEJMc1800748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beckerman P., Bi-Karchin J., Park A.S., et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat. Med. 2017;23(4):429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayek S.S., Koh K.H., Grams M.E., et al. A tripartite complex of suPAR, APOL1 risk variants and α(v)β(3) integrin on podocytes mediates chronic kidney disease. Nat. Med. 2017;23(8):945–953. doi: 10.1038/nm.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosset S., Tzur S., Behar D.M., et al. The population genetics of chronic kidney disease: Insights from the MYH9-APOL1 locus. Nat. Rev. Nephrol. 2011;7(6):313–326. doi: 10.1038/nrneph.2011.52. [DOI] [PubMed] [Google Scholar]
- 36.Drawz P.E., Sedor J.R. The genetics of common kidney disease: A pathway toward clinical relevance. Nat. Rev. Nephrol. 2011;7(8):458–468. doi: 10.1038/nrneph.2011.85. [DOI] [PubMed] [Google Scholar]
- 37.Genovese G., Friedman D.J., Pollak M.R. APOL1 variants and kidney disease in people of recent African ancestry. Nat. Rev. Nephrol. 2013;9(4):240–244. doi: 10.1038/nrneph.2013.34. [DOI] [PubMed] [Google Scholar]
- 38.Sidaway P. Glomerular disease: Innate immunity-APOL1 interaction. Nat. Rev. Nephrol. 2014;10(10):543. doi: 10.1038/nrneph.2014.158. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg A.Z., Naicker S., Winkler C.A., et al. HIV-associated nephropathies: Epidemiology, pathology, mechanisms and treatment. Nat. Rev. Nephrol. 2015;11(3):150–160. doi: 10.1038/nrneph.2015.9. [DOI] [PubMed] [Google Scholar]
- 40.Nobakht E., Cohen S.D., Rosenberg A.Z., et al. HIV-associated immune complex kidney disease. Nat. Rev. Nephrol. 2016;12(5):291–300. doi: 10.1038/nrneph.2015.216. [DOI] [PubMed] [Google Scholar]
- 41.Brown K.D., Campbell C., Roberts G.V. Precision medicine in kidney disease: The patient's view. Nat. Rev. Nephrol. 2020;16(11):625–627. doi: 10.1038/s41581-020-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adeyemo A.A., Shriner D., Bentley A.R., et al. Evolutionary genetics and acclimatization in nephrology. Nat. Rev. Nephrol. 2021;17(12):827–839. doi: 10.1038/s41581-021-00483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genovese G., Friedman D.J., Ross M.D., et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X.Q., Li M., Zhang H., et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat. Genet. 2011;44(2):178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- 45.Kiryluk K., Li Y., Scolari F., et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 2014;46(11):1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Rodriguez E., Southard C.T., Kiryluk K. GWAS-Based discoveries in IgA nephropathy, membranous nephropathy, and Steroid-Sensitive nephrotic syndrome. Clin J Am Soc Nephrol. 2021;16(3):458–466. doi: 10.2215/CJN.14031119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Köttgen A., Pattaro C. The CKDGen Consortium: Ten years of insights into the genetic basis of kidney function. Kidney Int. 2020;97(2):236–242. doi: 10.1016/j.kint.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 48.Gorski M., Jung B., Li Y., et al. Meta-analysis uncovers genome-wide significant variants for rapid kidney function decline. Kidney Int. 2021;99(4):926–939. doi: 10.1016/j.kint.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillies C.E., Putler R., Menon R., et al. An eQTL landscape of kidney tissue in human nephrotic syndrome. Am. J. Hum. Genet. 2018;103(2):232–244. doi: 10.1016/j.ajhg.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu C., Huang S., Park J., et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 2018;24(11):1721–1731. doi: 10.1038/s41591-018-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doke T., Huang S., Qiu C., et al. Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis. J. Clin. Invest. 2021;131(10) doi: 10.1172/JCI141801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ko Y.A., Yi H., Qiu C., et al. Genetic-Variation-Driven Gene-Expression changes highlight genes with important functions for kidney disease. Am. J. Hum. Genet. 2017;100(6):940–953. doi: 10.1016/j.ajhg.