Abstract
The difficulty of converting scientific research findings into novel pharmacological treatments for rare and life-threatening diseases is enormous. Biomarkers related to multiple biological processes involved in cell growth, proliferation, and disease occurrence have been identified in recent years with the development of immunology, molecular biology, and genomics technologies. Biomarkers are capable of reflecting normal physiological processes, pathological processes, and the response to therapeutic intervention; as such, they play vital roles in disease diagnosis, prevention, drug response, and other aspects of biomedicine. The discovery of valuable biomarkers has become a focal point of current research. Numerous studies have identified molecular biomarkers based on the differential expression/concentration of molecules (e.g., genes/proteins) for disease state diagnosis, characterization, and treatment. Although technological breakthroughs in molecular analysis platforms have enabled the identification of a large number of candidate biomarkers for rare diseases, only a small number of these candidates have been properly validated for use in patient treatment. The traditional molecular biomarkers may lose vital information by ignoring molecular associations/interactions, and thus the concept of network biomarkers based on differential associations/correlations of molecule pairs has been established. This approach promises to be more stable and reliable in diagnosing disease states. Furthermore, the newly-emerged dynamic network biomarkers (DNBs) based on differential fluctuations/correlations of molecular groups are able to recognize pre-disease states or critical states instead of disease states, thereby achieving rare disease prediction or predictive/preventative medicine and providing deep insight into the dynamic characteristics of disease initiation and progression.
Keywords: Rare disease, Molecular biomarker, Network biomarker, Dynamic network biomarker, Diagnosis, Prognosis, Prediction
Graphical Abstract
1. Introduction
Rare disease patients face unique challenges in accessing diagnosis, medical care, and access to support services [1]. There is currently no uniform definition of a rare disease, but it is given that the incidence of these diseases is extremely low [2]. Most rare diseases are genetic, but at least one in five are caused by infections, allergies, or environmental factors [3]. Of course, this is not set in stone. Rare disease patients face two conundrums. First, a correct diagnosis often takes a long time [4]. Rare disease patients often go through a long diagnostic journey that can take years or even decades. Understandably, most doctors do not understand each and every rare disease, and to make matters worse, the same rare disease may manifest in different ways in different patients [5]. Second, even when a patient is accurately diagnosed, they may not have many treatment options and may end up resorting to therapies designed to treat the associated condition or to simply relieve some of the symptoms rather than the actual cause [6]. Currently, the US Food and Drug Administration (FDA) has no specifically approved treatments for more than 95% of rare diseases [7].
Biomarkers refer to biochemical indicators that can mark the changes or possible changes in the structure and function of cells and subcellular structures of systemic organs and tissues. Biomarkers can be used in disease diagnosis to determine the disease stage or to evaluate the safety and efficacy of new drugs or therapies in target populations. Currently, a large number of methods have been developed in the search for biomarkers, including molecular biomarkers using expression/concentration information, network biomarkers using additional network information, and dynamic network biomarkers further using the dynamics of molecular information. Our aim herein is to present a summary of the methods used to find various types of biomarkers; we further consider their strengths and examine their applicability.
In rare diseases, due to difficulties in diagnosis and limited development of specific drugs it is necessary to find markers for diagnosis and treatment, and this is a current research focus. Fabry disease (FD) is an X-linked lysosomal storage disorder caused by mutations in the GLA gene encoding the enzyme α-galactosidase A (agalA). Enzyme replacement therapy (ERT) is a commonly used treatment. The enzyme cleaves the last sugar unit of glycosphingolipids such as globotriaosylceramide (Gb3), globotriaosylsphingosine (lyso-Gb3), and galabiosylceramide (Ga2) [8]. Urinalysis for Gb3 isoforms (different fatty acid moieties) as well as for lyso-Gb3 and its analogs is a reliable method used in monitoring and treatment. The efficacy of treatment can be followed through the levels of these markers [9]. A recent study employed circulating microRNAs (miRNAs) to examine their association with multiple sclerosis (MS) and disability progression. Using serum RT-PCR detection technology to analyze the expression of circulating miRNAs in 100 subjects, it was found that miR-24-3p and miR-128-3p showed trends related to disability accumulation and disease activity in MS patients [10]. For anal squamous cell carcinoma (SCCA), a systematic review of current prognostic markers in SCCA has been performed. A series of biomarkers have been identified that are correlated with survival after chemoradiotherapy for anal cancer. The tumor suppressor genes p53 and p21 are markers that have been shown to have important prognostic value in several studies [11]. However, researchers have yet to identify a biomarker that consistently predicts the outcomes of this disease.
There are more than 8000 rare diseases that affect >5% of the world's population, but only about 5% of rare diseases currently have licensed treatments, and many rare diseases have no effective treatment. Drug development for rare diseases is plagued by multiple challenges, and clinical trial failures are not uncommon. Personalized medicine allows for early warning, diagnosis, and guide treatment of human disease. In this context, the discovery of biomarkers for rare diseases is critical for timely prevention and effective treatment. A biomarker is defined as a biological feature that can be objectively measured and assessed as a signal of normal biological or pathological processes, and thus has the potential to improve diagnosis, predict disease manifestation, and monitor the response to therapeutic intervention. Because 80% of rare diseases are of genetic origin, the identification of novel genes and disease-causing mutations is central to identifying valuable biomarkers. In addition, molecular markers such as expressed genes, metabolites, and proteins are also important for diagnosis and treatment. Current advances in platform technology for molecular analysis can identify a broad range of candidate biomarkers for rare diseases, but few of these candidates have been sufficiently validated to be integrated into clinical management programs for patients.
2. Molecular biomarkers
Genes, RNAs, proteins, and metabolites can serve as molecular biomarkers, playing important roles in the diagnosis, prognosis, prediction, and therapeutic treatment of diseases. With the accelerated development of high-throughput sequencing technologies, large amounts of omics data have been accumulated and further exploited to identify molecular biomarkers. Molecular biomarkers are defined as one or a group of individual molecules measured by differential expression/concentration between a disease state and a normal control state. For a credible molecular biomarker, it should be objectively measured and significantly abnormal in a specific disease and thus should distinguish a disease state from a normal state [12].
2.1. Methods for identifying molecular biomarkers
Methods to identify molecular biomarkers have been widely explored (Table 1). DESeq2 [13] and edgeR [14] are two well-known methods used to identify differentially expressed genes (DEGs) from RNA-sequencing data. Thousands of differentially expressed genes for a specific disease have been found using these two methods, but the number of molecular biomarkers for a certain disease should be small from the perspective of clinical applications [12]. Thus, additional statistical and data mining approaches are also applied to identify reliable molecular biomarkers of diseases; these include support vector machines (SVM) [15, 16], partial least square-discriminant analysis (PLS-DA) [17], [18], [19], least absolute shrinkage and selection operator (LASSO) [20] and recursive feature elimination (RFE) [21] to screen for potential biomarkers.
