Abstract
Semliki Forest virus (SFV) is a mosquito-transmitted pathogen of small rodents, and infection of adult mice with SFV4, a neurovirulent strain of SFV, leads to lethal encephalitis in a few days, whereas mice infected with the avirulent A7(74) strain remain asymptomatic. In adult neurons, A7(74) is unable to form virions and hence does not reach a critical threshold of neuronal damage. To elucidate the molecular mechanisms of neurovirulence, we have cloned and sequenced the entire 11,758-nucleotide genome of A7(74) and compared it to the highly neurovirulent SFV4 virus. We found several sequence differences and sought to localize determinants conferring the neuropathogenicity by using a panel of chimeras between SFV4 and a cloned recombinant, rA774. We first localized virulence determinants in the nonstructural region by showing that rA774 structural genes combined with the SFV4 nonstructural genome produced a highly virulent virus, while a reciprocal recombinant was asymptomatic. In addition to several amino acid mutations in the nonstructural region, the nsp3 gene of rA774 displayed an opal termination codon and an in-frame 21-nucleotide deletion close to the nsp4 junction. Replacement in rA774 of the entire nsp3 gene with that of SFV4 reconstituted the virulent phenotype, whereas an arginine at the opal position significantly increased virulence, leading to clinical symptoms in mice. Completion of the nsp3 deletion in rA774 did not increase virulence. We conclude that the opal codon and amino acid mutations other than the deleted residues are mainly responsible for the attenuation of A7(74) and that the attenuating determinants reside entirely in the nonstructural region.
Semliki Forest virus (SFV) is an enveloped positive-stranded RNA virus of the family Togaviridae. The SFV prototype was isolated in 1942 from a pool of mosquitoes in Uganda (40). The A7(74) strain (4) is a derivative of an SFV strain isolated from mosquitoes in Mozambique in 1959 (26). Several strains of SFV, such as L10 (4) and SFV4 (24), cause lethal encephalitis in mice of all ages, leading to death of the animals in a few days (3, 11), whereas A7(74) infection of adult mice is asymptomatic per se but leads to axonal demyelination mediated by CD8+ T cells (2). However, the severity of A7(74) infection is strictly age dependent, being lethal for neonatal mice less than 2 weeks old (8, 27), probably because the strain is capable of virion formation in propagating neurons, unlike in mature neurons (27).
SFV nonstructural proteins (nsP) are translated as a polyprotein (nsP1234) from the genomic 42S RNA, and they form essential components of viral RNA replication and transcription complexes (14). The nsP1 protein is a methyl- and guanylyltransferase (1, 18), whereas the nsP2 is a proteinase (45) and nucleoside triphosphatase (33). The nsp3 gene product is a phosphoprotein, and it has been proposed to function together with nsP1 in anchoring the replication complex proteins to cytoplasmic membrane structures (30, 31). In Sindbis virus (SIN), p123 and p1234 are produced first and then cleaved proteolytically. p123 and nsP4 function in minus-strand RNA synthesis, but cleaved products from p123 are required for efficient plus-strand RNA synthesis (38). Mutations in the SIN nsP3 protein have been shown to result in blockage of RNA synthesis, indicating the importance of this protein or the polyprotein component in replication, although the exact mechanism of action remains unknown (21). The nsp4 gene displays high similarity to the RNA-dependent polymerase sequences of other RNA viruses (13, 15). Recently, enzymatically active RNA replication machinery was reconstructed for SIN in vitro by introducing together the single components of the multiprotein complex (22).
In several alphaviruses, such as SIN, Middelburg virus (43), and Ross River virus (42), as well as Venezuelan (16) and western and eastern (48) equine encephalitis viruses, an opal (UGA) termination codon interrupts the polygenic RNA at the 3′ end of the nsp3 gene. In contrast, in the SFV prototype (44) and in SFV4, an arginine codon is found at the analogous position. For the related O'Nyong-nyong virus, strains with either an arginine (42) or an opal codon (19) have been characterized. In RNA viruses generally, readthrough of an in-frame termination codon is often employed to regulate the synthesis of a viral polymerase or reverse transcriptase (23). In SIN, the opal readthrough proceeds with about 20% efficiency in vitro, leading to lower nsP4 amounts (25), but rather than total nsP4, the relative amounts of nsP3, nsP34, and nsP4 seem to be important for efficient alphavirus replication (23).
Although in many alphaviruses mutations in the virion proteins or nucleocapsid have been found to alter virulence (11, 36), often having a synergistic effect (32), the results presented in this paper strongly suggest that the replicase complex nsp3 gene is the main pathogenic determinant conferring the avirulent phenotype of A7(74) and provide a rare example of the presence of an opal termination codon in one alphavirus strain but not in another. In contrast, the structural genes of A7(74) do not seem to limit viral replication.
MATERIALS AND METHODS
Cell cultures.
Cerebellar granule neurons were isolated from 7-day-old Harlan Spraque Dawley rats (Harlan Laboratories) as described before (5). Briefly, the pups were decapitated, and the cerebella were removed into phosphate-buffered saline (PBS). The meninges were carefully removed, and the tissue was chopped with a razor blade, trypsinized, and subsequently resuspended by titurating in DNase containing trypsin inhibitor to separate the cells. The cells were cultured in Eagle's minimal essential medium (MEM; Gibco-BRL) containing 2 mM glutamine, 50 U of penicillin per ml, and 50 μg of streptomycin per ml, supplemented with 10% (vol/vol) fetal calf serum (Gibco), 20 mM KCl, and 30 mM glucose. After 1 day in culture, the medium was replaced with fresh medium containing 10 μM arabinocytidine (Sigma) to inhibit the replication of nonneuronal cells, thus maintaining the purity of the culture. The cultures were maintained at 37°C in a humidified atmosphere of 5% CO2–95% air on poly-l-lysine-coated cell culture plates until infected.
BHK-21 cells (American Type Culture Collection) were maintained in BHK-21 medium (Glasgow-MEM) supplemented with 5% fetal calf serum, 5% tryptose phosphate, 2 mM glutamine, and 10 mg of gentamicin per liter.
MBA-13 cells were obtained through transformation by Epstein-Barr virus of mouse brain cells and possess the antigenic properties of oligodendrocytes (unpublished data). The cells were maintained in Eagle's MEM supplemented with 5% fetal calf serum and 10 mg of gentamicin per liter.