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martini S., Nair V., Keller B.J., et al. Integrative biology identifies shared transcriptional networks in CKD. J. Am. Soc. Nephrol. 2014;25(11):2559–2572. doi: 10.1681/ASN.2013080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ledo N., Ko Y.A., Park A.S., et al. Functional genomic annotation of genetic risk loci highlights inflammation and epithelial biology networks in CKD. J. Am. Soc. Nephrol. 2015;26(3):692–714. doi: 10.1681/ASN.2014010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., Haug S., Schlosser P., et al. Integration of GWAS summary statistics and gene expression reveals target cell types underlying kidney function traits. J. Am. Soc. Nephrol. 2020;31(10):2326–2340. doi: 10.1681/ASN.2020010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xhonneux L.P., Knight O., Lernmark Å., et al. Transcriptional networks in at-risk individuals identify signatures of type 1 diabetes progression. Sci. Transl. Med. 2021;13(587) doi: 10.1126/scitranslmed.abd5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang H.M., Ahn S.H., Choi P., et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21(1):37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu X., Jiang L., Long M., et al. Metabolic reprogramming and renal fibrosis. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.746920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menez S., Ju W., Menon R., et al. Urinary EGF and MCP-1 and risk of CKD after cardiac surgery. JCI Insight. 2021;6(11) doi: 10.1172/jci.insight.147464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ju W., Nair V., Smith S., et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci. Transl. Med. 2015;7(316):193r–316r. doi: 10.1126/scitranslmed.aac7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teteris S.A., Menahem S.A., Perry G., et al. Dysregulated growth factor gene expression is associated with tubulointerstitial apoptosis and renal dysfunction. Kidney Int. 2007;71(10):1044–1053. doi: 10.1038/sj.ki.5002176. [DOI] [PubMed] [Google Scholar]
- 62.Josefsberg Z., Ross S.A., Lev-Ran A., et al. Effects of enalapril and nitrendipine on the excretion of epidermal growth factor and albumin in hypertensive NIDDM patients. Diabetes Care. 1995;18(5):690–693. doi: 10.2337/diacare.18.5.690. [DOI] [PubMed] [Google Scholar]
- 63.Barwinska D., El-Achkar T.M., Melo F.R., et al. Molecular characterization of the human kidney interstitium in health and disease. Sci. Adv. 2021;7(7) doi: 10.1126/sciadv.abd3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuppe C., Ibrahim M.M., Kranz J., et al. Decoding myofibroblast origins in human kidney fibrosis. Nature. 2021;589(7841):281–286. doi: 10.1038/s41586-020-2941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt I.M., Colona M.R., Kestenbaum B.R., et al. Cadherin-11, Sparc-related modular calcium binding protein-2, and Pigment epithelium-derived factor are promising non-invasive biomarkers of kidney fibrosis. Kidney Int. 2021 doi: 10.1016/j.kint.2021.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu H., Malone A.F., Donnelly E.L., et al. Single-Cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 2018;29(8):2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y., Hu J., Liu D., et al. Single-cell analysis reveals immune landscape in kidneys of patients with chronic transplant rejection. Theranostics. 2020;10(19):8851–8862. doi: 10.7150/thno.48201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Der E, Ranabothu S., Suryawanshi H., et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight. 2017;2(9) doi: 10.1172/jci.insight.93009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson P.C., Wu H., Kirita Y., et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A. 2019;116(39):19619–19625. doi: 10.1073/pnas.1908706116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Sullivan E.D., Mylonas K.J., Hughes J., et al. Complementary roles for Single-Nucleus and Single-Cell RNA sequencing in kidney disease research. J. Am. Soc. Nephrol. 2019;30(4):712–713. doi: 10.1681/ASN.2019020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng Y., Lu P., Deng Y., et al. Single-Cell transcriptomics reveal immune mechanisms of the onset and progression of IgA nephropathy. Cell Rep. 2020;33(12) doi: 10.1016/j.celrep.2020.108525. [DOI] [PubMed] [Google Scholar]