Table 1.
Overview of analysis methods for molecular biomarkers.
| Methods | Description | Applications | Reference |
|---|---|---|---|
| DESeq2 | A method using shrinkage estimation for differential expression analysis of count data. | Quantitative analysis of comparative RNA-seq data using shrinkage estimators for dispersion and fold change. | [13] |
| edgeR | Examining differential expression of replicated count data. | Can be used in experiments that generate counts. |
[14] |
| SVM | A powerful method to build a classifier. | Cancer genomic classification or subtyping. | [15, 16] |
| PLS-DA | A discrimination method based on PLS regression. | Predictive and descriptive modeling as well as for discriminative variable selection. | [17], [18], [19] |
| LASSO | A shrinkage and selection method for linear regression. | Estimating parameters and selecting variables. | [20] |
| RFE | Method of gene selection utilizing SVM methods based on recursive feature elimination. | Selecting features by recursively considering smaller sets of features. | [21] |
2.2. Application of molecular biomarkers in rare diseases
Mutations, gene polymorphisms, RNAs, proteins, and metabolites that become altered as diseases progress may be valuable molecular biomarkers for characterizing cell pathophysiology in rare diseases.
The most advanced omics technology is genomics, and research on the genome typically focuses on gene sequences and genetic variation [22]. In a genetic study of 124 people with FD, Niemann et al. discovered that the biomarker lyso-Gb3 may identify the clinically related agalA mutations leading to the disease [23]. Cerasuolo et al. developed a genetic screening test for the STK11 gene to investigate the molecular basis of the genotype-phenotype correlation of Peutz-Jeghers syndrome (PJS) and highlighted the importance of early genetic testing in young PJS patients [24].
Transcriptomics is a qualitative and quantitative omics involving RNA at the overall level. The qualitative analysis comprises studying the existence of transcripts and identifying alternative splicing events and RNA editing sites, while the quantitative analysis considers the expression level of each transcript [25]. Apart from messenger RNA (mRNA), which can encode proteins, transcriptome sequencing technology can also quantify some non-coding RNAs, including miRNAs, long non-coding RNAs (lncRNAs), and circular RNAs (cirRNAs). Many studies have shown that these non-coding RNAs play regulatory roles in the occurrence and development of multiple rare diseases [10, [26], [27], [28]]. Gupta et al. performed RNA-seq on 20 patients with MS and employed RT-qPCR on a validation cohort of 44 participants. Four lncRNAs with significant differential expression in both cohorts were regarded as biomarkers for distinguishing the severity of the MS phenotype [26]. Torroglosa et al. screened the expression of 84 lncRNAs and identified three (SOCS2-AS, MEG3, and NEAT1) associated with enteric nervous system (ENS) development in Hirschsprung disease (HSCR) [27]. Expression levels of CASC2 and miR-21 were evaluated in a study on malignant gliomas, and it was found that the lncRNA CASC2 that acts as a tumor suppressor gene was downregulated in gliomas and likely interacted with miR-21 [28].
Metabolomics is an emerging discipline that includes qualitative and quantitative analyses of small molecular weight metabolites in a certain organism or cell. The methods can help us to better understand the process of pathological change. Because body fluids can be used for metabolomics, this is a noninvasive method that can lead to the diagnosis and grading of diseases [22]. A series of published studies have shown that metabolomics can be applied to identify molecular biomarkers for rare diseases [29], [30], [31]. For example, Auray-Blais et al. identified novel urinary Fabry disease biomarkers using a time-of-flight mass spectrometry metabolomic approach [30]. Menkovic et al. performed a semi-targeted plasma metabolomic study using an ultra-performance liquid chromatography system coupled to a time-of-flight mass spectrometer to identify novel Gaucher disease (GD) biomarkers [31].
Proteins are biomolecules composed of amino acids, and they are responsible for cell functions. Proteomics studies the characteristics of proteins on a large scale to quantify their abundance and to detect the interactions between different proteins. The in-depth study of proteomics has advantages for a comprehensive understanding of the molecular mechanisms of disease, investigation of early diagnosis biomarkers, and development of new treatment schemes. Hollander et al. utilized an unbiased proteomic screening approach and identified gender-specific plasma protein biomarker panels for Anderson-Fabry disease (AFD) [32]. Benabdelkamel et al. performed serum proteomics profiling in 28 cystic fibrosis (CF) patients and identified 15 proteins as potential biomarkers for CF [33]. Cigna et al. assessed proteome changes in peripheral blood mononuclear cells to pinpoint FD-specific biomarkers and to better understand the pathophysiological changes involved in FD [34]. An overview of application of molecular biomarkers in rare diseases mentioned above is given in Table 2.
Table 2.
Application of molecular biomarkers in rare diseases.
| Data type | Sequencing technology | Examples | Reference |
|---|---|---|---|
| Genomics | WGS, WES | Identify agalA mutations determining clinically relevant FD. | [23] |
| Perform STK11 genetic screening test and investigate the genotype-phenotype correlation in PJS. | [24] | ||
| Transcriptomics | RNA-seq, RT-qPCR | Investigate possible lncRNA biomarkers to differentiate phenotypic severity in MS. | [26] |
| Perform qRT-PCR on a set of lncRNAs to identify lncRNAs associated with ENS development in HSCR. | [27] | ||
| Assess levels of CASC2 and miR-21 and their interplay in glioma. | [28] | ||
| Metabolomics | NMR, MS | Detect lyso-Gb3-related FD biomarkers in urine using mass spectrometry metabolomic approach. | [30] |
| Perform metabolomic study to identify novel GD biomarkers. | [31] | ||
| Proteomics | MS | Identify gender-specific plasma protein biomarker panels for AFD. | [32] |
| Perform serum proteomics profiling in CF patients and healthy subjects. | [33] | ||
| Assess proteome changes in peripheral blood mononuclear cells from FD patients and healthy controls. | [34] |
WGS: whole-genome sequencing; WES: whole-exome sequencing; NMR: nuclear magnetic resonance; MR: mass spectrometry
2.3. Deficiencies of molecular biomarkers
Although traditional molecular biomarkers are widely applied in clinical practice, their use has some limitations and shortcomings, including accuracy and reliability. First and foremost, the interactions or associations between biomarkers are usually not considered, although this may be critical for a robust diagnosis and for understanding the molecular mechanisms of complex diseases [35]. Many complex or chronic diseases result not from the dysfunction of individual genes or molecules, but rather from the dysfunction of molecular networks/pathways, and thus can be viewed as network diseases. Moreover, disease progression is not a static phenomenon but rather a dynamic process. In addition, identification of traditional molecular biomarkers is often achieved from the differential analysis between disease and normal states while ignoring the dynamic changes during the development and progression of the disease [36]. Therefore, biomarkers with networks and with dynamic information are essential in order to characterize complex biological systems [37].