Virus strains and passage histories.
The SFV4 clone, a prototype derivative, was obtained by in vitro transcription of the full-length cDNA clone pSP6-SFV4, kindly donated by P. Liljeström (Stockholm, Sweden). The A7(74) strain used for molecular cloning in the present study was originally derived by Bradish et al. (4) from strain AR2066 through seven passages in neonatal mouse brain and two plaque purifications. We obtained this strain from H. E. Webb (London, United Kingdom), and it has since then been passaged several times in MBA-13 cells and subsequently plaque-purified three times on the same cells for later molecular analyses and construction of the full-length genome. The CA7 strain analyzed previously by Tarbatt et al. (46) was separated from the Bradish isolate after the initial seven passages in mouse brain (4) and passaged once more in mouse brain before three plaque-purifications and five or fewer passages in BHK cells (G. Atkins, personal communication).
Sequence analysis.
Sequence analysis of the recombinant A7(74), named rA774, and SFV4 nsp region genes was performed with a Perkin-Elmer ABI Prism automatic sequencer, model 377.
Construction of full-length and chimeric RNA expression plasmids.
The chimeric plasmids and viruses obtained are listed in Fig. 1. The validity of the intermediate plasmids was confirmed by restriction fragment analysis or sequencing. The cDNA of the A7(74) nsp region was obtained by PCR following reverse transcription of the purified viral RNA (Ultraspec; Biotecx Laboratories). The primers were designed on the basis of the SFV prototype (44) sequence. Overlapping PCR fragments were cloned in the pGEM-T vector (Promega) to produce intermediate plasmids. For construction of the structural (str) region, cDNA was obtained from a lambda phage cDNA library used recently for sequencing (34, 35).
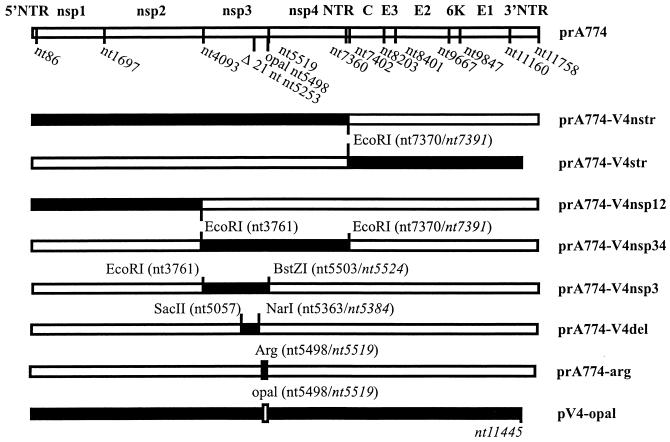
FIG. 1.
Schematic illustration of the full-length SFV cDNA constructs used as a source for infectious RNA. Solid bars indicate SFV4 cDNA, and open bars represent the A7(74) genome. The pGEM1-derived vector (24) is not shown. The 21-nt deletion in prA774 alters the numbering of nucleotides with regard to SFV4. SFV4-specific nucleotide position numbers are given in italics. The 3′ NTR of SFV4 is 334 nt shorter than that of rA774 (34). prA774 is the cDNA clone of A7(74). prA774-V4nstr was obtained by combining the 5′NTR and nonstructural genes of SFV4 with the structural genes and 3′NTR of rA774. prA774-V4str is reciprocal to prA774-V4nstr. In prA774-V4nsp12, a fragment containing the 5′ NTR, nsp1, and nsp2 of SFV4 was introduced into prA774, and similarly, in prA774-V4nsp34 and prA774-V4nsp3, the indicated genes were used to replace the corresponding regions in rA774. In prA774-V4del, a short fragment of SFV4 nsp3 containing the 21 nt deleted in rA774 has been inserted into the corresponding region of rA774. prA774-arg differs from rA774 only by having an arginine codon at the opal position. pV4-opal is identical to SFV4 except that the arginine has been mutated to opal.
To exactly link the rA774 5′-nontranslated region (5′NTR) cDNA with the promoter region of pSP6-SFV4, site-directed mutagenesis was used. The 5′NTR of rA774 was first cloned into the NcoI site at the vector junction region of pSP6-SFV3 (24), followed by removal of the two C residues from the NcoI site by site-directed mutagenesis to obtain a transcriptionally more active construct (24). The procedure for connecting the rA774 3′NTR to the pSP6-SFV4 vector has been described previously (36).
To link the nsp3 gene with nsp4 in the prA774-V4nsp3 construct, PCR was used to create a BstZI site at nucleotide (nt) 5524. pV4-opal, a derivative of pSP6-SFV4 containing an arginine-to-opal (CGA to UGA) mutation (nt 5519), and a reciprocal, rA774-arg, with an arginine codon at the opal position in rA774, were obtained by in vitro mutagenesis (17). DNA sequence analysis was used to verify the sequences of the mutant clones.
Virus production.
Intact full-length cDNA clones were transcribed in vitro using reagents from Boehringer, and the RNA transcripts were electroporated into BHK-21 cells (Bio-Rad gene pulser) (24). For large-scale preparation, viruses obtained from the primary cultures were propagated in MBA-13 cells in roller bottles as described previously (35).
Treatment of mice.
Four- to seven-week-old, specific-pathogen-free female BALB/c AnNHsd mice (Harlan Laboratories) were used throughout this study and treated and housed at the animal facility of the University of Turku in accordance with the guidelines of the University Ethics Committee.
For mortality studies, 15 to 20 mice were inoculated intraperitoneally (i.p.) with 106 PFU of virus in 100 μl of PBS and observed daily for 20 days. Another group of 16 mice were infected as above for virus titration and immunohistochemistry. Two mice were sampled daily for a period of 8 days. Mice were anaesthesized with CO2, and blood was collected with a heparinized pipette from the right ventricle, after which the mice were perfused with cold PBS. Brains and spinal cords were removed, and the brains were divided sagittally. One half of the brain and the spinal cord were immersed in 4% PBS–Formalin for at least 24 h before being embedded in paraffin wax. Deparaffinized sections (4 μm thick) were stained for SFV antigens with a polyclonal A7(74)-specific antibody and the avidin-biotin-peroxidase detection method (Vectastain ABC kit; Vector Laboratories). The SFV-specific antibody was produced in a rabbit immunized with purified, inactivated A7(74). Appropriate antigen-negative controls were obtained from mice inoculated with PBS, while negative staining controls were provided by processing SFV4-positive samples and omitting the primary antibody. The other half of the brain was taken in PBS and homogenized. The homogenate was centrifuged, and the supernatant was used for virus titration. The blood samples were stored at 4°C until centrifuged at low speed and diluted 1:10 in PBS. Virus titers were obtained by analyzing plaque formation in MBA-13 cells as described previously (35).
