Abstract
Since the recent discovery of cancer cell-intrinsic programmed cell death protein-1 (PD-1), the mechanisms that manipulate PD-1 functions in tumor development beyond its immune checkpoint roles have become attractive research topics in oncology. Our previous study validated that PD-1 exists in lung cancer cells and is directly transactivated by p53 in a DNA-binding domain (DBD) acetylation-dependent manner. Here, we report that the carboxyl-terminal domain (CTD) of p53 likewise participates in PD-1 transcriptional regulation in cancer cells under different regulatory mechanisms. By mutating the lysine residues within the CTD to mimic either acetylation-deficient or fully acetylated status, we proved that acetylated CTD dramatically impeded p53-mediated transactivation of PD-1. Furthermore, we identified bromodomain-containing protein 4 (BRD4) as a transcriptional coactivator of p53 that facilitates p53-mediated PD-1 transcription. Mechanistically, BRD4 specifically bound to the unacetylated CTD of p53, while CTD acetylation almost completely destroyed the BRD4-p53 interaction and thus led to compromised PD-1 expression. Collectively, this study unveils an alternative mechanism of p53 acetylation-directed PD-1 transcriptional regulation, which would broaden our current understanding of the molecular regulatory network of cancer cell-intrinsic PD-1.
Keywords: p53, Acetylation, PD-1, BRD4, Carboxyl-terminal Domain (CTD)
Graphical abstract
1. Introduction
As the most frequently inactivated tumor suppressor in human cancers, p53 functions primarily through transcriptional activation of a plethora of target genes that are extensively involved in the control of cell proliferation, apoptosis, senescence, DNA repair and metabolic reprogramming, all of which confer anticancer characteristics to p53 [1,2]. Upon various cellular stresses (including DNA damage, hypoxia, hyperproliferative signals, etc.), p53 transactivates targets in a context-specific manner to orchestrate a wide range of downstream processes [3]. Multiple mechanisms have been proposed for the selectivity of the p53-mediated transcriptional profile, among which posttranslational modifications (PTMs), such as phosphorylation, acetylation, and ubiquitination, act as refined regulatory modes that modulate the functional spectrum of p53 [4,5].
Acetylation occurs at multiple lysine residues of p53, which plays general or site-specific roles in influencing the stability, DNA binding affinity and protein–protein interactions of p53. The acetylation status of the lysine residue cluster (K370, K372, K373, K381, K382 and K386) within the CTD governs both the stability and transactivity of p53 [6,7]. Mechanistically, distinct acetylation pattern (hyper or hypo-) creates unique docking sites for recruiting specific reader proteins, which either induce or repress p53 transactivity. Several bromodomain-containing proteins were discovered to recognize acetylated lysine residues of p53 [8,9]. A previous study from our group identified a class of acid domain (AD)-containing proteins (including SET, PELP1, VPRBP and DAXX) that specifically recognize unacetylated p53 and function as transcriptional cofactors of p53 in unstressed cells [10,11]. In addition to the CTD, the DBD also contains several lysine residues (such as K101, K120 and K164), whose acetylation status dramatically determines the transcriptional selectivity of p53. Specifically, K120/164 acetylation was tightly associated with the growth inhibitory and pro-apoptotic role of p53, whereas K101 acetylation exclusively induced SLC7A11-mediated ferroptosis [12], [13], [14], [15], [16]. In addition to the individual regulating behaviors, multisite acetylation may interplay cooperatively to guide exquisite p53 responses, which form up a more complex layer of PTMs regulation characteristics. Taken together, site-specific acetylation and their crosstalk may endow p53 adaptive modulations in response to complex cellular conditions, yet the acetylation-directed target selection and regulation of p53 remain incompletely resolved.
As one of the most promising targets in cancer immunotherapy, PD-1 was initially detected in immune cells by serving as a coinhibitory checkpoint, and subsequent studies uncovered its existence in certain cancer cells [17], [18], [19]. Our recent study revealed intrinsic PD-1 expression in lung cancer cells and verified the direct transcriptional activating role of p53 on cancer cell-intrinsic PD-1. Notably, K120 and K164 acetylation are indispensable for p53-mediated PD-1 transcriptional expression and, consequently, the suppression of tumor growth in an immunity-independent manner [20]. These findings suggest a potential role for cancer cell-intrinsic PD-1 as a therapeutic target; however, the comprehensive signaling pathways controlling PD-1 expression still need to be completely investigated.
Here, we report an opposite role of p53 CTD acetylation in modulating PD-1 transcription in lung cancer cells. Unlike the DBD acetylation that dramatically enhanced p53-driven PD-1 expression, CTD acetylation completely abolished p53-dependent induction of PD-1. Mechanistically, CTD acetylation prevented p53 from interacting with BRD4, a critical p53 coactivator that facilitated p53-mediated PD-1 transcriptional expression. Together with our previous findings focusing on DBD acetylation, our studies replenish the regulatory rules by which distinct PTMs may crosstalk mutually to modulate p53-mediated transcription of cancer cell-intrinsic PD-1.