3. Network biomarkers
Although molecules are fundamental components of cells and that molecular biomarkers play a vital role in the occurrence and progression of diseases, complex diseases are generally affected by a group of molecules or a molecular network rather than an individual molecule [38]. As a matter of fact, the causes of diseases are diverse; alterations in cell signaling, chromatin, epigenome regulation, RNA splicing, protein homeostasis, or metabolism are all likely to trigger diseases [39]. These alterations are influenced by differential associations/correlations of molecule pairs and thus cannot be elucidated via single molecules. Regarding the disease state in the organism as a dynamic system gives rise to systems biology, defined as the study of all components in a biological system and the associations between these units under particular circumstances [40]. In other words, a disease is generally caused not by the dysfunction of individual genes or molecules but by the dysfunction of molecular networks/pathways, and thus can be viewed as a network disease. Hence, in addition to expression information, a biomarker with network information is of greater value. Network biomarkers or modular biomarkers composed of several interacting molecules with similar performance could supply a more quantifiable and stable approach to characterizing biomedical phenotypes or diseases instead of individual molecular biomarkers, thus motivating the investigation at a network level in systems biology [41]. Consequently, utilizing high-throughput omics data to reveal the interactions among molecules or to construct their networks and integrating biological data through computational modeling will contribute to a deeper understanding of the biological system. To address the difficulties in obtaining samples, network biomarkers can be employed to improve the study of biomarkers for rare diseases. In addition, a multi-dimensional omics analysis of a single sample can efficiently identify network biomarkers.
3.1. Methods for network biomarkers
Network biomarkers can be used to comprehend how a biological system operates, determine personalized medicine or treatment plans, and predict the disease progression and prognosis of individual patients, as well as the potential targets of established or novel drugs. Various calculation methods based on omics data have been developed to achieve accurate categorization of patients and reliable diagnosis of diseases. Methods of network biomarkers can be divided into two categories: a) networks constructed based on significant molecular biomarkers; b) networks built directly and used to find the various biomarkers. The first method relies on existing networks such as the KEGG and PPI networks. While this type of method is widely used, it is difficult to develop new specific network biomarkers using this approach due to the limitations of existing knowledge. At present, many methods of the second type have been developed. For example, Zhang et al. proposed the EdgeBiomarker method to diagnose the phenotype of each individual by integrating the edge and node markers of a biological network [42], and this method can construct person-specific networks from the expression profile of a single sample, meaning that it is suitable for small sample sizes. Furthermore, Liu et al. introduced the sample-specific network (SSN) method to construct individual networks based on a set of reference samples [43]. Through this method, one can elucidate the molecular mechanisms of complex diseases for each patient at the system level. To exclude indirect interactions in the SSN method, a partial correlation-based single-sample network (P-SSN) was proposed that only retained direct relations and could competently subtype complex diseases as well as cluster single cells via the network distance between two samples [44]. Integration of data from different platforms is an important issue in systems biology, and the transcriptional regulated flux balance analysis (TRFBA) algorithm is an approach that focuses on integrating transcriptional regulatory and metabolic models for multiple perturbations [45]. In addition, by using mutation and gene expression data, a single-sample controller strategy (SCS) was developed to accurately identify personalized driver mutation spectra from the perspective of network controllability, thereby providing new ideas for personalized medicine and targeted treatment of cancer [46]. The technology of single-cell sequencing has made tremendous progress in the last decade, while the high noise level and low coverage have led many of the traditional computational methods to fail. Hence, new methods for single-cell data have emerged; for example, the cell-specific network (CSN) was developed to quantify the overall relationship among genes at a single-cell resolution level [47]. Subsequently, the conditional cell-specific network (c-CSN) method has improved CSN through eliminating indirect associations to better measure direct interactions between genes [48]. In the field of drug development, a disease-specific network-based sparse Bayesian machine (NBSBM) method is able to predict drug sensitivity and reveal the underlying mechanism of drug action by selecting the most predictive sub-network from a network [49]. An overview of the analysis methods for the network biomarkers mentioned above is given in Table 3.
Table 3.
Overview of analysis methods for network biomarkers.
| Methods | Description | Applications | Reference |
|---|---|---|---|
| PPI | Estimate edge biomarkers according to differential expressed genes and protein-protein interactions. | Classify patients accurately and integrate protein-protein interaction information. | [50] |
| EdgeBiomarker | Construct an individual specific network through the expression profile of a single sample and integrating the edge and node markers of the biological network. | Diagnose the phenotype of each individual. | [42] |
| SSN | Construct individual specific networks based on the molecular expression of a single sample. | Clarify the molecular mechanisms of complex diseases of each individual at the system level. | [43] |
| P-SSN | Construct single-sample network and retain the direct interactions by excluding indirect interactions. | Predict driver mutation genes based on single-sample data, subtype complex diseases and cluster single cells. | [44] |
| TRFBA | Integrate transcriptional regulatory and metabolic models using a set of expression data for various perturbations. | Integrate transcriptome and metabolome data and improve the quantitatively prediction of growth rate. | [45] |
| SCS | Use mutation data and expression data to identify personalized driver mutation profiles from the perspective of network controllability. | Predict personalized driver mutation spectrum. | [46] |
| CSN | Construct a cell-specific network (CSN) for each single cell from scRNA-seq data. | Cluster and pseudo-trajectory at network level; find important non-differential genes. | [47] |
| c-CSN | Eliminate indirect associations to measure direct associations between genes. | Resolve the direction of differentiation trajectories by quantifying the potency of each single cell. | [48] |
| NBSBM | Method of sparse Bayesian machine based on network. | Predict drug sensitivity and reveal the underlying mechanism of drug action. | [49] |
3.2. Application of network biomarkers in diseases
The application of network biomarkers in rare diseases is still relatively limited, and most studies have focused on regulatory networks of molecular biomarkers instead of taking the edge or network as an independent biomarker. Villalba-Benito et al. concentrated on exploring the interaction network of PAX6 during ENS formation to better outline HSCR etiology [51]. Nuzziello et al. have constructed a comprehensive miRNA-TF co-regulatory network for MS and recognized that NF-κB and STAT3 collaboratively regulate the immune response genes [52].