- 72.Ronco P., Debiec H. Molecular pathogenesis of membranous nephropathy. Annu Rev Pathol. 2020;15:287–313. doi: 10.1146/annurev-pathol-020117-043811. [DOI] [PubMed] [Google Scholar]
- 73.Ronco P., Debiec H. Pathophysiological advances in membranous nephropathy: Time for a shift in patient’s care. Lancet. 2015;385(9981):1983–1992. doi: 10.1016/S0140-6736(15)60731-0. [DOI] [PubMed] [Google Scholar]
- 74.Beck L.J., Bonegio R.G., Lambeau G., et al. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N. Engl. J. Med. 2009;361(1):11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stanescu H.C., Arcos-Burgos M., Medlar A., et al. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N. Engl. J. Med. 2011;364(7):616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 76.Cui Z., Xie L.J., Chen F.J., et al. MHC class II risk alleles and amino acid residues in idiopathic membranous nephropathy. J. Am. Soc. Nephrol. 2017;28(5):1651–1664. doi: 10.1681/ASN.2016020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lv J., Hou W., Zhou X., et al. Interaction between PLA2R1 and HLA-DQA1 variants associates with anti-PLA2R antibodies and membranous nephropathy. J. Am. Soc. Nephrol. 2013;24(8):1323–1329. doi: 10.1681/ASN.2012080771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H.Y., Cui Z., Xie L.J., et al. HLA class II alleles differing by a single amino acid associate with clinical phenotype and outcome in patients with primary membranous nephropathy. Kidney Int. 2018;94(5):974–982. doi: 10.1016/j.kint.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Xie J., Liu L., Mladkova N., et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat. Commun. 2020;11(1):1600. doi: 10.1038/s41467-020-15383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matías-García P., Wilson R., Guo Q., et al. Plasma proteomics of renal function: A trans-ethnic meta-analysis and mendelian randomization study. J. Am. Soc. Nephrol. 2021 doi: 10.1681/ASN.2020071070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park S., Lee J., Yang S.H., et al. Comprehensive metabolomic profiling in early IgA nephropathy patients reveals urine glycine as a prognostic biomarker. J. Cell. Mol. Med. 2021;25(11):5177–5190. doi: 10.1111/jcmm.16520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steinbrenner I., Schultheiss U.T., Kotsis F., et al. Urine metabolite levels, adverse kidney outcomes, and mortality in CKD patients: A metabolome-wide association study. Am. J. Kidney Dis. 2021 doi: 10.1053/j.ajkd.2021.01.018. [DOI] [PubMed] [Google Scholar]
- 83.Wu I.W., Gao S.S., Chou H.C., et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. 2020;10(12):5398–5411. doi: 10.7150/thno.41725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X., Yang S., Li S., et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. 2020;69(12):2131–2142. doi: 10.1136/gutjnl-2019-319766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cardozo L.E., Russo P., Gomes-Correia B., et al. WebCEMiTool: Co-expression modular analysis made easy. Front Genet. 2019;10:146. doi: 10.3389/fgene.2019.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fukushima A. DiffCorr: An R package to analyze and visualize differential correlations in biological networks. Gene. 2013;518(1):209–214. doi: 10.1016/j.gene.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 88.Zhou G., Soufan O., Ewald J., et al. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic. Acids. Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Z.P., Wu C., Miao H., et al. RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford) 2015;2015 doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou G., Xia J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic. Acids. Res. 2018;46(W1):W514–W522. doi: 10.1093/nar/gky510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zoppi J., Guillaume J.F., Neunlist M., et al. MiBiOmics: An interactive web application for multi-omics data exploration and integration. BMC Bioinformatics. 