2. Materials and methods
2.1. Cell culture, plasmid construction and cell transfection
The H1299 or U2OS cell line was purchased from the Cell Resource Center of the Chinese Academy of Medical Science (Beijing, China) and cultured in Dulbecco's modified Eagle's (DEME) medium (Corning, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, USA). The Tet-on inducible H1299 stable cell line was cultured in DMEM with 10% Tet System Approved FBS (Clontech, USA). Doxycycline (Doxy; Sigma–Aldrich, USA) was used at 1 μg/ml to induce p53 expression in the stable cell line. All cells were maintained in 37 °C incubators supplemented with 5% CO2. p53-expressing plasmids were generated by subcloning human p53 cDNA (including full-length, CTD portion and CTD-truncated) into pWG-F-HA. The point mutant(s) p53-expressing plasmids were generated using a site-directed mutagenesis kit (Stratagene, USA) according to the manufacturer's protocol. Expressing plasmid and siRNA transfection were conducted by using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's protocol. The control siRNA (UUCUCCGAACGUGUCACGU) was purchased from GenePharma, China. The pooled siBRD4 (including 4 oligos. #1: AAACCGAGAUCAUGAUAGU; #2: CUACACGACUACUGUGACA; #3: AAACACAACUCAAGCAUCG; #4: CAGCGAAGACUCCGAAACA) and the pooled siPD-1 (including 4 oligos. #1: GGGCGUGACUUCCACAUGA; #2: UCGGAGAGCUUCGUGCUAA; #3: CCAGCAACCAGACGGACAA; #4: GCACGAGGGACAAUAGGAG) were purchased from Dharmacon, USA.
2.2. RT–qPCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, USA). cDNA was synthesized from 1 μg RNA using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen, USA) according to the manufacturer's protocol. The relative expression of gene targets was measured in an Applied Biosystems 7500 Fast Real-Time PCR (polymerase chain reaction) System. All values were normalized to human β-actin or B2M. Primers: human PD-1 (forward, CCAGGATGGTTCTTAGACTCCC; reverse, TTTAGCACGAAGCTCTCCGAT), human p21 (forward, CTGTCACTGTCTTGTACCCTTGT; reverse, GGTAGAAATCTGTCATGCTGGT), human TIGAR (forward, GACTTCGGGAAAGGAAATACG; reverse, CACTCTTCCCTGGCTGCTTTG), human β-actin (forward, GGCCAACCGCGAGAAGAT; reverse, GCCAGAGGCGTACAGGGATA) and human B2M (forward, GAGGCTATCCAGCGTACTCCA; reverse, CGGCAGGCATACTCATCTTTT).
2.3. Western blotting assay
Cells were harvested and lysed in NP-40 buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, and 1× protease inhibitor (Sigma–Aldrich, USA)] for 30 minutes on ice. Cell lysates were centrifuged at 15,000 rpm for 15 minutes at 4 °C, and the supernatant was collected. Equal amounts of protein from each sample were denatured with 1× loading buffer for 5 minutes at 95 °C, separated by SDS–polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose membranes. The membranes were blocked in 5% nonfat milk for 15 minutes at room temperature and incubated with primary antibodies overnight at 4 °C. The membranes were then washed in TBS-T buffer [20 mM Tris-HCl (pH 7.6), 137 mM NaCl and 0.05% Triton X-100] and incubated in secondary antibodies for 1 hour at room temperature. Finally, the membranes were incubated with enhanced chemiluminescence (ECL) substrate (Pierce, USA) and exposed.
2.4. Luciferase assay
H1299 p53 Tet-on cells were cotransfected with a firefly reporter containing the p53-binding element of the PD-1 promoter plasmid and a Renilla control reporter plasmid with or without BRD4 depletion for 48 hours and treated with or without doxycycline, as indicated, for the last 24 hours. The relative luciferase activity was measured by the Dual-Luciferase Reporter Assay System according to the manufacturer's protocol (Promega, E1910).
2.5. Chromatin immunoprecipitation (ChIP)
Cells were fixed with 1% formaldehyde for 10 minutes at room temperature and lysed in ChIP Lysis Buffer [50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 1% SDS and 1× protease inhibitor] for 10 minutes at 4 °C. After sonication, the lysates were centrifuged at 15,000 rpm for 10 minutes at 4 °C, and the supernatants were collected and precleaned in dilution buffer [(20 mM Tris-HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 1× protease inhibitor] incubated with salmon sperm DNA–saturated protein A agarose (Millipore, USA) for 1 hour at 4 °C. The precleaned lysates were aliquoted equally and incubated with the indicated antibodies overnight at 4 °C. Then, each sample was incubated with saturated Protein A agarose for 2 h at 4 °C. After incubation, the agarose was washed sequentially with TSE I [(20 mM Tris-HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 0.1% SDS and 1% Triton X-100)], TSE II [(20 mM Tris-HCl (pH 8.0), 2 mM EDTA, 500 mM NaCl, 0.1% SDS and 1% Triton X-100)], Buffer III [(10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.25 M LiCl, 1% DOC and 1% NP40)] and buffer TE [(10 mM Tris-HCl (pH 8.0) and 1 mM EDTA)]. The agarose-binding complex was eluted by elution buffer [1% SDS and 0.1 M NaHCO3] and reverse cross-linked for at least 6 h at 65 °C. Purified DNA was extracted by a PCR purification kit (QIAGEN, Germany), and qPCR was performed to detect the relative enrichment of each protein. Primers: human PD-1 (forward: AGGGAAGGAGAGTGGGTGACA; reverse: GCGGGCACAGGGAGAAA), human p21 (forward: AGCAGGCTGTGGCTCTGATT; reverse: CAAAATAGCCACCAGCCTCTTCT), human TIGAR (forward: CACTTTACGTTCCGTGTTTCAGATAAAGTG; reverse: CCACCAAGTGCCTACGAACC).