Virus RNA detection.
Confluent BHK-21 cell layers and 72-h-cultured primary rat cerebellar granule neurons on 12-well dishes were infected with virus at a multiplicity of infection (MOI) of 20 in 0.2-ml Glasgow-MEM and conditioned medium (MEM), respectively, and incubated at 37°C for 1 h. The medium was replaced with 0.3 ml of warm medium containing 1 μg of actinomycin D per ml for 2 h at 37°C and incubated for a further 3 h in the same conditions except for the addition of 20 μCi of [5,6-3H]uridine (Amersham) per ml. Wells were washed twice with ice-cold serum-free medium and incubated on ice twice with 1 ml of 10% trichloroacetic acid (TCA) for at least 5 min. TCA precipitates were solubilized in 1 ml of 10% sodium dodecyl sulfate (SDS). One fifth was mixed with 3 ml of scintillation cocktail (OptiPhase HiSafe3), and the activity was determined with a liquid scintillation counter (1216 Rackbeta; Wallac, Turku, Finland).
Western analysis.
Rat cerebellar granule neurons cultured for 72 h were infected on 12-well plates at an MOI of 10 in 0.2 ml of conditioned medium (MEM). After 0.5 h at 37°C, the medium was replaced with 0.8 ml of conditioned medium. The cells were incubated at 37°C, and at 0, 8, and 24 h, samples were collected by removal of medium and addition of 100 μl of Laemmli sample buffer. The samples were sonicated briefly with a needle sonicator, and the proteins were separated by SDS–12% polyacrylamide gel electrophoresis (SDS-PAGE). One tenth of the sample was run and stained with Coomassie to control for the amount of protein, while an identical gel was blotted on nitrocellulose (Schleicher & Schuell) with a Hoefer Semi-Phor blotter (Hoefer Scientific Instruments), 0.8 mA/cm2 for 30 min at room temperature. The filter was blocked in PBS–0.4% Tween containing 10% dry milk and incubated subsequently in the presence of SFV-specific rabbit serum (1:6,000 dilution) in PBS–0.4% Tween containing 3% dry milk and peroxidase-conjugated goat anti-rabbit antibody for 1 h at room temperature. The filter was developed using enhanced chemiluminescence (ECL) reagents from Amersham. Exposure times on Kodak X-OMAT AR film varied between 1 and 3 min.
Confluent BHK-21 cells in 60-mm plates and 72-h-cultured rat cerebellar granule neurons on 35-mm plates were infected at an MOI of 10 with SFV chimeras in Glasgow-MEM and conditioned MEM, respectively, at 37°C for 30 min. Virus was removed, and after replacement of medium, incubation was continued for 3 h at 37°C. The medium was then removed, and the cells were suspended in 0.2 or 0.1 ml of reducing sample buffer and sonicated briefly to shear the DNA. Samples of 3.5 to 20 μl were run on SDS–10 to 12% PAGE and blotted onto a nitrocellulose membrane, and nsP3 and nsP4 were detected with monospecific polyclonal antibodies made in rabbits (generously provided by L. Kääriäinen, Helsinki, Finland) by the ECL method (Amersham).
Nucleotide sequence accession numbers.
The EMBL, GenBank, and DDBJ accession numbers for rA774 are: 5′NTR/nsp1/2, Y17207; nsp3, Y12518; nsp4, Y14761; capsid, X78109; E3, X78110; E2, X78111; 6K, X78112; E1, X74425; 3′NTR, X74423. The SFV4 sequence number is AJ251359, and the SFV prototype number is X04129.
RESULTS
Sequence analysis.
Given the differences in the pathogenesis of rA774 and SFV4, we sequenced their entire nsp regions. Comparisons were made between these viruses and with CA7 (46), another attenuated strain of SFV which originates from the same isolate of viruses as A7(74) (4) (see Materials and Methods). For nsp1, 99.4 and 98.1% nucleotide identity was observed compared with CA7 and SFV4, respectively, while the deduced amino acid sequences differed at five and six residues, respectively (Table 1). The nsp2 of rA774 shows 98.5 and 98.1% identity to CA7 and SFV4, respectively, which predicts 20 and 4 amino acid differences, respectively.
TABLE 1.
Nucleotide substitutions in SFV nonstructural protein genes resulting in amino acid changesa
| Gene | Nucleotides, rA774/CA7/SFV4 (position) | Amino acids, rA774/CA7/SFV4 (position) |
|---|---|---|
| nsp1 | TAT/ATA/TAT (369–371) | VC/DS/VC (95–96) |
| C/G/C (660) | A/G/A (192) | |
| T/T/A (794) | S/S/C (237) | |
| C/C/T (1007) | H/H/Y (308) | |
| G/G/A (1244) | V/V/I (387) | |
| A/G/G (1365) | K/R/R (427) | |
| C/C/T (1536) | A/A/V (484) | |
| GA/TG/GA (1650–1651) | G/V/G (522) | |
| G/G/A (1686) | R/R/H (534) | |
| nsp2 | A/C/A (2021) | N/H/N (109) |
| A/A/G (2120) | I/I/V (142) | |
| A/T/A (2193) | Y/F/Y (166) | |
| (2374–2398)b | — | |
| C/A/C (2472) | S/Y/S (259) | |
| CAG/AAA/CAG (2879) | Q/K/Q (395) | |
| A/A/T (3240) | E/E/V (515) | |
| G/A/G (3359) | G/R/G (555) | |
| C/A/C (3428) | Q/K/Q (578) | |
| A/T/A (3466) | R/S/R (590) | |
| C/T/C (3660) | S/L/S (655) | |
| C/A/C (3675) | T/K/T (660) | |
| A/T/A (3693) | Y/F/Y (666) | |
| A/C/G (3730) | R/S/R (678) | |
| AC/TA/TC (3732) | Y/L/F (679) | |
| A/A/G (3858) | N/N/S (721) | |
| nsp3 | G/A/A (4124) | V/I/I (11) |
| C/C/A (4236) | A/A/E (58) | |
| G/G/C (4302) | G/G/A (70) | |
| C/A/T (4346) | L/M/L (85) | |
| T/C/T (4644) | L/P/L (184) | |
| G/T/T (4696) | L/F/F (201) | |
| G/A/A (4838) | D/N/N (239) | |
| Δ21 nt (5253–5273)c | Δ7 aa (387–393)d | |
| C/A/A (5274) | A/D/D (394) | |
| G/C/G (5371) | E/D/E (426) | |
| C/A/C (5375) | P/T/P (428) | |
| G/T/T (5391) | R/I/R (433) | |
| A/G/G (5417) | T/A/A (442) | |
| T/G/G (5440) | F/L/L (449) | |
| T/C/C (5519) | Opal/R/R (476) | |
| nsp4 | G/T/T (6007) | E/D/D (156) |
| A/T/A (6959) | I/F/I (474) | |
| A/A/G (7371) | K/K/R (611) |
The accession numbers for rA774 are given in the text. The accession number for CA7 is Z48163, and the accession number for SFV4 is AJ251359. Nucleotides and amino acids (aa) are numbered according to the SFV4 sequence.