2021;22(1):6. doi: 10.1186/s12859-020-03921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hernández-de-Diego R., Tarazona S., Martínez-Mira C., et al. PaintOmics 3: A web resource for the pathway analysis and visualization of multi-omics data. Nucleic. Acids. Res. 2018;46(W1):W503–W509. doi: 10.1093/nar/gky466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chong J., Soufan O., Li C., et al. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic. Acids. Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otasek D., Morris J.H., Bouças J., et al. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eales J.M., Jiang X., Xu X., et al. Uncovering genetic mechanisms of hypertension through multi-omic analysis of the kidney. Nat. Genet. 2021;53(5):630–637. doi: 10.1038/s41588-021-00835-w. [DOI] [PubMed] [Google Scholar]
- 96.Hodgin J.B., Nair V., Zhang H., et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013;62(1):299–308. doi: 10.2337/db11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang H., Nair V., Saha J., et al. Podocyte-specific JAK2 overexpression worsens diabetic kidney disease in mice. Kidney Int. 2017;92(4):909–921. doi: 10.1016/j.kint.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tao J., Mariani L., Eddy S., et al. JAK-STAT activity in peripheral blood cells and kidney tissue in IgA nephropathy. Clin J Am Soc Nephrol. 2020;15(7):973–982. doi: 10.2215/CJN.11010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tao J., Mariani L., Eddy S., et al. JAK-STAT signaling is activated in the kidney and peripheral blood cells of patients with focal segmental glomerulosclerosis. Kidney Int. 2018;94(4):795–808. doi: 10.1016/j.kint.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He J., Baxter S.L., Xu J., et al. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019;25(1):30–36. doi: 10.1038/s41591-018-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Medicine in the digital age. Nat. Med. 2019;25(1):1. doi: 10.1038/s41591-018-0322-1. [DOI] [PubMed] [Google Scholar]
- 102.Norgeot B., Glicksberg B.S., Butte A.J. A call for deep-learning healthcare. Nat. Med. 2019;25(1):14–15. doi: 10.1038/s41591-018-0320-3. [DOI] [PubMed] [Google Scholar]
- 103.Topol E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019;25(1):44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 104.Esteva A., Robicquet A., Ramsundar B., et al. A guide to deep learning in healthcare. Nat. Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 105.Ngiam K.Y. Braving the new world of artificial intelligence. Nat. Med. 2019;25(1):13. doi: 10.1038/s41591-018-0317-y. [DOI] [PubMed] [Google Scholar]
- 106.Gottesman O., Johansson F., Komorowski M., et al. Guidelines for reinforcement learning in healthcare. Nat. Med. 2019;25(1):16–18. doi: 10.1038/s41591-018-0310-5. [DOI] [PubMed] [Google Scholar]
- 107.Price W.N., Cohen I.G. Privacy in the age of medical big data. Nat. Med. 2019;25(1):37–43. doi: 10.1038/s41591-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mincholé A., Rodriguez B. Artificial intelligence for the electrocardiogram. Nat. Med. 2019;25(1):22–23. doi: 10.1038/s41591-018-0306-1. [DOI] [PubMed] [Google Scholar]
- 109.Gurovich Y., Hanani Y., Bar O., et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat. Med. 2019;25(1):60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 110.Abbott B. Deeper learning. Nat. Med. 2019;25(1):9–11. doi: 10.1038/s41591-018-0313-2. [DOI] [PubMed] [Google Scholar]
- 111.Hannun A.Y., Rajpurkar P., Haghpanahi M., et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019;25(1):65–69. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Attia Z.I., Kapa S., Lopez-Jimenez F., et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 2019;25(1):70–74. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 113.Arnaout R. Toward a clearer picture of health. Nat. Med. 2019;25(1):12. doi: 10.1038/s41591-018-0318-x. [DOI] [PubMed] [Google Scholar]