In common complex diseases that have been more intensively studied (e.g., cancer), network biomarkers have been used to analyze the problems of biomedical phenotypes as well as the occurrence and development of diseases that traditional molecular biomarkers cannot explain. For example, in order to examine the discrepancy between estrogen receptor (ER)-negative and ER-positive breast cancer, one study evaluated the correlations between molecular pairs in the kinase regulatory network to find kinase-substrate nodes and edge biomarkers and to demonstrate their ability for prognosis [53]. A Chinese colorectal cancer (CRC) cohort study employed kinase network analysis to reveal significant heterogeneity between primary colorectal tumors and associated liver metastases, and through kinase-substrate network analysis, it was possible to obtain personalized responses of tumors via in vivo xenograft drug testing [54]. In addition to research on disease mechanisms, network biomarkers can also be used for genetic robustness research. Zhang et al. constructed a gene modular network (GMN) using expression data from Drosophila and performed an asymptotic dynamics analysis on the GMN showing that a morphogen-directed GMN could tolerate most genetic disturbances and was essential to ensure proper tissue patterning [55].
3.3. Advantages and disadvantages of network biomarkers
From a systems perspective, network biomarkers are formed by integrating multiple omics data; as a result, network biomarkers are expected to diagnose disease states more accurately than conventional molecular biomarkers. The network biomarkers determined by a systems analysis or differential association/correlation analysis may include non-differentially expressed genes, called "dark genes", that nonetheless contain abundant disease information and can make up for the lack of molecular biomarkers based on differential expression analysis. The requirement for biomarker standardization and the association of biomarkers with clinical and other quantitative characteristics are fundamental recommendations in rare diseases. More research into the underlying molecular mechanisms of disease pathology as well as the integration of biomarker studies into controlled clinical trials will allow for improvement in an area that has many needs. Perhaps network biomarkers can contribute to resolving this issue. However, the early warning signals of disease occurrence or deterioration are vital indicators for the prevention and treatment of complex diseases. Accurate early warning signal measurements have far-reaching implications for disease prevention. Generally, disease development is a dynamic process, and thus forecasting disease requires dynamic data. Although network biomarkers may reliably diagnose diseases, they cannot foresee diseases in the same way that biological biomarkers can. As a result, novel biomarkers should be developed by further studying the dynamic information contained in the data. Patients with rare diseases may have more complete electronic medical records that will aid in the application of dynamic network approaches to the analysis of biomarkers.
4. Dynamic network biomarkers
Molecular biomarkers and network biomarkers are largely restricted to comparisons between disease states and normal states. However, normally there is little or almost no change at the molecular level in the pre-disease state before the disease occurs compared to the normal state. Increasing evidence shows that non-smooth or sudden state changes exist in various biological processes, including the disease system [56, 57]. In the progression of a complex disease, the system gradually shifts from the normal state to the pre-disease state and then rapidly passes to an irreversible disease state. Hence, in order to realize an early diagnosis of a complex disease, it would be necessary to detect the critical state or the pre-disease state to prevent the immediate deterioration to the full disease state [12, 56]. To circumvent this difficulty, a method based on a model-free concept called dynamic network biomarker (DNB) was developed [58] based on nonlinear dynamic systems theory by exploiting the information contained in both the network and the dynamic patterns in the data. A DNB is a group of associations/correlations between molecules and their fluctuations, or among a group of collectively correlated molecules. DNB was identified based on differential fluctuations/correlations/distributions of a group of molecules, and as a model-free method, it is able to detect critical states just before the bifurcation point from the normal to the disease state, even with a small amount of high-throughput sequencing data [37, 56].
4.1. Methods and applications of dynamic network biomarkers
Theoretically, it can be shown that when the system is about to approach a critical point or tipping point, there exists a dominant group of molecules that comprise the so-called DNB meeting the following three necessary conditions. First and foremost, variation (standard deviations) within the DNB group drastically increases. Secondly, correlations among the DNB group also increase. Finally, correlations between the DNB group and non-DNB molecules decrease [58]. According to these statistical criteria, DNBs are measurable sub-networks composed of several specific molecules. When the system is near the tipping point, the molecules within the sub-network are strongly correlated and show fluctuations. Furthermore, DNB elements/members tend to be separated from non-DNB molecules. For quantitatively identifying DNB molecules as well as detecting the critical point, a composite index (CI) combining two or three features was proposed as follows [58]:
In the last several decades, DNB theory has been successfully employed in numerous cases and has revealed crucial states and their key molecules in the progression of various diseases. Tong et al. utilized DNB to reveal the critical transitions of CRC, finally concluding that MYC was associated with tumor amplification, immune cells, and survival [59]. Epithelial-mesenchymal transition (EMT) plays an essential role in cancer metastasis, and via DNB Jiang et al. discovered the early-warning signals of EMT along with two key genes, SMAD7 and SERPINE1, that could promote EMT by acting as switches in their regulatory networks [60]. Moreover, Chen et al. succeeded in recognizing the early signals prior to the catastrophic transition to an influenza pandemic using the DNB method [61]. In addition to the prediction of disease progression, DNB can also be applied to study other biological processes. For example, Tang et al. detected the critical periods of infant brain development by taking advantage of DNB [62].
The original DNB method was based on population samples. Thus, single-sample DNB (sDNB) was developed to extend the method to individual samples and enable DNB to be applied in clinical practice [63]. The sDNB method was generated based on the SSN method [29]. In the beginning, a set of control/reference samples are employed to construct a reference network, and then the difference in correlation information on each sample relative to the reference network is calculated to achieve personalized disease prediction. Theoretically, when the number of control samples is sufficiently large, the single-sample network can be uniquely determined, even if there is genetic heterogeneity among samples. The CI of sDNB can be described as follows [63]:
Furthermore, a new methodology called landscape DNB (l-DNB) was designed through constructing a DNB landscape of the observed variables for efficiently determining the DNB molecules in a single sample [64]. Consistent with sDNB, l-DNB also takes advantage of SSN theory, the difference being that l-DNB exploits SSN to establish a local module for each gene and its first-order neighbors, and then estimates the DNB composite index of each local module in order to rank the genes [29]. l-DNB can not only reliably detect the critical point prior to serious disease deterioration but also identify dynamic network biomarkers for disease prediction within each person. It has been demonstrated that l-DNB works well for various diseases and biological processes, including breast cancer cell differentiation, coronary atherosclerosis, hepatocellular carcinoma progression, malignant development of gallbladder cancer and skin response to repetitive ultraviolet B irradiation [65], [66], [67], [68], [69]. Additional optimization methods for DNB from different aspects include a method taking advantage of biological pathways [70], a method of identifying DNBs from a multi-objective optimization viewpoint [71], and a method for predicting tipping point-like transitions in single cell resolution data [72, 73], i.e., CIDNB= <PCC(gi,gj)> / <PCC(ck,cl)> where gi is a vector of gene-i for all cells, ck is a vector of all genes at cell k, and <PCC> denotes the average of all Pearson’s correlation coefficients of respective pairs of vectors, which is actually equivalent to the original DNB index but at a single cell level. An overview of the analysis methods for the dynamic network biomarkers mentioned above is given in Table 4.