The nucleotide sequence at nt 2374 to 2398 is controversial, as an insertion of two nucleotides was found in CA7 followed, however, by two single nucleotide deletions reconstituting the original reading frame. Also, one nucleotide substitution was found, C/G (nt 2388).
Nucleotides deleted in rA774 but present in CA7 and SFV4: GTGCACCCTGAACCCGCAG.
Amino acids deleted in rA774 but present in CA7 and SFV4: GIADLAA.
In nsp3 of rA774, an in-frame deletion of 21 nt was detected at nt 5253 to 5273, abolishing seven amino acid residues (GIADLAA), which was retained in both SFV4 and CA7. The deletion site was flanked on both sides by the motif 5′-GTGCACCCTGAACCCGCAG-3′, present at this position in all three SFV strains. We showed by reverse transcription-PCR that the viral RNA species with the deletion was the only RNA present in our original A7(74) virus stock. Sequence analysis further revealed an opal (UGA) termination codon in rA774 nsp3 at nt 5498, close to the nsp3-nsp4 junction, which was rather unexpected, as SFV4 and CA7 (46) both display an arginine codon at the equivalent position. On the other hand, in several other alphaviruses, such as SIN, Middelburg virus (43), Ross River virus (42), and Venezuelan (16) and western and eastern (48) equine encephalitis viruses, an opal codon has been found. The presence of the opal codon in the parental rA774 was confirmed by sequencing three independent PCR clones obtained from our original plaque-purified virus, and it could also be shown in the stock virus. Ignoring the 21-nt deletion, the nsp3 gene of rA774 displayed 97.4 and 96.6% nucleotide identity with CA7 and SFV4, respectively, the mutations predicting 12 and 9 amino acid changes, respectively (Table 1). The nsp4 gene of rA774 exhibited 99.1 and 97.4% nucleotide identity with CA7 and SFV4, respectively, with the deduced amino acid sequences differing by only two amino acids in both (Table 1). Interestingly, in both rA774 and CA7 (46), there was an adenine residue in nsp4 (nt 7350 and 7371, respectively) immediately preceding the transcription start site of 26S RNA, while SFV4 had a guanidine at this site (44). As this could have been critical for transcription efficiency, we replaced in rA774 the whole 26S RNA-encoding region, including the preceding 28 nt, with the equivalent SFV4 sequence. The recombinant did not, however, display increased virulence (not shown). The NTR between nsp4 and the capsid gene was identical in all three virus strains.
A7(74) genome-length clone.
The A7(74) virus is avirulent for adult mice but lethal for neonatal mice, whereas SFV4 is lethal for mice of all ages (8, 27). It has been suggested that virulent strains reach a critical threshold titer in the central nervous system (CNS), leading to neuronal damage by either necrosis or apoptosis (10, 37). Virus chimeras have proven a powerful tool in identification of pathogenic sites of viral genomes (32, 35). In order to facilitate the construction of recombinant SFV for identification of genome loci involved in attenuation of A7(74), a full-length cDNA of A7(74) was synthesized and used to replace the prototype SFV insert in the pSP6-SFV4 plasmid (24). This plasmid, prA774, gave rise to rA774 virus after transcription with SP6 RNA polymerase (see Materials and Methods). rA774 replicated to comparable titers in mouse brain with the plaque-purified A7(74) (Fig. 2), although somewhat lower titers were measured in the blood at days 1 and 2. At day 3 the values in blood reached comparable levels in both strains (Fig. 2), and no virus could be detected in the samples at 4 days postinfection or later. When administered i.p. (106 PFU), rA774 was asymptomatic for adult BALB/c mice (Fig. 3), except that 1 mouse out of 20 showed opticus neuritis and another had slight, transient hind limb paralysis. For neonatal mice, rA774 was lethal (unpublished results). In BHK-21 cells and cultured rat cerebellar granule neurons, the rA774 virus displayed a phenotype similar to that of the parental A7(74) (see below). The vehicles prA774 and pSP6-SFV4 (24) were then used to construct a panel of recombinant viral RNA expression plasmids which, upon in vitro transcription and transfection of mammalian cells, produced a collection of virus chimeras (Fig. 1).