2.6. GST pull-down assay
The GST or GST-fusion protein containing Escherichia coli was treated with 0.1 mM IPTG overnight at 25 °C to induce protein expression. The proteins were purified by GST•Bind™ Resin (Novagen, USA), and equal amounts of immobilized GST or GST-fusion proteins were incubated with purified BRD4, BID or PDID for 1 hour at 4 °C. Then, the cells were washed with BC100 buffer [20 mM Tris-HCl (pH 7.3), 100 mM NaCl, 10% glycerol, 2 mM EDTA and 0.1% Triton X-100], and the binding components were eluted by boiling with 1× loading buffer and subjected to western blotting analysis.
2.7. In vitro peptide binding assay
Equal amounts of synthesized biotin-conjugated peptide were incubated with purified BRD4 for 1 hour overnight at 4 °C. After washing with BC100 buffer, the binding components were eluted with 2× Laemmli buffer for further western blotting analysis.
2.8. In vitro cell proliferation assay
For each sample, cells were seeded at a density of 1 × 105 into six-well plates with three replicates. The cells were cultured for three or five consecutive days. Then, the cell number was analyzed by crystal violet staining: the cells were fixed with 4% PFA for 20 min at room temperature and stained with 0.1% crystal violet for 30 min at room temperature. The cells were washed with double-distilled H2O three times, and pictures were taken. Cell-containing crystal violet was extracted with 10% acetic acid at room temperature for 30 min. The relative cell number was analyzed by measuring the extracted crystal violet absorption at 590 nm.
2.9. Ethics approval
All C57BL/6 mice were purchased from the Charles River Laboratory (Beijing, China). All mice were housed with 16/8 h light/dark cycles at 25 °C and allowed free access to water and food. The maintenance and experimental procedures of mice were approved by the Institutional Animal Care and Use Committee (IACUC) of CAMS&PUMC.
2.10. Xenograft tumor growth
MC38 cells (2 × 106) were suspended in a volume of 200 µl in PBS per injection. Inject cells subcutaneously (s.c.) into 4-week-old female C57BL/6 mice. When the tumors reached an average volume of ∼200 mm3, therapy was started and treated with anti-PD-1 antibody (CD279; Bio X Cell, USA), JQ1 (S7110; Selleck, USA), anti-PD-1 antibody plus JQ1, and vehicle (4% DMSO+30% PEG300+5% Tween 80+61% ddH2O), respectively. JQ1 was injected daily at a dose of 25 mg/kg, and anti-PD-1 antibody was injected every three days at a dose of 1.25 mg/kg. The tumor diameters were measured with digital calipers every other day, and the tumor volume was calculated as follows: Volume = (Width)2 × Length/2.
2.11. Statistical analysis
All data are presented as the means ± SD. The difference was analyzed using a two-tailed, unpaired Student t-test or one-way analysis of variance (ANOVA) with Bonferroni post hoc test. All statistical analyses were performed using GraphPad Prism software. p < 0.05 was considered statistically significant.
3. Results and discussion
3.1. CTD acetylation inhibits p53-mediated PD-1 transcriptional activation
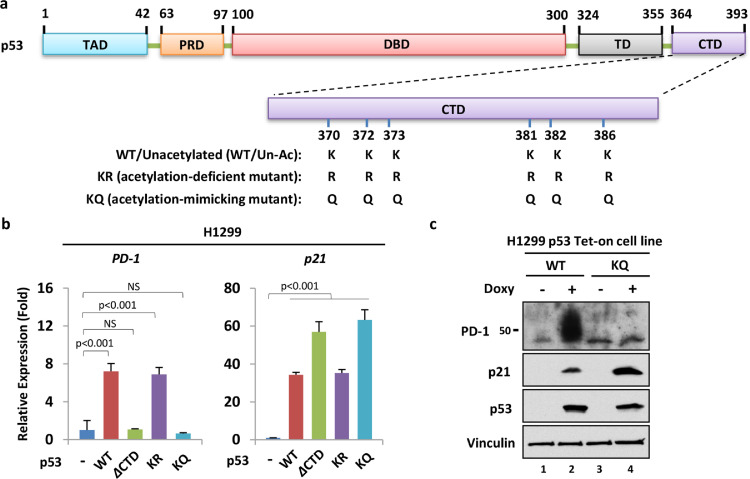
PD-1 is primarily discovered on the surface of immune cells and cooperates subtly with its tumor cell-intrinsic ligand PD-L1 (programmed cell death ligand 1) to inhibit the T lymphocyte response, which may assist tumor cells in evading microenvironmental immune defense [17]. In recent years, accumulating studies have reported the existence of PD-1 in melanoma, hepatic carcinoma cells, non-small lung cancer cells and some other cancer cells, while the signaling pathway driving the expression and activation of the cancer cell-intrinsic PD-1 remains incompletely understood [21]. Our recent study revealed that p53 directly activates PD-1 transcription in lung cancer cells, largely relying on its DBD acetylation [20]. Notably, various PTMs established interweaved yet redundant regulatory mechanisms that controlled p53 biological activities in a context-dependent manner, whereas individual modifications rarely play a decisive role in determining p53 functions. Rather, homogeneous modifications located at discrete residues, or even heterogeneous modifications, interplayed cooperatively in fine-tuning p53 activities [5,22]. To evaluate whether the CTD acetylation status, which represents another critical PTM regulatory module of p53, plays a vital role in PD-1 transactivation, we generated two mutant p53 by replacing the six lysine residues within the CTD with either arginine (KR) or glutamine residues (KQ), respectively (Fig. 1a). Functionally, the KR mutant mimics CTD acetylation-deficient p53, while the KQ mutant mimics the CTD fully acetylated p53.
Fig. 1.