- 114.Rajkomar A., Dean J., Kohane I. Machine learning in medicine. N. Engl. J. Med. 2019;380(14):1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 115.Ngiam K.Y., Khor I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–e273. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 116.Schwalbe N., Wahl B. Artificial intelligence and the future of global health. Lancet. 2020;395(10236):1579–1586. doi: 10.1016/S0140-6736(20)30226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu X., Li N., Lv L., et al. Improving precision of glomerular filtration rate estimating model by ensemble learning. J. Transl. Med. 2017;15(1):231. doi: 10.1186/s12967-017-1337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Molitoris B.A. Beyond biomarkers: Machine learning in diagnosing acute kidney injury. Mayo Clin. Proc. 2019;94(5):748–750. doi: 10.1016/j.mayocp.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 119.Almansour N.A., Syed H.F., Khayat N.R., et al. Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study. Comput. Biol. Med. 2019;109:101–111. doi: 10.1016/j.compbiomed.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 120.Brier M.E., Gaweda A.E., Aronoff G.R. Personalized anemia management and precision medicine in ESA and iron pharmacology in End-Stage kidney disease. Semin. Nephrol. 2018;38(4):410–417. doi: 10.1016/j.semnephrol.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 121.Tang J., Liu R., Zhang Y.L., et al. Application of Machine-Learning models to predict tacrolimus stable dose in renal transplant recipients. Sci. Rep. 2017;7:42192. doi: 10.1038/srep42192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rashidi P., Bihorac A. Artificial intelligence approaches to improve kidney care. Nat. Rev. Nephrol. 2020;16(2):71–72. doi: 10.1038/s41581-019-0243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kellum J.A., Bihorac A. Artificial intelligence to predict AKI: Is it a breakthrough? Nat. Rev. Nephrol. 2019;15(11):663–664. doi: 10.1038/s41581-019-0203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barisoni L., Lafata K.J., Hewitt S.M., et al. Digital pathology and computational image analysis in nephropathology. Nat. Rev. Nephrol. 2020;16(11):669–685. doi: 10.1038/s41581-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bera K., Schalper K.A., Rimm D.L., et al. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alnazer I., Bourdon P., Urruty T., et al. Recent advances in medical image processing for the evaluation of chronic kidney disease. Med. Image Anal. 2021;69 doi: 10.1016/j.media.2021.101960. [DOI] [PubMed] [Google Scholar]
- 127.Uchino E., Suzuki K., Sato N., et al. Classification of glomerular pathological findings using deep learning and nephrologist-AI collective intelligence approach. Int. J. Med. Inform. 2020;141 doi: 10.1016/j.ijmedinf.2020.104231. [DOI] [PubMed] [Google Scholar]
- 128.Bülow R.D., Kers J., Boor P. Multistain segmentation of renal histology: First steps toward artificial intelligence-augmented digital nephropathology. Kidney Int. 2021;99(1):17–19. doi: 10.1016/j.kint.2020.08.025. [DOI] [PubMed] [Google Scholar]
- 129.Jayapandian C.P., Chen Y., Janowczyk A.R., et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 2021;99(1):86–101. doi: 10.1016/j.kint.2020.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ginley B., Lutnick B., Jen K.Y., et al. Computational segmentation and classification of diabetic glomerulosclerosis. J. Am. Soc. Nephrol. 2019;30(10):1953–1967. doi: 10.1681/ASN.2018121259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ginley B., Jen K.Y., Han S.S., et al. Automated computational detection of interstitial fibrosis, tubular atrophy, and glomerulosclerosis. J. Am. Soc. Nephrol. 2021;32(4):837–850. doi: 10.1681/ASN.2020050652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reeve J., Böhmig G.A., Eskandary F., et al. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am. J. Transplant. 2019;19(10):2719–2731. doi: 10.1111/ajt.15351. [DOI] [PubMed] [Google Scholar]