Table 4.
Overview of analysis methods for dynamic network biomarkers.
| Methods | Description | Applications | Reference |
|---|---|---|---|
| DNB | The original DNB method to detect a pre-disease state based on correlations and deviations among molecules. | Detect critical states from population samples. | [58] |
| sDNB | Single sample DNB based on the SSN method. | Expand adoption of DNB on individual sample. | [63] |
| l-DNB | Construct DNB landscape of the observed variables by exploiting SSN to establish a local module. | Reliably detect the critical point and identify real network biomarkers for disease prediction in individual samples. | [64] |
| Pathway-Induced DNB | Induce sparsity on the adjacency matrix of the genes by considering the biological pathways | Detect the DNBs responsible for catastrophic transition into the disease stage based on optimization-based algorithms. | [70] |
| A metaheuristic multi-objective optimization method for DNB | Filter the relevant genes in the dataset and identify DNBs from a multi-objective optimization viewpoint. | Identify DNBs that are the smallest gene network while showing the strongest signal at the earliest time-point and best correlate with the phenotype. | [71] |
| Predict critical state transitions in single-cell gene expression data | Propose a new type of early warning signal: an index Ic based on changes in high-dimensional cell population structure obtained from single-cell data. | Predict tipping point-like transitions in multicellular systems. | [72] |
To characterize the early warning signs of the diseases or block the further deterioration to the disease state, it is essential to investigate the critical points in the occurrence and progression of complex diseases. Because there is little or no difference in molecular/phenotypic levels between the critical state and the normal state, neither traditional molecular biomarkers nor network biomarkers can assess the tipping point exactly. Therefore, a model-free approach based on nonlinear dynamic systems theory called DNB was developed to overcome this difficulty by exploiting the information from both dynamic data and networks [41]. DNB is based on second-order statistics (i.e., correlations and deviations), in contrast to the first-order statistics of the molecular biomarkers (i.e., average values). Because it considers the perturbations of correlations among a group of molecules, the false positives or false negatives caused by the high expression levels of individual molecules will be effectively controlled. DNB is able to accurately estimate the tipping points in complex diseases as well as their corresponding disease-associated molecules. Further interfering with these molecules may contribute to the prevention of disease worsening. In addition, using SSN theory, DNB can detect pre-diseases state and to predict the disease based only on a single sample, thus providing a foundation for the realization of personalized treatment. According to the methods of DNB, standardization of health management can be established, e.g., (1) quantifying the health status of individuals by DNB in addition to disease status quantified by traditional molecular biomarkers; (2) quantifying complex diseases based on the distance from their critical states measured by DNB from the available time-series data. Although there is presently no study on dynamic network indicators for rare diseases, early warning of rare diseases could be achieved if long-term biological data records of patients are successfully exploited in conjunction with the idea of dynamic disease development.
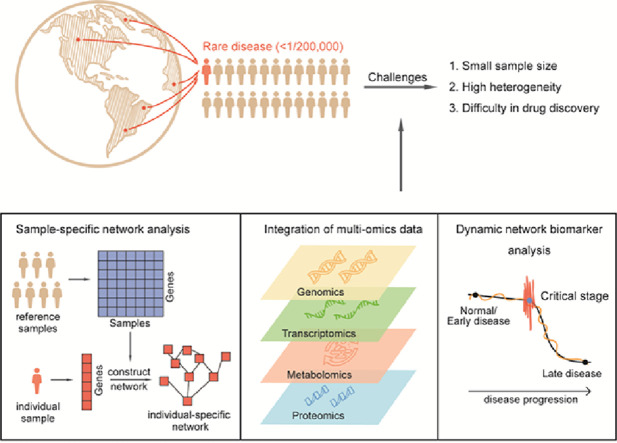
In summary, we have comprehensively reviewed recent advances and applications of molecular biomarkers, network biomarkers, and dynamic network biomarkers in diagnosis, prediction, and therapy for rare diseases. The molecular biomarkers are primarily based on differential expression/concentration, while network biomarkers are largely based on differential associations/correlations, and dynamic network biomarkers are a collection of associations/correlations between molecules accompanied by their patterns of fluctuation (Fig. 1).
Fig. 1.
Schematic of biomarkers in diseases. (a) Brief description on methods of molecular and network biomarkers. (b) Brief description of DNB theory (left) and dynamic change of the network (right).