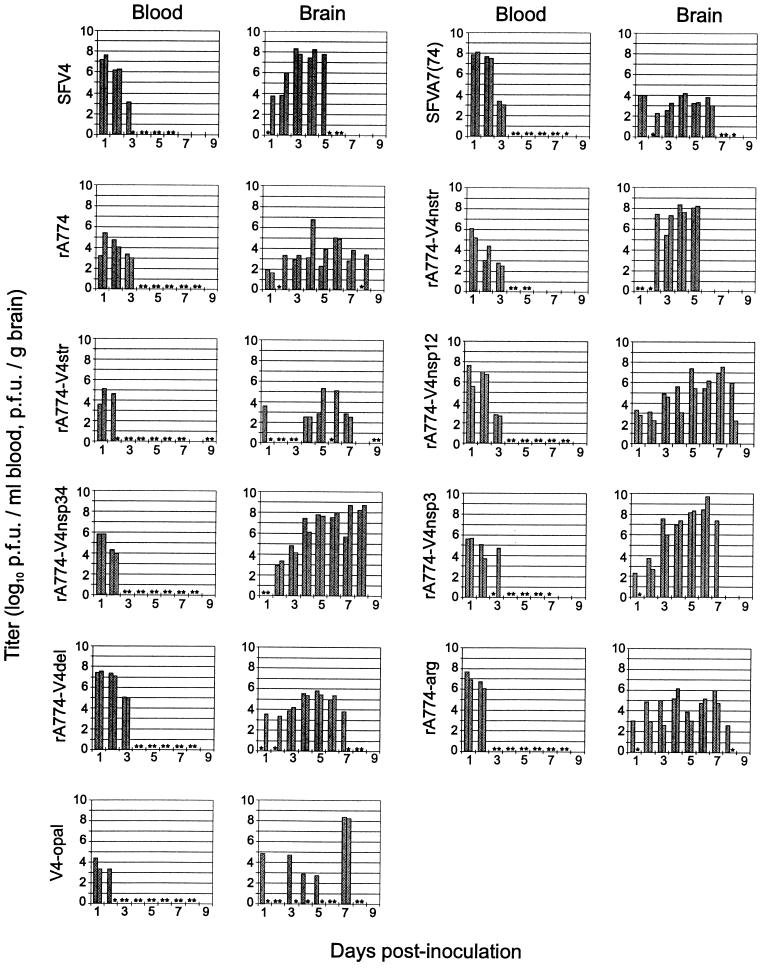
FIG. 2.
Virus titers in the blood and brain of mice infected with SFV. Adult BALB/c mice were infected i.p. with 106 PFU of virus in 100 μl of PBS. Serum and brains from two mice were taken each day. Each bar represents an individual animal. Stars indicate animals with titers under the detection limit (50 PFU/ml or 200 PFU/g). Note that the high titers at day 7 for both V4-opal-infected mice represent reversion of the mutants.
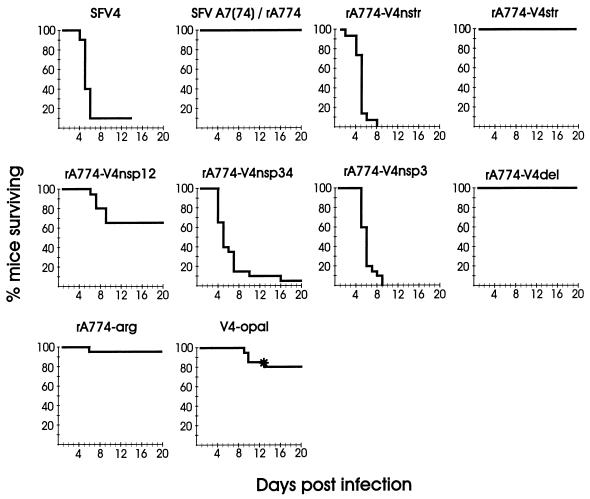
FIG. 3.
Kinetics of mortality in SFV infection. Four- to seven-week-old female BALB/c AnNHsd mice were infected i.p. with 106 PFU of virus in 100 μl of PBS. The star indicates a V4-opal-infected mouse with irreversible hind limb paralysis that was sacrificed for histological analysis on day 13.
A7(74) replicase complex mediates attenuation.
For the localization of virulence determinants, we first analyzed the function of the SFV4 replicase complex attached to the complete set of rA774 capsid and envelope glycoproteins including the 3′NTR, which differs from SFV4 in that it contains a 101-nt insertion as well as a repeated motif (34). For this purpose we constructed a chimeric rA774-V4nstr virus (Fig. 1), which expressed the nonstructural proteins of SFV4 and the proteins encoded by the subgenomic 26S RNA of rA774. rA774-V4nstr was highly virulent upon intraperitoneal (i.p.) infection, with 106 PFU of virus killing all of the 15 infected female mice (Fig. 3) (15 of 15 male mice tested in parallel died as well [data not shown]), suggesting that the structural proteins of rA774 do not restrict virus replication. rA774-V4nstr reached brain titers up to 7 × 108 PFU/g, comparable to SFV4 (Fig. 2). This experiment also showed that the viral NTRs are functionally compatible when originating from avirulent and virulent strains. To analyze the reciprocal case, rA774-V4str virus (Fig. 1) containing the rA774 replicase complex genes, including the 5′NTR combined with SFV4 structural genes was constructed, and the clone was shown to grow to only moderate titers (104 to 105 PFU/g) in murine brains, similar to rA774 (Fig. 2) in being asymptomatic for adult animals (Fig. 3). This result confirmed that the avirulence of A7(74) derives from the viral replicase complex.
Search for virulence loci within the replicase complex genes.
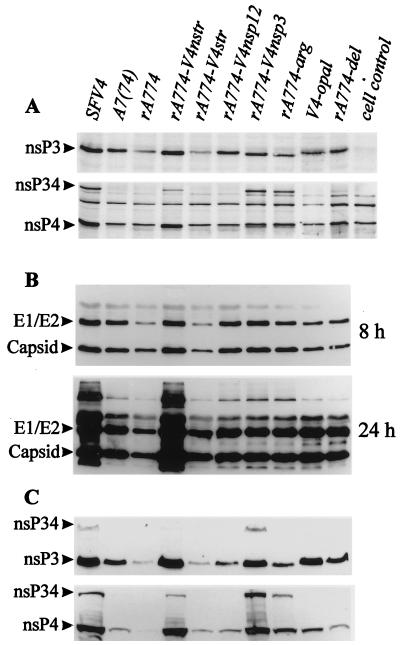
Mutations introduced at multiple genome loci may interfere with the correct secondary structure of viral RNA or the functional assembly of viral proteins, thus abolishing virulence, but transforming an avirulent virus to a virulent one requires introduction of true virulence determinants. We therefore sought to identify loci in the SFV4 nsp region which, when inserted into the avirulent rA774, would confer virulence. We first replaced rA774 nt 1 to 3761, comprising the 5′NTR, nsp1, and nsp2, and nt 3761 to 7370, containing genes nsp3 and nsp4, with the corresponding segments of SFV4. The chimeras obtained were designated rA774-V4nsp12 and rA774-V4nsp34, respectively (Fig. 1). rA774-V4nsp12 showed increased virulence, killing 7 of 20 mice (Fig. 3) and causing paralytic symptoms in an additional 6 animals, proving the involvement of this region in regulation of pathogenicity. rA774-V4nsp34 replicated to high titers in the CNS of mice (Fig. 2) and killed 95% of the infected animals (Fig. 3). As sequence analysis had revealed major modifications in nsp3, the effect on virulence of this gene was analyzed next. A highly neurovirulent virus with a lethal phenotype comparable to that of parental SFV4 and rA774-V4nstr was obtained by replacement of the nsp3 gene in rA774 with the corresponding SFV4 nsp3 gene. The phenotype of this recombinant, designated rA774-V4nsp3 (Fig. 1), was confirmed by histological analysis, and the virus showed wide spreading in the cortex and cerebellum, with Purkinje neurons and granule cells strongly affected (Fig. 4C; also see below). In rA774-V4nsp3-infected BHK-21 cells, the amount of nsP4 was higher than that measured in the chimeras expressing rA774 nsP3 (see Fig. 6C). However, the corresponding viral RNA amounts in both BHK-21 cells and cultured neurons were clearly lower than the amounts produced by other virulent clones (see Fig. 6).