CTD acetylation inhibits p53-mediated PD-1 activation. (a) Schematic diagram of p53 and the lysine residues for acetylation within the CTD. TAD, transactivation domain; PRD, proline-rich domain; DBD, DNA-binding domain; TD, tetramerization domain; CTD, Carboxyl-terminal domain. (b) RT–qPCR analysis of the relative mRNA levels of PD-1 and p21 in H1299 cells transfected with the empty vector or the construct expressing the wild-type, CTD-truncated (ΔCTD), acetylation-deficient (KR) and acetylation-mimicking (KQ) versions of p53 for 24 hours (hrs). (c) Western blotting analysis of the protein level of PD-1 in H1299 p53 Tet-on stable cell lines. The stable cell lines were treated with or without 1 μg/ml doxycycline (Doxy) for 24 hrs to induce wild-type and mutant p53 expression, respectively.
Intriguingly, the KR mutant displayed transcriptional activity of PD-1 similar to that of wild-type p53 upon introduction into H1299, a p53-null lung cancer cell line (Fig. 1b). However, both CTD-truncated p53 (p53-ΔCTD) and the KQ mutant were incapable of triggering PD-1 transcription (Fig. 1b). In contrast, the transcription of cyclin-dependent kinase inhibitor 1A (CDKN1A, also known as p21), one of the canonical downstream target genes of p53, was easily induced by all four types of p53 and particularly showed elevated expression in the p53-ΔCTD and KQ mutant groups (Fig. 1b). These findings implied that both the intact CTD structure and the unacetylated CTD status are necessary for p53-mediated transactivation of PD-1, which is relatively distinct from the commonly accepted conception that full acetylation of CTD universally boosts the transcription of p53 downstream target genes, such as p21 [6,7].
Moreover, we employed a Tet-on system in H1299 cells to investigate p53 regulation of PD-1 protein levels. Following doxycycline (Doxy) treatment, ectopically expressed wild-type p53 induced significant upregulation of PD-1 protein levels, while this effect was sharply compromised upon KQ mutant involvement (Fig. 1c). Taken together, our data demonstrate that CTD acetylation significantly inhibits p53-induced PD-1 expression in cancer cells and suggest that CTD acetylation could also be involved in regulating p53-dependent transcriptional selectivity.
Apparently, our findings about p53-dependent PD-1 expression may represent a paradigm of a unique group of p53 downstream targets whose regulation is largely reliant on the patterns and, to some extent, the potential crosstalk of acetylation within both the CTD and DBD. This multiple residue acetylation-cooperated regulation of p53 was also observed by Tang et al.; loss of acetylation at each individual site failed to hinder p21 transcription, whereas synchronous loss of acetylation at eight lysine residues completely abrogated p53-mediated transactivation of p21 [15]. The profound crosstalk among acetyl-lysine residues could jointly lead to altered p53 binding affinity to certain target genes, although the underlying mechanisms still need expanding investigation. Hence, beyond single-site acetylation, a full view of the multisite combined acetylation status of p53, even covering other modifications such as phosphorylation and ubiquitination, should be comprehensively considered to evaluate the global influence of these PTMs on particular target gene transactivation.
3.2. BRD4 contributes to p53-mediated PD-1 activation by serving as a coactivator
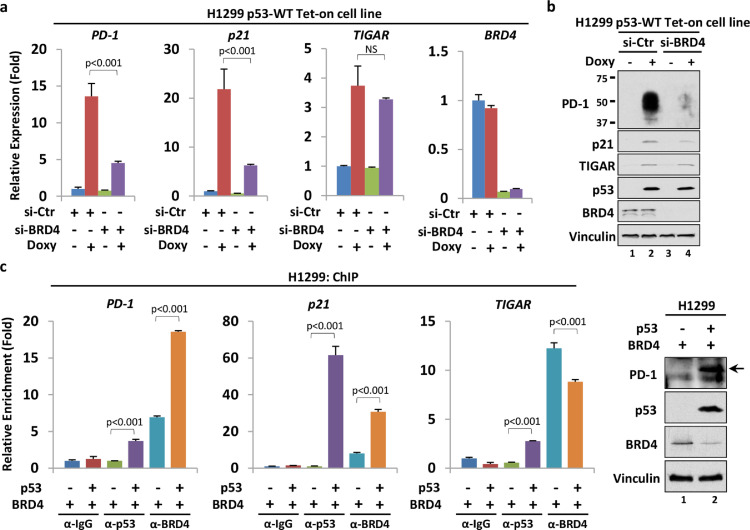
Generally, PTM patterns influence p53 functions by transforming its protein–protein interactions, with outcomes that either reinforce or weaken cofactor-binding affinity [4,5]. For example, we previously proved that DBD acetylation of p53 benefited the recruitment of acetyltransferases, including p300, CBP and TIP60, onto chromatin surrounding the PD-1 promoter, which further promoted its transcription by editing histone markers [20]. Regarding the underlying mechanism, we next attempted to explore CTD acetylation-related cofactors that may participate in the p53-transactivating modulation of PD-1. BRD4 is a typical transcriptional coactivator that recognizes acetylated histones or transcription factors and facilitates gene transcription [23]. It has been reported that BRD4 directly interacts with p53 and regulates the DNA-binding activities of p53 [24]. To evaluate the potential role of BRD4 in the p53-mediated transcription of PD-1, we knocked down BRD4 expression in H1299 p53-WT Tet-on cells by using a pooled small interfering RNA (siRNA). Following BRD4 depletion, p53-induced transcription of both PD-1 and p21 was significantly attenuated, while another p53 target gene, TP53-inducible glycolysis and apoptosis regulator (TIGAR), maintained a similar induction level (Fig. 2a). This BRD4-involved PD-1 transcriptional regulation by p53 was further confirmed when individual BRD4 siRNAs were used (Fig. S1a). As expected, induced p53 also failed to upregulate PD-1 protein levels when BRD4 was depleted (Fig. 2b), suggesting that BRD4 is critically involved in p53-mediated transcription of certain target genes.