- 133.Barbour S.J., Coppo R., Zhang H., et al. Evaluating a new international Risk-Prediction tool in IgA nephropathy. JAMA Intern Med. 2019;179(7):942–952. doi: 10.1001/jamainternmed.2019.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Endre Z.H., Mehta R.L. Identification of acute kidney injury subphenotypes. Curr. Opin. Crit. Care. 2020;26(6):519–524. doi: 10.1097/MCC.0000000000000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tomašev N., Harris N., Baur S., et al. Use of deep learning to develop continuous-risk models for adverse event prediction from electronic health records. Nat. Protoc. 2021;16(6):2765–2787. doi: 10.1038/s41596-021-00513-5. [DOI] [PubMed] [Google Scholar]
- 136.Tomašev N., Glorot X., Rae J.W., et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572(7767):116–119. doi: 10.1038/s41586-019-1390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Churpek M.M., Carey K.A., Edelson D.P., et al. Internal and external validation of a machine learning risk score for acute kidney injury. JAMA Netw Open. 2020;3(8) doi: 10.1001/jamanetworkopen.2020.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu Z., Chou J., Zhang X.S., et al. Identifying sub-phenotypes of acute kidney injury using structured and unstructured electronic health record data with memory networks. J. Biomed. Inform. 2020;102 doi: 10.1016/j.jbi.2019.103361. [DOI] [PubMed] [Google Scholar]
- 139.Sandokji I., Yamamoto Y., Biswas A., et al. A Time-Updated, parsimonious model to predict AKI in hospitalized children. J. Am. Soc. Nephrol. 2020;31(6):1348–1357. doi: 10.1681/ASN.2019070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.De Vlieger G., Kashani K., Meyfroidt G. Artificial intelligence to guide management of acute kidney injury in the ICU: A narrative review. Curr. Opin. Crit. Care. 2020;26(6):563–573. doi: 10.1097/MCC.0000000000000775. [DOI] [PubMed] [Google Scholar]
- 141.Koyner J.L., Carey K.A., Edelson D.P., et al. The development of a machine learning inpatient acute kidney injury prediction model. Crit. Care Med. 2018;46(7):1070–1077. doi: 10.1097/CCM.0000000000003123. [DOI] [PubMed] [Google Scholar]
- 142.Tseng P.Y., Chen Y.T., Wang C.H., et al. Prediction of the development of acute kidney injury following cardiac surgery by machine learning. Crit. Care. 2020;24(1):478. doi: 10.1186/s13054-020-03179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sabanayagam C., Xu D., Ting D., et al. A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet Digit Health. 2020;2(6):e295–e302. doi: 10.1016/S2589-7500(20)30063-7. [DOI] [PubMed] [Google Scholar]
- 144.Kuo C.C., Chang C.M., Liu K.T., et al. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ Digit Med. 2019;2:29. doi: 10.1038/s41746-019-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Galloway C.D., Valys A.V., Shreibati J.B., et al. Development and validation of a Deep-Learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol. 2019;4(5):428–436. doi: 10.1001/jamacardio.2019.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shang N., Khan A., Polubriaginof F., et al. Medical records-based chronic kidney disease phenotype for clinical care and "big data" observational and genetic studies. NPJ Digit Med. 2021;4(1):70. doi: 10.1038/s41746-021-00428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Inaguma D., Kitagawa A., Yanagiya R., et al. Increasing tendency of urine protein is a risk factor for rapid eGFR decline in patients with CKD: A machine learning-based prediction model by using a big database. PLoS One. 2020;15(9) doi: 10.1371/journal.pone.0239262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Raynaud M., Aubert O., Reese P.P., et al. Trajectories of glomerular filtration rate and progression to end stage kidney disease after kidney transplantation. Kidney Int. 2021;99(1):186–197. doi: 10.1016/j.kint.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 149.Schena F.P., Anelli V.W., Trotta J., et al. Development and testing of an artificial intelligence tool for predicting end-stage kidney disease in patients with immunoglobulin a nephropathy. Kidney Int. 2021;99(5):1179–1188. doi: 10.1016/j.kint.2020.07.046. [DOI] [PubMed] [Google Scholar]