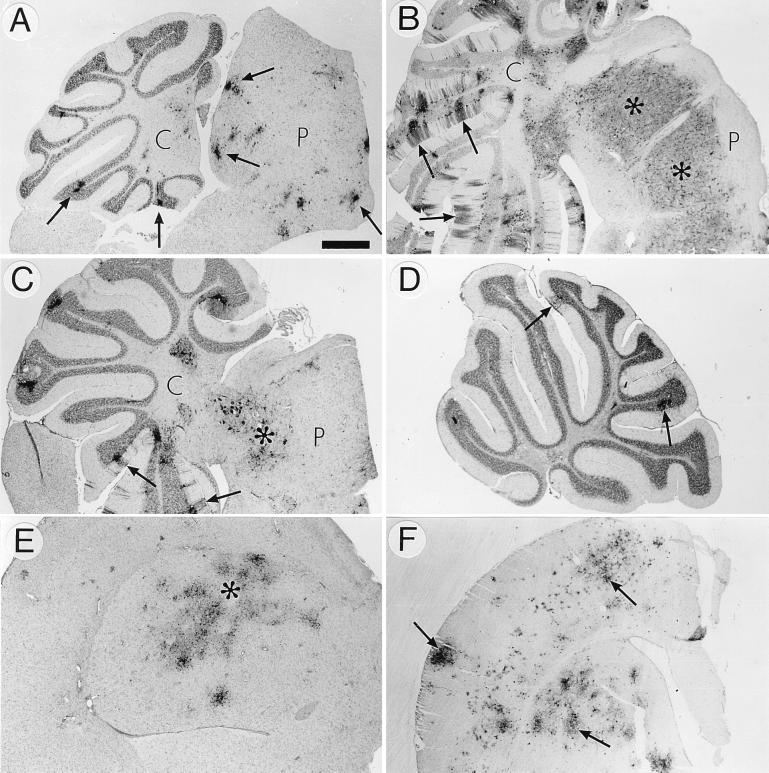
FIG. 4.
SFV polyclonal antibody-stained brain sections from mice infected i.p. with 106 PFU of virus. (A) rA774 infection. Multiple small antigen-positive foci of virus are visible in the pons (P) and cerebellum (C) (arrows). Bar, 0.5 mm. (B) Widespread rA774-V4nstr infection of the pons (P), indicated with asterisks. Arrows, infected granule cells and cerebellar (C) Purkinje neurons with antigen-positive dendrites. Bar, 0.7 mm. (C) rA774-V4nsp3-infected areas of pons (P, asterisk) and cerebellum (C) with affected Purkinje cells, including dendrites (arrows). Bar, 0.6 mm. (D) rA774-V4del-infected cerebellum with a few viral foci (arrows). Bar, 0.6 mm. (E) Extensive rA774-arg invasion (asterisk) of the cerebral nucleus (putamen). Bar, 0.6 mm. (F) Widespread infection with V4-opal of brain hemisphere (arrows) of a mouse infected with the V4-opal construct which reverted to arginine. Bar, 0.7 mm.
FIG. 6.
Expression of viral proteins in cell cultures infected with parental and chimeric SFV. At given time points, the cells were lysed in Laemmli sample buffer and sonicated briefly to shear DNA prior to SDS-PAGE. The proteins were blotted onto nitrocellulose and visualized with specific antibody and peroxidase ECL detection. (A) Production of the nonstructural proteins nsP3 and nsP4 was measured 3.5 h after infection of 72-h-cultured rat cerebellar granule neurons (10 PFU/cell). Polyclonal monospecific antibodies (provided by L. Kääriäinen) were used in detection. (B) Synthesis of capsid protein and E1 and E2 envelope glycoproteins (visible as one overlapping band) in cultured rat cerebellar granule neurons. Samples were taken at the indicated time points postinfection. Polyclonal SFV-specific rabbit antibody was used for detection. (C) Production of the nonstructural proteins nsP3 and nsP4 in SFV-infected monolayers of BHK-21 cells was measured as described above for neurons. The applied amounts of total protein were highly similar, as confirmed by Coomassie staining (not shown).
Completion of the nsp3 deletion.
To more closely analyze whether the deleted residues are responsible for the reduced CNS replication and pathogenicity of rA774, the seven deleted amino acids (GIADLAA) in the nonconserved region of nsP3 were reconstructed. Given that a membrane-binding function has been ascribed to the alphavirus nsP3 protein, it was possible that the absence of such a hydrophobic domain could reduce its binding properties and thus could affect transcription. Reconstruction in rA774 of this deleted site with a 327-nt fragment from SFV4 that restored the amino acid sequence resulted in recombinant rA774-V4del virus (Fig. 1). This chimera was, however, unable to replicate in mice to titers high enough to cause clinical symptoms (Fig. 2), and it could not be phenotypically distinguished from the parental rA774 virus or other avirulent clones (Fig. 3).
In vitro mutagenesis of the in-frame opal codon.