Fig. 2.
BRD4 contributes to p53-mediated PD-1 activation by serving as a coactivator. (a) RT–qPCR analysis of the p53-induced relative mRNA levels of PD-1, p21 and TIGAR upon BRD4 knockdown in H1299 p53-WT Tet-on cells. The cells were transfected with control siRNA or siRNA targeting BRD4 for 96 hrs, followed by or without 1 μg/ml Doxy treatment for the last 24 hrs. (b) Western blotting analysis of the p53-induced protein level of PD-1 upon BRD4 depletion in H1299 p53 Tet-on cells. The cells were transfected with control siRNA or siRNA targeting BRD4 for 96 hours, followed by or without 1 μg/ml Doxy treatment for the last 24 hrs. (c) ChIP-qPCR analysis of p53 and BRD4 recruitment to the promoters of PD-1, p21 and TIGAR. The BRD4-expressing plasmid was cotransfected into H1299 cells with or without the wild-type p53-expressing plasmid for 48 hrs. The relative expression levels of PD-1, p53 and BRD4 were detected by Western blotting.
Since depletion of BRD4 apparently alleviated p53-driven luciferase reporter activity of the PD-1 promoter (Fig. S1b), we speculated that BRD4 may act as a coactivator of p53 to regulate PD-1 transcriptional activation. To this end, we performed a chromatin immunoprecipitation (ChIP) assay in H1299 cells. As expected, p53 exhibited decent enrichment on the promoter of PD-1, as well as other targets, including p21 and TIGAR (Fig. 2c). More importantly, BRD4 recruitment was markedly increased to the PD-1 and p21, but not TIGAR, promoters upon p53 coexpression (Fig. 2c). Since BRD4 alone was able to bind chromatin, we tested whether BRD4 affects p53 recruitment to its target promoter. Indeed, we observed that BRD4 overexpression enhanced p53 occupancy on the p21 promoter (Fig. S2). However, under the same experimental conditions, BRD4 showed no statistically significant effect on p53 binding with the PD-1 and TIGAR promoters (Fig. S2). These data suggest that BRD4 may selectively regulate the chromatin recruitment of p53. Collectively, these results reveal that BRD4 functions closely with p53 as a coactivator to propel the expression of PD-1 in cancer cells.
In addition to serving as an acetyl-histone reader that functions as a scaffold for modifying chromatin, BRD4 was recently found to recognize several acetylated transcription factors (TFs), including NF-κB and P-TEFb, prompting the recruitment of these TFs onto chromatin and facilitating the transcription of their responsive targets [25,26]. Although the direct physical interaction between BRD4 and p53 has been proposed by several groups, their mutual effects remain context-dependent. For example, BRD4 bound p53 and drove it to the target promoter in HEK293 cells, whereas their interaction conferred transcriptionally repressive activity toward p53 targets in acute myeloid leukemia (AML) cells [24,27]. Our current findings extended the provoking role of BRD4 in p53-mediated transcription of certain target genes, including p21 and PD-1, in lung cancer cells, which may imply the application of emerging preclinical BRD4 inhibitors as candidate approaches for modulating cancer cell-intrinsic PD-1.
3.3. CTD acetylation blocks the p53-BRD4 interaction
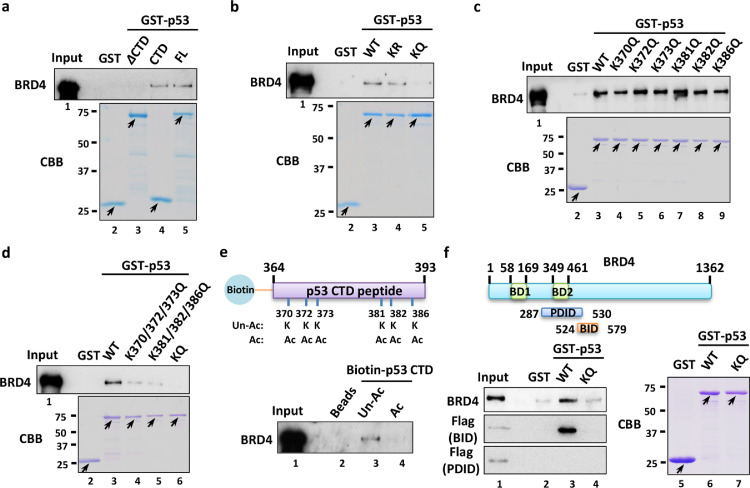
Mechanistically, the CTD of p53 acts as a docking site that selectively attracts coregulators, positively or negatively, according to the modification patterns. For instance, acetylated K370 and K373 of p53 recruit TAF1 to the p21 promoter and activate its transcription [9]. Conversely, the unacetylated CTD of p53 is specifically recognized by a group of AD-containing proteins, such as SET, which serves as a transcriptional corepressor [10]. We then assumed that p53 CTD acetylation-directed PD-1 regulation was achieved by manipulating the recruitment of the coactivator BRD4. To this aim, a GST pull-down assay verified a direct physical interaction between p53 and BRD4 (Fig. 3a). Notably, the CTD fragment was necessary and required for BRD4 binding, as deletion of the CTD completely abolished the p53 interaction with BRD4 (Fig. 3a). In addition, we examined the interaction between BRD4 and CTD acetylation-deficient or CTD acetylation-mimicking mutants of p53. Remarkably, the KR mutant exhibited a normal binding ability to BRD4, whereas the KQ mutant almost completely lost its interaction with BRD4 (Fig. 3b). Interestingly, the single KQ mutation in the CTD (K370Q, K372Q, K373Q, K381Q, K382Q or K386Q) did not affect the p53-BRD4 interaction, indicating that acetylation of a single lysine residue of the p53 CTD is insufficient to block p53 binding with BRD4 (Fig. 3c). In contrast, we found that the combined KQ mutations on the p53 CTD (K370/372/373Q or K381/382/386Q) indeed attenuated the interaction between p53 and BRD4 to some extent (Fig. 3d), supporting that acetylation of multiple lysine residues of the CTD works together to regulate p53-BRD4 binding affinity. Consistent to this notion, p53 maintaining all KQ mutations within its CTD almost completely lost the binding affinity to BRD4 (Fig. 3d).