- 150.Diciolla M., Binetti G., Di Noia T., et al. Patient classification and outcome prediction in IgA nephropathy. Comput. Biol. Med. 2015;66:278–286. doi: 10.1016/j.compbiomed.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 151.Pesce F., Diciolla M., Binetti G., et al. Clinical decision support system for end-stage kidney disease risk estimation in IgA nephropathy patients. Nephrol. Dial. Transplant. 2016;31(1):80–86. doi: 10.1093/ndt/gfv232. [DOI] [PubMed] [Google Scholar]
- 152.Liu Y., Zhang Y., Liu D., et al. Prediction of ESRD in IgA nephropathy patients from an asian cohort: A random forest model. Kidney Blood Press. Res. 2018;43(6):1852–1864. doi: 10.1159/000495818. [DOI] [PubMed] [Google Scholar]
- 153.Bülow R.D., Dimitrov D., Boor P., et al. How will artificial intelligence and bioinformatics change our understanding of IgA Nephropathy in the next decade? Semin. Immunopathol. 2021 doi: 10.1007/s00281-021-00847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen T., Li X., Li Y., et al. Prediction and risk stratification of kidney outcomes in IgA nephropathy. Am. J. Kidney Dis. 2019;74(3):300–309. doi: 10.1053/j.ajkd.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 155.de Gonzalo-Calvo D., Martínez-Camblor P., Bär C., et al. Improved cardiovascular risk prediction in patients with end-stage renal disease on hemodialysis using machine learning modeling and circulating microribonucleic acids. Theranostics. 2020;10(19):8665–8676. doi: 10.7150/thno.46123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang J., Huth C., Covic M., et al. Machine learning approaches reveal metabolic signatures of incident chronic kidney disease in individuals with prediabetes and type 2 diabetes. Diabetes. 2020;69(12):2756–2765. doi: 10.2337/db20-0586. [DOI] [PubMed] [Google Scholar]
- 157.Burlacu A., Iftene A., Jugrin D., et al. Using artificial intelligence resources in dialysis and kidney transplant patients: A literature review. Biomed Res. Int. 2020;2020 doi: 10.1155/2020/9867872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hurvitz N., Azmanov H., Kesler A., et al. Establishing a second-generation artificial intelligence-based system for improving diagnosis, treatment, and monitoring of patients with rare diseases. Eur. J. Hum. Genet. 2021 doi: 10.1038/s41431-021-00928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hirsch M.C., Ronicke S., Krusche M., et al. Rare diseases 2030: How augmented AI will support diagnosis and treatment of rare diseases in the future. Ann. Rheum. Dis. 2020;79(6):740–743. doi: 10.1136/annrheumdis-2020-217125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Amiri H., Kohane I.S. Machine learning of patient characteristics to predict admission outcomes in the undiagnosed diseases network. JAMA Netw Open. 2021;4(2) doi: 10.1001/jamanetworkopen.2020.36220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bergier H., Duron L., Sordet C., et al. Digital health, big data and smart technologies for the care of patients with systemic autoimmune diseases: Where do we stand? Autoimmun. Rev. 2021;20(8) doi: 10.1016/j.autrev.2021.102864. [DOI] [PubMed] [Google Scholar]
- 162.Brasil S., Pascoal C., Francisco R., et al. Artificial intelligence (AI) in rare diseases: Is the future brighter? Genes (Basel) 2019;10(12) doi: 10.3390/genes10120978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Das S., Huang S., Lo A.W. Acceleration of rare disease therapeutic development: A case study of AGIL-AADC. Drug Discov. Today. 2019;24(3):678–684. doi: 10.1016/j.drudis.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 164.Liu Z., Zhu L., Roberts R., et al. Toward clinical implementation of Next-Generation Sequencing-Based genetic testing in rare diseases: Where are we? Trends Genet. 2019;35(11):852–867. doi: 10.1016/j.tig.