In SIN, an opal termination codon in the nsp3 gene close to the nsp4 junction dramatically reduces the amount of nsP4 mRNA (25). In order to analyze the role of the opal codon in rA774 replication, two mutant viruses were constructed. First, the opal codon in rA774 was changed to an arginine (CGA) codon, and second, the arginine (amino acid 476) CGA (nt 5516) in SFV4 was mutated to a UGA stop codon. The resulting viruses, rA774-arg and V4-opal, respectively (Fig. 1), were then tested in adult BALB/c mice. The rA774-arg clone with nonrestricted readthrough displayed markedly increased virulence compared to rA774, killing 1 of 20 mice (Fig. 3) and causing paralysis in 5 of 20 mice, being, however, far less virulent than the parental SFV4 and the lethal rA774-V4nstr or rA774-V4nsp3 chimeras. Upon i.p. infection with the V4-opal mutant, 3 mice out of 20 died (Fig. 3), and a fourth mouse showed hind limb paralysis. In another experiment, we isolated virus from the brains of 2 V4-opal-infected mice (total, 16) who were moribund at day 7, which showed a high virus titer in the brain (Fig. 2) and a high virus load in the CNS (Fig. 4F) at the time of sampling. We showed with RT-PCR and sequencing that in these mice the opal codon had undergone mutation, giving rise to an arginine (CGA) codon in one and a tryptophan (UGG) codon in the other mouse, indicating that V4-opal is unstable and most probably even less virulent than what was indicated by the mortality numbers. The viruses in the brains of the affected mice in the mortality study were not sequenced, but it is possible that reversion was also responsible for the reoccurrence of virulence in these. The late mean day of death (9.7) (Table 2) in V4-opal-infected mice probably indicates slow evolution of revertants. Thus, the results with the V4-opal mutant suggested an important role for the opal codon in regulation of virulence.
TABLE 2.
Mortality after SFV infectiona
| Virus | No. of mice infected | No. dead | Mean day of death (SEM) |
|---|---|---|---|
| A7(74) | 10 | 0 | NAb |
| SFV4 | 10 | 9 | 5.2 (0.2) |
| rA774 | 20 | 0 | NA |
| rA774-V4nstr | 15 | 15 | 4.9 (0.3) |
| rA774-V4str | 20 | 0 | NA |
| rA774-V4nsp12 | 20 | 7 | 7.7 (0.4) |
| rA774-V4nsp34 | 20 | 19 | 6.8 (0.6) |
| rA774-V4nsp3 | 20 | 20 | 6.1 (0.3) |
| rA774-V4del | 20 | 0 | NA |
| rA774-arg | 20 | 1 | 6.0 (0.0) |
| V4-opal | 20 | 3 | 9.7 (0.3) |
Adult BALB/c AnNHsd mice were inoculated i.p. with 106 PFU of virus in 100 μl of PBS.
NA, not applicable.
The stability of the opal codon in A7(74) and rA774 might be explained by lack of replication advantage of any sense mutation, as observed for SIN, in which sense mutants of opal displayed a lower RNA synthesis rate than the opal (23). Although introduction of arginine at the opal site did not markedly increase lethality, the observed paralytic symptoms indicate a considerable increase in virulence, which suggests a significant role for opal as an attenuating factor in rA774.
Replication of virus in blood and brain.
Generally, mice infected with SFV chimeras or mutants constructed in this study developed plasma viremia at day 1 postinfection (Fig. 2). No virus could be detected in the blood after 2 days in rA774-V4str- or V4-opal-infected mice, whereas rA774 and rA774-V4nstr had been cleared by day 4. In accordance with the mortality studies, the virulent clones SFV4, rA774-V4nstr, rA774-V4nsp34, and rA774-V4nsp3 replicated to high titers in the brain (108 to 109 PFU/g of brain tissue), whereas the avirulent A7(74), rA774, rA774-V4str, rA774-arg, and rA774-V4del reached moderate values (103 to 107 PFU/g of brain tissue). We frequently observed no virus in the brains of mice infected with rA774-V4str and V4-opal. In a recent study in our laboratory (36) and that of Grieder et al. (12), similar observations were made for alphavirus E2 in vitro mutants. This phenomenon could be explained, for example, by a reduced capacity of the virus to migrate to and/or penetrate the CNS, and indeed, infected mice lacking CNS pathology had developed serum antibody titers comparable to those measured in mice showing effective virus dissemination in the brain (36). It is of interest that while the majority of the V4-opal-infected mice displayed virus titers in the brain lower than those of A7(74)-infected mice, with little or no virus detected in histology, the two mice displaying high virus titers (108 to 109 PFU/g) (see above) at day 7 were those infected with arginine and tryptophan revertants.
Histochemical analysis.
In general, the degree of virulence of the SFV chimeras and mutants, expressed as percent survival (Fig. 3), correlated well with the spreading and distribution of the virus within the CNS, as demonstrated by the histochemical analysis (Fig. 4). Like the plaque-purified parental A7(74), the rA774 clone was unable to efficiently disseminate in brain tissue, forming only a few scattered foci, mainly in the perivascular areas (Fig. 4A). In contrast, the virulent rA774-V4nstr virus effectively spread along the cerebellar Purkinje cell dendrites, occupying practically the entire brain (Fig. 4B), similar to SFV4 (41). Of mice infected with the reciprocal rA774-V4str, only one was virus antigen positive (not shown). The rA774-V4nsp3 chimera showed no restriction in dissemination within the spinal cord and brain, and abundant virus was found in the cerebellum, cortex, and pons areas (Fig. 4C). In contrast, only a few foci were detected in the brains of rA774-V4del-infected animals (Fig. 4D), while in rA774-arg-infected brains (Fig. 4E), the virus load was markedly higher, indicating improved spread.
In V4-opal infection, virus spread in the neural tissue of the clinically asymptomatic animals was generally extremely limited (not shown), comparable to rA774, except for one asymptomatic mouse which at day 7 postinfection displayed a high virus load in the cortex and pons as well as in the periventricular areas but with no virus detected in the cerebellum (not shown). Both moribund mice sampled in this group were shown to be infected with opal revertants, and they displayed extensive virus spreading. The arginine revertant shown in Fig. 4F invaded the cortex and cerebrum. We also undertook histological studies on one affected mouse from the mortality study group, indicated with an asterisk in Fig. 3, which displayed left hind limb paralysis although otherwise clinically healthy. Severe myelin destruction was manifested, with numerous macrophages in the spinal cord, both phenomena similar to those observed during Wallerian degeneration (not shown).