5. Prospects for biomarkers in rare diseases
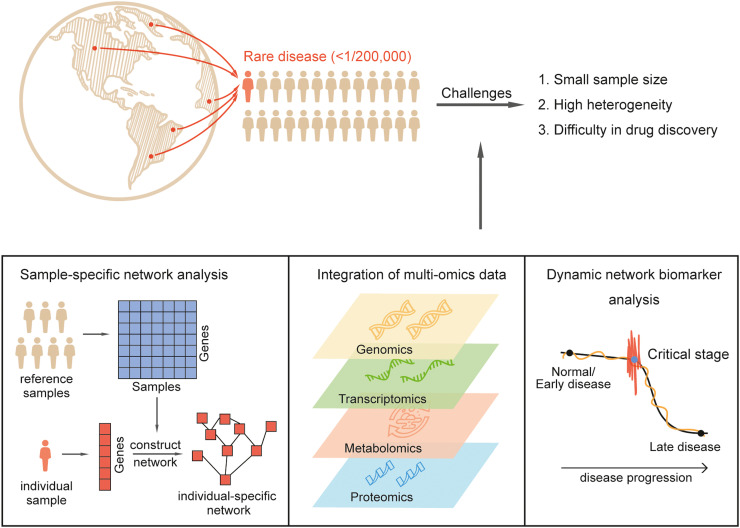
Due to the difficulty in the diagnosis and treatment of rare diseases, the use of high-throughput sequencing technology and advances in algorithm development will enable the identification of better diagnostic and prognostic biomarkers that will aid patients with such diseases. Characterizing diseases at the molecular level helps reveal biochemical pathways for potential drug targets. However, the main problem at present is that due to the limited number of rare disease patients, it is very difficult to obtain a large sample size. This problem can be overcome through national and international cooperative efforts and multi-omics data sharing. At the same time, the small sample size problem could also be solved by using a network approach that can be realized with small amounts of data or even individual data such as in a single-sample network. The main advantage of network biomarkers over molecular biomarkers is that a network biomarker further considers associations/correlations among molecular biomarkers at a system level or exploits the network information from the data, and thus is better able to accurately diagnose complex diseases. Furthermore, a dynamic network biomarker considers both network and dynamic information from the data, i.e., correlations/fluctuations of biological processes, and thus can robustly predict complex diseases. Dynamic network biomarker theory can also be used in the small-sample analysis. Therefore, if long-term data records of rare disease patients are properly used in combination with the concept of dynamic disease progression, early warning of rare diseases can be achieved. With the development of multi-omics technology, more efficient methods of integrating data to construct biomarkers for predicting disease states may appear in the future rather than being limited to a single type of data [74]. Compared with single-gene testing alone, whole-genome and exome sequencing can improve the identification of causal variants with a diagnostic rate of about 50% for inherited diseases, and integrated multi-omics analysis can further improve the rare disease diagnosis rate. In addition, compared with single-omics analysis, multi-omics analysis can better deal with genetic and phenotypic heterogeneity. Moreover, while edge information has been used in network biomarkers to obtain a more systematic understanding of the disease, the measurement of edge information is largely restricted to correlations. Recently, several methods have utilized entropy to characterize edges, and superior indicators may emerge in the future. In brief, molecular biomarkers using expression information, network biomarkers using additional network information, and dynamic network biomarkers further using additional dynamic information have been effectively applied in the diagnosis, prediction, and treatment of diseases. It is possible to achieve accurate disease diagnosis and prognosis via network biomarkers, and robust prediction of disease can be obtained from dynamic network information via DNB. All in all, customized personalized treatment will be guided by an individual's specific biomarker panel, especially in rare diseases where early diagnosis and treatment remain a bottleneck. A personalized medicine approach will have a major impact on improving the quality of medical care for patients with rare diseases (Fig. 2). In addition, with an increasingly accurate diagnosis by various biomarkers, a common disease can be subtyped into more detailed subtypes or subclasses, each of which can be considered as a rare disease from the perspective of the numbers and mechanisms of such a disease subtype. Hence, developing accurate biomarkers is of great importance for both common and rare diseases.
Fig. 2.
Prospects for biomarkers in rare diseases.
6. Conclusion
Recent developments and applications of molecular biomarkers, network biomarkers, and dynamic network biomarkers in the diagnosis, prediction, and therapy of rare diseases have been thoroughly explored. Much research on molecular markers in rare diseases has been conducted. Unfortunately, the majority of these findings have not been successfully implemented in clinical practice. The limited sample size, high level of phenotypic heterogeneity, and difficulty in drug discovery remain three major challenges for reliable biomarker identification in rare diseases. Several network biomarker and dynamic network biomarker approaches have performed well with small data sets, and thus are suitable for identifying rare disease biomarkers. Moreover, network biomarkers consider the relationships of molecular markers at the system level to extract network information from data and thereby more accurately diagnose complex rare diseases. Furthermore, dynamic network biomarkers incorporate the dynamic changes in the relevant biological processes and thus can reliably predict complex diseases at the early stages. Additionally, the utilization of different forms of omics data can be used to analyze a complex biological process from many aspects for non-single gene rare diseases. Exploring the interplay between multiple levels of data will provide more trustworthy and comprehensive information for exploring the occurrence and development mechanisms of rare diseases.
Author contributions
Original draft preparation, S.T., K.Y.; writing—review and editing, S.T., K.Y., L.C. ALL authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
National Key Research and Development Program of China [2017YFA0505500]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38040400]; National Natural Science Foundation of China (NSFC) [12131020, 31930022, 12026608]; Special Fund for Science and Technology Innovation Strategy of Guangdong Province [2021B0909050004, 2021B0909060002]; Major Key Project of Peng Cheng Laboratory [PCL2021A12]; JST Moonshot R&D [JPMJMS2021].
Biographies

Shijie Tang is a Ph.D. student at CAS Center for Excellence in Molecular Cell Science, the Chinese Academy of Sciences, Shanghai, China. She finished her bachelor's degree from Tongji University in 2016. Her current research focuses on the application of multi-omics research in the disease.

Luonan Chen has been a professor, and executive director of Key Laboratory of Systems Biology, CAS Center for Excellence in Molecular Cell Science, the Chinese Academy of Sciences, since 2009. He received BS degree in Electrical Engineering from Huazhong University of Science and Technology in 1984, and the M.E. and Ph.D. degrees in electrical engineering from Tohoku University in 1988 and 1991, respectively. In 2009, He joined CAS Center for Excellence in Molecular Cell Science. In recent years, he published over 400 journal papers and two monographs in the area of bioinformatics, nonlinear dynamics and machine learning.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fmre.2022.07.011.