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 165.Li Q., Zhao K., Bustamante C.D., et al. Xrare: A machine learning method jointly modeling phenotypes and genetic evidence for rare disease diagnosis. Genet. Med. 2019;21(9):2126–2134. doi: 10.1038/s41436-019-0439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McInnes G., Sharo A.G., Koleske M.L., et al. Opportunities and challenges for the computational interpretation of rare variation in clinically important genes. Am. J. Hum. Genet. 2021;108(4):535–548. doi: 10.1016/j.ajhg.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang X., Wang Y., Byrne R., et al. Concepts of artificial intelligence for Computer-Assisted drug discovery. Chem. Rev. 2019;119(18):10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 168.Schneider P., Walters W.P., Plowright A.T., et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2020;19(5):353–364. doi: 10.1038/s41573-019-0050-3. [DOI] [PubMed] [Google Scholar]
- 169.Ekins S., Puhl A.C., Zorn K.M., et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 2019;18(5):435–441. doi: 10.1038/s41563-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bravo À., Piñero J., Queralt-Rosinach N., et al. Extraction of relations between genes and diseases from text and large-scale data analysis: Implications for translational research. BMC Bioinformatics. 2015;16:55. doi: 10.1186/s12859-015-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim J., Kim J.J., Lee H. An analysis of disease-gene relationship from Medline abstracts by. DigSee. Sci Rep. 2017;7:40154. doi: 10.1038/srep40154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jeon J., Nim S., Teyra J., et al. A systematic approach to identify novel cancer drug targets using machine learning, inhibitor design and high-throughput screening. Genome Med. 2014;6(7):57. doi: 10.1186/s13073-014-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tunyasuvunakool K., Adler J., Wu Z., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021 doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Senior A.W., Evans R., Jumper J., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 176.Hey T., Butler K., Jackson S., et al. Machine learning and big scientific data. Philos Trans A Math Phys Eng Sci. 2020;378(2166) doi: 10.1098/rsta.2019.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Senior A.W., Evans R., Jumper J., et al. Protein structure prediction using multiple deep neural networks in the 13th Critical Assessment of Protein Structure Prediction (CASP13) Proteins. 2019;87(12):1141–1148. doi: 10.1002/prot.25834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.AlQuraishi M. AlphaFold at CASP13. Bioinformatics. 2019;35(22):4862–4865. doi: 10.1093/bioinformatics/btz422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Coelho T., Mossotto E., Gao Y., et al. Immunological profiling of paediatric inflammatory bowel disease using unsupervised machine learning. J. Pediatr. Gastroenterol. Nutr. 2020;70(6):833–840. doi: 10.1097/MPG.0000000000002719. [DOI] [PubMed] [Google Scholar]
- 180.Mossotto E., Ashton J.J., Coelho T., et al. Classification of Paediatric Inflammatory Bowel Disease using. Machine Learning. Sci Rep. 2017;7(1):2427. doi: 10.1038/s41598-017-02606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hueso M., de Haro L., Calabia J., et al. Leveraging data science for a personalized haemodialysis. Kidney Dis (Basel) 2020;6(6):385–394. doi: 10.1159/000507291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Barbieri C., Molina M., Ponce P., et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int. 2016;90(2):422–429. doi: 10.1016/j.kint.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 183.Bierzynska A., McCarthy H.J., Soderquest K., et al. Genomic and clinical profiling of a national nephrotic syndrome cohort advocates a precision medicine approach to disease management. Kidney Int. 2017;91(4):937–947. doi: 10.1016/j.kint.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 184.Ong E., Wang L.L., Schaub J., et al. Modelling kidney disease using ontology: Insights from the Kidney Precision Medicine Project. Nat. Rev. Nephrol. 2020;16(11):686–696. doi: 10.1038/s41581-020-00335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]