Fig. 3.
CTD acetylation blocks the p53-BRD4 interaction. (a) In vitro binding analysis of BRD4 with the p53-ΔCTD fragment, p53-CTD fragment or full-length p53. Immobilized GST or GST-fusion proteins, as indicated, were incubated with purified BRD4, and then western blotting was performed to detect their interaction. (b) In vitro binding analysis of BRD4 with wild-type, KR mutant and KQ mutant p53. (c) In vitro binding analysis of BRD4 with wild-type or mutant p53 (K370Q, K372Q, K373Q, K381Q, K382Q or K386Q), as indicated. (d) In vitro binding analysis of BRD4 with the combinational lysine residue mutant p53 (K370/372/373Q, K381/382/386Q or KQ). (e) In vitro peptide binding analysis of BRD4 with unacetylated (Un-Ac) or fully acetylated (Ac) CTD of p53. Synthesized biotin-conjugated lysine residue-modified peptides (as described in the schematic diagram) were incubated with purified BRD4, and then western blotting was performed to detect their interaction. (f) In vitro binding analysis of full-length (aa 1-1362), BID (aa 524-579) or PDID (aa 287-530) of BRD4 with wild-type or KQ mutant p53.
To further validate that BRD4 specifically binds to the acetylated lysine residues of the p53 CTD, we synthesized biotin-conjugated unacetylated (Un-Ac) and fully acetylated (Ac) CTD peptides. The enrichment of BRD4 was easily observed on the unacetylated CTD peptide, while the fully acetylated CTD failed to bind to BRD4 (Fig. 3e). These observations strongly suggest that the acetylation of CTD abrogates the p53 interaction with its coactivator BRD4, which further blocks the transactivation of PD-1.
Considering BRD4 as a well-established reader of acetylated histones, it is simply presumed that BRD4 may recognize the acetylated CTD of p53 via its bromodomain as well. However, the following study proved that the association between BRD4 and p53 was irrelevant to bromodomain binding with acetyl-lysine residues, since JQ1, a potent BRD4 inhibitor that competitively binds with the bromodomain of BRD4, failed to disrupt their interaction [27]. Instead, Wu et al. suggested that BRD4 directly contacted the CTD of p53 through a basic residue-enriched interaction domain (BID) and/or a phosphorylation-dependent interaction domain (PDID) [24]. Along these clues, our observations found that the BID, but not the PDID, of BRD4 strongly bound with p53, and more importantly, the BID almost completely lost the binding affinity to the CTD acetylation-mimicking p53-KQ mutant (Fig. 3f). Taken together, our data indicate that full acetylation of CTD lysine residues potently blocks the interaction of p53 with BRD4.
3.4. Functional implications of CTD acetylation in regulating the p53-PD-1 axis
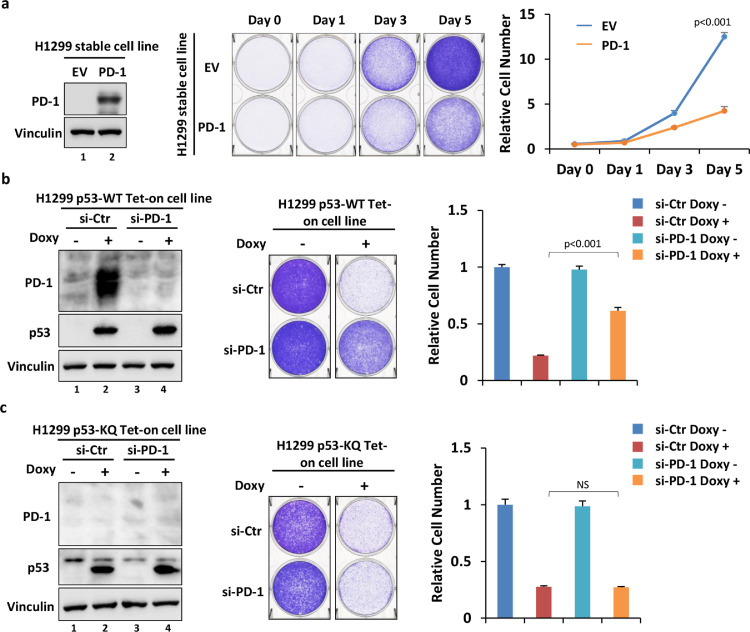
Compared with its well-known immune checkpoint role in T cells, the functional character of the novel cancer cell-intrinsic PD-1 still remains enigma. Since the basal level of PD-1 was low in H1299 cells, we generated a stable H1299 cell line that constitutively expresses ectopic PD-1. Consistent with our previous finding [20], PD-1 re-expression markedly attenuated cell proliferation compared with the control cells (Fig. 4a). Additionally, the wild-type p53-induced proliferative suppression of cancer cells was apparently alleviated when PD-1 induction by p53 was abolished, indicating that PD-1 is involved in the p53-induced growth arrest pathway in cancer cells (Fig. 4b). Notably, similar to wild-type p53, the induction of p53-KQ, which mimics CTD-acetylated p53, also potently inhibited cancer cell growth (Fig. 4c). However, the p53-KQ-mediated suppressive effect on cell growth was not changed upon PD-1 depletion (Fig. 4c). This was not surprising, since p53-KQ did not upregulate PD-1 expression (Figs. 1c and 4c). Taken together, our data support the notion that the mechanism by which CTD acetylation regulates p53 functions is complex and diversified. On one hand, CTD acetylation may impede the stimulation of the p53-PD-1 axis, which is considered to impair one way of p53-mediated anti-proliferation of cancer cells. On the other hand, CTD acetylation may enhance tumor suppressive actions in manners rather than activating cancer cell-intrinsic PD-1.