Appendix. Supplementary materials
Reference
- 1.Bax B.E. Biomarkers in rare diseases. Int J Mol Sci. 2021;22(2):673. doi: 10.3390/ijms22020673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter T., et al. Rare disease terminology and definitions-A systematic global review: report of the ISPOR rare disease special interest group. Value Health. 2015;18(6):906–914. doi: 10.1016/j.jval.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Gülbakan B., et al. Discovery of biomarkers in rare diseases: innovative approaches by predictive and personalized medicine. Epma j. 2016;7(1):24. doi: 10.1186/s13167-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbade S.F., et al. Quantitative retrospective natural history modeling for orphan drug development. J Inherit Metab Dis. 2021;44(1):99–109. doi: 10.1002/jimd.12304. [DOI] [PubMed] [Google Scholar]
- 5.Blöß S., et al. Diagnostic needs for rare diseases and shared prediagnostic phenomena: results of a German-wide expert Delphi survey. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0172532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesselheim A.S., Treasure C.L., Joffe S. Biomarker-defined subsets of common diseases: policy and economic implications of orphan drug Act coverage. PLoS Med. 2017;14(1) doi: 10.1371/journal.pmed.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gromova M., et al. Biomarkers: opportunities and challenges for drug development in the current regulatory landscape. Biomarker Insights. 2020;15 doi: 10.1177/1177271920974652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auray-Blais C., et al. Urinary Globotriaosylsphingosine-related biomarkers for fabry disease targeted by metabolomics. Anal Chem. 2012;84(6):2745–2753. doi: 10.1021/ac203433e. [DOI] [PubMed] [Google Scholar]
- 9.Boutin M., et al. Diurnal variation of urinary fabry disease biomarkers during enzyme replacement therapy cycles. Int J Mol Sci. 2020;21(17):6114. doi: 10.3390/ijms21176114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vistbakka J., et al. Evaluation of serum miR-191-5p, miR-24-3p, miR-128-3p, and miR-376c-3 in multiple sclerosis patients. Acta Neurol Scand. 2018;138(2):130–136. doi: 10.1111/ane.12921. [DOI] [PubMed] [Google Scholar]
- 11.Lampejo T., et al. Prognostic biomarkers in squamous cell carcinoma of the anus: a systematic review. Br J Cancer. 2010;103(12):1858–1869. doi: 10.1038/sj.bjc.6605984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu R., et al. Early diagnosis of complex diseases by molecular biomarkers, network biomarkers, and dynamical network biomarkers. Med Res Rev. 2014;34(3):455–478. doi: 10.1002/med.21293. [DOI] [PubMed] [Google Scholar]
- 13.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S., et al. Applications of Support Vector Machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics. 2018;15(1):41–51. doi: 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boser B. ACM Press; 1992. A training algorithm for optimal margin classifiers. [Google Scholar]
- 17.Lee L., Liong C.Y. Partial Least Squares-Discriminant Analysis (PLS-DA) for classification of high-dimensional (HD) data: a review of contemporary practice strategies and knowledge gaps. Analyst. 2018:143. doi: 10.1039/c8an00599k. [DOI] [PubMed] [Google Scholar]
- 18.Barker M., Rayens W. Partial least squares for discrimination. J Chemom. 2003;17(3):166–173. [Google Scholar]
- 19.Wold S., Sjöström M., Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst. 2001;58(2):109–130. [Google Scholar]
- 20.Tibshirani R. Regression Shrinkage and Selection via the LASSO. J Royal Statist Soc. 1996;73:273–282. [Google Scholar]
- 21.Guyon I., et al. Gene Selection for Cancer Classification using support vector machines. Mach Learn. 2002;46(1):389–422. [Google Scholar]
- 22.de Anda-Jáuregui G., Hernández-Lemus E. Computational oncology in the multi-omics era: state of the art. Front Oncol. 2020;10(423) doi: 10.3389/fonc.2020.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niemann M., et al. Gene mutations versus clinically relevant phenotypes. Circulation: Cardiovascular Genetics. 2014;7(1):8–16. doi: 10.1161/CIRCGENETICS.113.000249. [DOI] [PubMed] [Google Scholar]
- 24.Cerasuolo A., et al. Implications of splicing alterations in the onset and phenotypic variability of a family with subclinical manifestation of Peutz–Jeghers syndrome: bioinformatic and molecular evidence. Int J Mol Sci. 2020;21(21) doi: 10.3390/ijms21218201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta M., et al. Long noncoding RNAs associated with phenotypic severity in multiple sclerosis. Mult Scler Relat Disord. 2019;36 doi: 10.1016/j.msard.2019.101407. [DOI] [PubMed] [Google Scholar]
- 27.Torroglosa A., et al. Identification of new potential LncRNA biomarkers in Hirschsprung disease. Int J Mol Sci. 2020;21(15) doi: 10.3390/ijms21155534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skiriute D., et al. The Role of CASC2 and miR-21 interplay in glioma malignancy and patient outcome. Int J Mol Sci. 2020;21(21) doi: 10.3390/ijms21217962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavoie P., Boutin M., Auray-Blais C. Multiplex analysis of novel urinary lyso-Gb3-related biomarkers for Fabry disease by tandem mass spectrometry. Anal Chem. 2013;85(3):1743–1752. doi: 10.1021/ac303033v. [DOI] [PubMed] [Google Scholar]
- 30.Auray-Blais C., et al. Urinary globotriaosylsphingosine-related biomarkers for Fabry disease targeted by metabolomics. Anal Chem. 2012;84(6):2745–2753. doi: 10.1021/ac203433e. [DOI] [PubMed] [Google Scholar]
- 31.Menkovic I., et al. Identification of a reliable biomarker profile for the diagnosis of Gaucher disease Type 1 patients using a mass spectrometry-based metabolomic approach. Int J Mol Sci. 2020;21(21) doi: 10.3390/ijms21217869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollander Z., et al. Gender-specific plasma proteomic biomarkers in patients with Anderson-Fabry disease. Eur J Heart Fail. 2015;17(3):291–300. doi: 10.1002/ejhf.230. [DOI] [PubMed] [Google Scholar]
- 33.Benabdelkamel H., et al. Serum-based proteomics profiling in adult patients with cystic fibrosis. Int J Mol Sci. 2020;21(19) doi: 10.3390/ijms21197415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cigna D., et al. Alteration of proteomic profiles in PBMC isolated from patients with Fabry disease: preliminary findings. Mol Biosyst. 2013;9(6):1162–1168. doi: 10.1039/c3mb25402j. [DOI] [PubMed] [Google Scholar]
- 35.Szklarczyk D., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucl Acids Res. 2017;45(D1):D362–d368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang Z., Chen L. Personalized prediction of human diseases with single-sample dynamic network biomarkers. Biomark Med. 2020;14(8):615–620. doi: 10.2217/bmm-2020-0066. [DOI] [PubMed] [Google Scholar]
- 37.Wu X., Chen L., Wang X. Network biomarkers, interaction networks and dynamical network biomarkers in respiratory diseases. Clin Transl Med. 2014;3:16. doi: 10.1186/2001-1326-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang X., Wang J. From molecules to cellular networks: past and future outlook. Phys Biol. 2017;14(1) doi: 10.1088/1478-3975/aa5b6a. [DOI] [PubMed] [Google Scholar]
- 39.Garraway L.A., Lander E.S. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Clarke R. Introduction: cancer gene networks. Methods Mol Biol. 2017;1513:1–9. doi: 10.1007/978-1-4939-6539-7_1. [DOI] [PubMed] [Google Scholar]