Fig. 4.
Functional implications of CTD acetylation in regulating the p53-PD-1 axis. (a) Cell growth assay of H1299-EV and H1299-PD-1 stable cell lines. The expression of ectopic PD-1 in stable cell lines was measured by western blotting. (b) Cell growth assay of H1299 p53-WT Tet-on cells treated with or without 1 μg/ml Doxy for 3 days upon control or PD-1 knockdown. The p53 induction and PD-1 knockdown efficiency were measured by western blotting. (c) Cell growth assay of H1299 p53-KQ Tet-on cells treated with or without 1 μg/ml Doxy for 3 days upon control or PD-1 knockdown. The p53 induction and PD-1 knockdown efficiency were measured by western blotting.
The BRD4 inhibitor JQ1 has been under multiple clinical trials for cancer therapy [28]. Interestingly, JQ1 did not interrupt the p53-BRD4 interaction (Fig. S3a), but it still markedly reduced the cellular PD-1 levels regardless of p53 status (Fig. S3b). Similar to the effect of anti-PD-1 immunotherapy, administration of JQ1 apparently alleviated tumor growth (Figs. S3c-e). However, we failed to observe a synergistic effect of the combination of anti-PD-1 and JQ-1 on tumor regression (Figs. S3c-e).
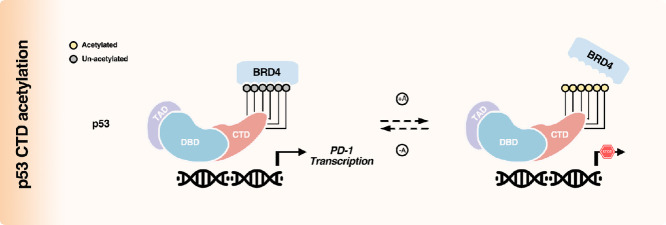
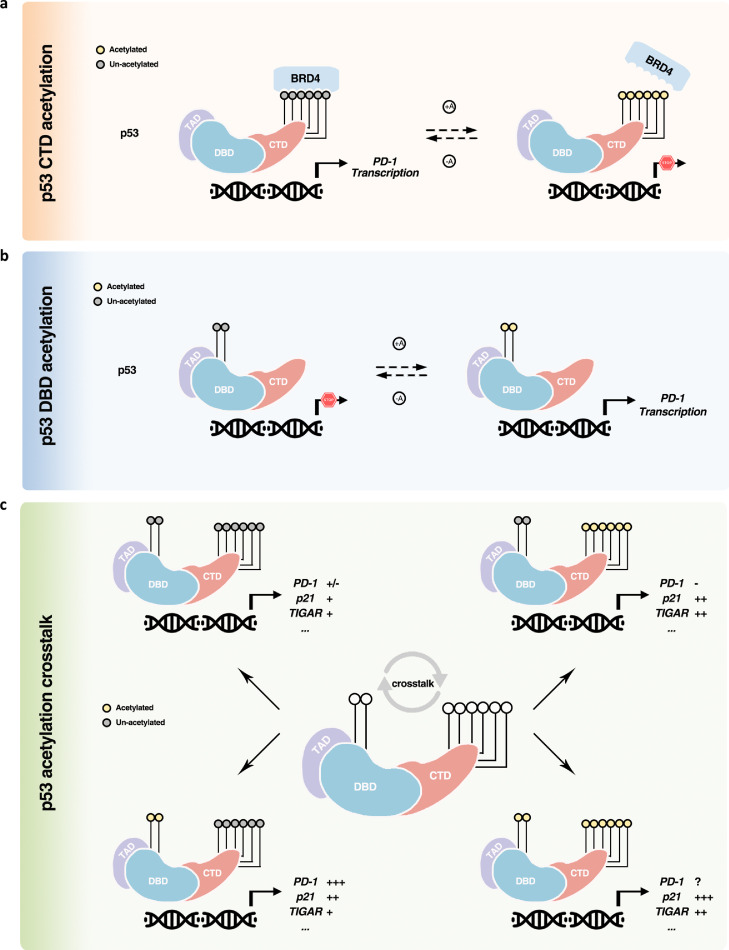
Collectively, our previous and current findings unveiled an intriguing phenomenon that the acetylation status from a distinct domain of p53 may instruct entirely opposing directions on certain target transcription, taking the cancer cell intrinsic PD-1 as an example (Fig. 5a and b). That is, the ultimate transcription output of p53 could probably be derived from the intensive and combined crosstalk among the dispersed acetylation groups, spatially and/or temporally, which might be determined by upstream stimuli (Fig. 5c). Thus, instead of exploring the general acetylation status (and other modification types) of p53 (or other transcription factors), the state-of-the-art PTMomic strategy is likely optimized for clarifying the physiochemical properties and functions of these cellular regulator proteins.