- 41.Aryee M.J., et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5(169):169ra10. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W., et al. Diagnosing phenotypes of single-sample individuals by edge biomarkers. J Mol Cell Biol. 2015;7(3):231–241. doi: 10.1093/jmcb/mjv025. [DOI] [PubMed] [Google Scholar]
- 43.Liu X., et al. Personalized characterization of diseases using sample-specific networks. Nucl Acids Res. 2016;44(22):e164. doi: 10.1093/nar/gkw772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y., et al. Disease characterization using a partial correlation-based sample-specific network. Brief Bioinform. 2021;22(3) doi: 10.1093/bib/bbaa062. [DOI] [PubMed] [Google Scholar]
- 45.Motamedian E., et al. TRFBA: an algorithm to integrate genome-scale metabolic and transcriptional regulatory networks with incorporation of expression data. Bioinformatics. 2017;33(7):1057–1063. doi: 10.1093/bioinformatics/btw772. [DOI] [PubMed] [Google Scholar]
- 46.Guo W.F., et al. Discovering personalized driver mutation profiles of single samples in cancer by network control strategy. Bioinformatics. 2018;34(11):1893–1903. doi: 10.1093/bioinformatics/bty006. [DOI] [PubMed] [Google Scholar]
- 47.Dai H., et al. Cell-specific network constructed by single-cell RNA sequencing data. Nucl Acids Res. 2019;47(11):e62. doi: 10.1093/nar/gkz172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., et al. c-CSN: Single-cell RNA sequencing data analysis by conditional cell-specific network. Genomics Proteomics Bioinform. 2021 doi: 10.1016/j.gpb.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Q., Muglia L.J., Huang L.F. Network as a biomarker: a novel network-based sparse bayesian machine for pathway-driven drug response prediction. Genes (Basel) 2019;10(8) doi: 10.3390/genes10080602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng M.J., et al. Identification of molecular marker associated with ovarian cancer prognosis using bioinformatics analysis and experiments. J Cell Physiol. 2019;234(7):11023–11036. doi: 10.1002/jcp.27926. [DOI] [PubMed] [Google Scholar]
- 51.Villalba-Benito L., et al. ChIP-Seq-based approach in mouse enteric precursor cells reveals new potential genes with a role in enteric nervous system development and Hirschsprung disease. Int J Mol Sci. 2020;21(23) doi: 10.3390/ijms21239061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nuzziello N., et al. Investigating the role of MicroRNA and transcription factor Co-regulatory networks in multiple sclerosis pathogenesis. Int J Mol Sci. 2018;19(11):3652. doi: 10.3390/ijms19113652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y., et al. Kinase-substrate Edge Biomarkers provide a more accurate prognostic prediction in er-negative breast cancer. Genomics Proteomics Bioinform. 2021 doi: 10.1016/j.gpb.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C., et al. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell. 2020;38(5):734–747. doi: 10.1016/j.ccell.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Zhang S., et al. Analysis on gene modular network reveals morphogen-directed development robustness in Drosophila. Cell Discov. 2020;6:43. doi: 10.1038/s41421-020-0173-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu R., Aihara K., Chen L. Dynamical network biomarkers for identifying critical transitions and their driving networks of biologic processes. Quan Biol. 2013;1(2):105–114. [Google Scholar]
- 57.Venegas J.G., et al. Self-organized patchiness in asthma as a prelude to catastrophic shifts. Nature. 2005;434(7034):777–782. doi: 10.1038/nature03490. [DOI] [PubMed] [Google Scholar]
- 58.Chen L., et al. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci Rep. 2012;2:342. doi: 10.1038/srep00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tong Y., et al. Theoretical and in silico analyses reveal MYC as a dynamic network biomarker in colon and rectal cancer. Front Genet. 2020;11 doi: 10.3389/fgene.2020.555540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Z., et al. SMAD7 and SERPINE1 as novel dynamic network biomarkers detect and regulate the tipping point of TGF-beta induced EMT. Science Bulletin. 2020;65(10):842–853. doi: 10.1016/j.scib.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 61.Chen P., et al. Detecting early-warning signals of influenza outbreak based on dynamic network marker. J Cell Mol Med. 2019;23(1):395–404. doi: 10.1111/jcmm.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang H., et al. Gene expression analysis reveals the tipping points during infant brain development for human and chimpanzee. BMC Genomics. 2020;21(Suppl 1):74. doi: 10.1186/s12864-020-6465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., et al. Quantifying critical states of complex diseases using single-sample dynamic network biomarkers. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X., et al. Detection for disease tipping points by landscape dynamic network biomarkers. Natl Sci Rev. 2019;6(4):775–785. doi: 10.1093/nsr/nwy162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y., et al. Single-cell RNA-sequencing atlas reveals an MDK-dependent immunosuppressive environment in ErbB pathway-mutated gallbladder cancer. J Hepatol. 2021;75(5):1128–1141. doi: 10.1016/j.jhep.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 66.Zhao H., Gao J. [Identifying critical state of breast cancer cell differentiation based on landscape dynamic network biomarkers] Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2020;37(2):304–310. doi: 10.7507/1001-5515.201908013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ge J., et al. Personalized early-warning signals during progression of human coronary atherosclerosis by landscape dynamic network biomarker. Genes (Basel) 2020;11(6) doi: 10.3390/genes11060676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Y., et al. Identifying critical states of hepatocellular carcinoma based on landscape dynamic network biomarkers. Comput Biol Chem. 2020;85 doi: 10.1016/j.compbiolchem.2020.107202. [DOI] [PubMed] [Google Scholar]
- 69.Zhang C., et al. Landscape dynamic network biomarker analysis reveals the tipping point of transcriptome reprogramming to prevent skin photodamage. J Mol Cell Biol. 2022;13(11):822–833. doi: 10.1093/jmcb/mjab060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torshizi A.D., Petzold L. Sparse pathway-induced dynamic network biomarker discovery for early warning signal detection in complex diseases. IEEE/ACM Trans Comput Biol Bioinf. 2018;15(3):1028–1034. doi: 10.1109/TCBB.2017.2687925. [DOI] [PubMed] [Google Scholar]
- 71.Coleto-Alcudia V., Vega-Rodríguez M.A. A metaheuristic multi-objective optimization method for dynamical network biomarker identification as pre-disease stage signal. Appl Soft Comput. 2021;109 [Google Scholar]
- 72.Mojtahedi M., et al. Cell fate decision as high-dimensional critical state transition. PLoS Biol. 2016;14(12) doi: 10.1371/journal.pbio.2000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao R., et al. Detecting the critical states during disease development based on temporal network flow entropy. Brief Bioinform. 2022;23(5) doi: 10.1093/bib/bbac164. [DOI] [PubMed] [Google Scholar]
- 74.Hu C., Jia W. Multi-omics profiling: the way toward precision medicine in metabolic diseases. J Mol Cell Biol. 2021;13(8):576–593. doi: 10.1093/jmcb/mjab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.