Fig. 5.
Schematic diagram of acetylation and crosstalk in the regulation of p53-directed selective transcription. (a) CTD acetylation negatively modulates p53-mediated PD-1 transactivation by dissociating the coactivator BRD4 from p53 (orange square). (b) Instead, DBD acetylation promotes p53-regulated transcription of PD-1 (blue square). (c) Multiform crosstalk among acetylated lysine residues cooperatively determines the transcriptional output of p53 (green square). The quantity of the plus sign represents the relative transcription intensity of each target gene.
4. Conclusion
In summary, our findings illustrate an alternative mechanism by which CTD acetylation determines p53-dependent transcriptional regulation of PD-1 in cancer cells. BRD4 critically contributes to p53-mediated PD-1 activation by acting as a target-specific coactivator of p53 in a CTD acetylation-sensitive manner.
Declaration of competing interest
The authors declare that they have no conflicts of interest in this work.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81872311, 82073132 and 82122054), Beijing Municipal Natural Science Foundation (7192126) and CAMS Innovation Fund for Medical Sciences (2021-I2M-1-016).
Biographies

Jia Wen received her B.S. and Ph.D. degrees from China Agricultural University. Now she works as a postdoctoral fellow at the Chinese Academy of Medical Sciences & Peking Union Medical College. Her current research interests focus on the understanding of the roles of post-translational modifications (PTMs) in regulating the activities of the tumor suppressor p53 during tumorigenesis.

Donglai Wang is a principle investigator of Department of Medical Genetics, Institute of Basic Medical Sciences & School of Basic Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College. He received his B.S. degree in China Medical University, and his Ph.D. degree in Peking University Health Science Center. His postdoctoral training was done in UT MD Anderson Cancer Center and Columbia University, respectively. His research interests are focused on protein posttranslational modifications (PTMs) and tumorigenesis. He has published 15 scientific research articles. He is a recipient of the “National Science Fund for Excellent Young Scholars in China”.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fmre.2022.03.012.
Appendix. Supplementary materials
References
- 1.Kastenhuber E.R., Lowe S.W. Putting p53 in Context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hafner A., Bulyk M.L., Jambhekar A., et al. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20(4):199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Tavana O., Gu W. p53 modifications: exquisite decorations of the powerful guardian. J Mol Cell Biol. 2019;11(7):564–577. doi: 10.1093/jmcb/mjz060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen J., Wang D. Deciphering the PTM codes of the tumor suppressor p53. J Mol Cell Biol. 2021;13(11):774–785. doi: 10.1093/jmcb/mjab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 7.Luo J., Li M., Tang Y., et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101(8):2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W., Su L., Liao L., et al. PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nat Commun. 2019;10(1):5800. doi: 10.1038/s41467-019-13608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li A.G., Piluso L.G., Cai X., et al. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28(3):408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Wang D., Kon N., Lasso G., et al. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538(7623):118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang D., Kon N., Tavana O., et al. The "readers" of unacetylated p53 represent a new class of acidic domain proteins. Nucleus. 2017;8(4):360–369. doi: 10.1080/19491034.2017.1313939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sykes S.M., Mellert H.S., Holbert M.A., et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24(6):841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y., Luo J., Zhang W., et al. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24(6):827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Rokudai S., Laptenko O., Arnal S.M., et al. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci U S A. 2013;110(10):3895–3900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Y., Zhao W., Chen Y., et al. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S.J., Li D., Ou Y., et al. Acetylation Is Crucial for p53-Mediated Ferroptosis and Tumor Suppression. Cell Rep. 2016;17(2):366–373. doi: 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussiotis V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med. 2016;375(18):1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du S., McCall N., Park K., et al. Blockade of Tumor-Expressed PD-1 promotes lung cancer growth. Oncoimmunology. 2018;7(4) doi: 10.1080/2162402X.2017.1408747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Yang X., Zhang C., et al. Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc Natl Acad Sci U S A. 2020;117(12):6640–6650. doi: 10.1073/pnas.1921445117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Z., Kon N., Liu Y., et al. An unexpected role for p53 in regulating cancer cell-intrinsic PD-1 by acetylation. Sci Adv. 2021;7(14) doi: 10.1126/sciadv.abf4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao H., Wang H., Li C., et al. Cancer Cell-Intrinsic PD-1 and Implications in Combinatorial Immunotherapy. Front Immunol. 2018;9:1774. doi: 10.3389/fimmu.2018.01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed S.M., Quelle D.E. p53 Acetylation: Regulation and Consequences. Cancers (Basel) 2014;7(1):30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White M.E., Fenger J.M., Carson W.E., 3rd Emerging roles of and therapeutic strategies targeting BRD4 in cancer. Cell Immunol. 2019;337:48–53. doi: 10.1016/j.cellimm.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu S.Y., Lee A.Y., Lai H.T., et al. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49(5):843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang B., Yang X.D., Zhou M.M., et al. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang M.K., Mochizuki K., Zhou M., et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 27.Stewart H.J., Horne G.A., Bastow S., et al. BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1. Cancer Med. 2013;2(6):826–835. doi: 10.1002/cam4.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi J. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol. 2014;6(12) doi: 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.