Analysis of RNA and protein synthesis in cultured cells.
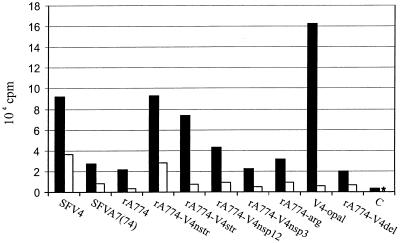
Viral RNA production was measured by infecting confluent monolayers of cultured BHK-21 cells and 72-h-cultured primary rat cerebellar granule neurons at an MOI of 20 with virus chimeras. [5,6-3H]uridine was used to label the RNA synthesized in the presence of actinomycin D (see Materials and Methods), and TCA-precipitated total RNA was analyzed by liquid scintillation counting. High relative RNA amounts were synthesized in BHK-21 cells infected with virulent SFV4, rA774-V4nsp12, and rA774-V4nstr, while avirulent A7(74), rA774, and rA774-V4del showed decreased expression (Fig. 5). However, elevated levels of viral RNA were also observed for the avirulent rA774-V4str and V4-opal mutants, while unexpectedly low amounts were synthesized by the virulent rA774-V4nsp3, which on the other hand produced nsP3 and nsP4 proteins in quantities comparable to those produced by SFV4 and rA774-V4nstr (see Fig. 6C). Moderate levels of RNA expression were obtained with the rA774-arg mutant. In neurons, only virulent SFV4 and rA774-V4nstr infections produced viral RNA amounts clearly above those seen in infections with the avirulent chimeras. These results show that virulence does not in all instances correlate with the amount of viral RNA.
FIG. 5.
Viral RNA synthesis during early infection measured by scintillation counting. BHK-21 monolayers (solid bars) and 72-h-cultured rat cerebellar granule neurons (open bars) were infected with parental strains and recombinants at 20 PFU per cell. [3H]uridine was added at 3 h postinfection, and cells were lysed 3 h later in 10% SDS. Actinomycin D was used to inhibit cellular RNA synthesis. Control cells (C) were uninfected. The star indicates a value below 500 cpm.
Given the phenotypic differences between the distinct SFV nsp chimeras, we sought to study the synthesis of nsP3 and nsP4 as well as the expression of the viral structural proteins during infection. Generally, the expression of the nonstructural proteins by the virus chimera in cultured rat cerebellar neurons was much lower than that measured in BHK-21 cells. As the amounts of nsP4 were extremely low and the antibody showed binding to cellular proteins, the estimation of nsP4 amounts was somewhat hampered. In cultured neurons, the virulent chimeras produced higher amounts of nsP3 and nsP4 than the avirulent clones, as shown by immunodetection (Fig. 6A), which is in accordance with the titers obtained in mouse brains (Fig. 2). However, plaque-purified A7(74) expressed nsP3 amounts similar to those expressed by rA774-V4nsp12 and rA774-V4nsp3. The same was generally true for BHK-21 cells, although here V4-opal also showed elevated levels (Fig. 6C). These results show that virulence does not in all instances correlate with protein amounts, and it is possible that, e.g., temporal differences in the viral protein levels are of importance. There was a tendency, however, for the chimeras with an arginine codon at the opal position in nsp3, including the avirulent rA774-arg mutant, to express higher amounts of both nsP34 and nsP4 in neurons (Fig. 6A). In BHK-21 cells, SFV4, rA774-V4nstr, and rA774-V4nsp3 showed clearly increased nsP4 expression compared with the apathogenic virus chimeras. In cultured rat neurons, the expression levels of the viral structural proteins were similar at 8 h postinfection, but differences became more pronounced at 24 h after infection, SFV4 and rA774-V4nstr showing clearly higher relative amounts compared with virulent rA774-V4nsp3 and rA774-V4nsp12 and the apathogenic chimeras (Fig. 6B).
DISCUSSION
In several alphaviruses, mutations in the glycoprotein genes alter virulence (6, 11, 47), and mutations in both E2 and E1 may have a synergistic effect (32). Alterations in E2 protein may affect SFV virulence, as shown by our recent analysis (35), in which an SFV chimera (CME2) carrying seven of a total of eight A7(74) E2 amino acid mutations was highly attenuated, and by two other studies, in which single E2 amino acid residues contributing to attenuation were identified (9, 36). The E2 gene does not, however, represent a natural avirulence determinant of A7(74) as demonstrated by this study, showing that the entity of the structural protein-encoding genes of rA774 in association with the SFV4 replicase complex in the rA774-V4nstr chimeric virus are capable of composing a highly virulent virus. The avirulent phenotype of the SFV4-derived CME2 (35) could derive from a functional failure of the E1-E2 complex with E2 originating from rA774. Amino acid mutations in this chimeric spike complex could prevent proper structure formation.
The complete nonstructural genome sequence of rA774 determined in this study revealed a large number of nucleotide mutations compared with SFV4, which often led to changes in the predicted amino acid sequence. The most notable differences were those detected in the nsp3 gene of rA774, of which the in-frame deletion of 21 nt in the nonconserved gene region abolished seven amino acid residues, five of which are hydrophobic. Although the role and interactions of nsP3 in alphavirus replication remain unclear, its role as a docking protein stabilizing the active replicase complex on cytopathic vacuoles has been suggested, and it has been shown to accumulate on cellular vacuole surfaces (28, 29). Thus, it was possible that this particular domain with the deletion could be involved in forming the putative contact surface for cytopathic vacuoles, and hence changes at this site that reduce hydrophobicity might also reduce the amount of vesicle-bound nsP3, thus lowering RNA synthesis efficiency. However, the fact that introduction into rA774 of the deleted residues comprising the heptapeptide GIADLAA did not increase virulence excluded this possibility and rather confirmed previous observations that the nonconserved C-terminal domain of alphavirus nsP3 allows remarkable changes in both length and sequence without affecting the phenotype (20).
The opal codon in nsp3 of rA774 was an unexpected finding, because the SFV4 and CA7 strains (46) both express an arginine codon at this site. Although most alphaviruses express an opal codon (16, 42, 43, 48), its significance in the regulation of alphavirus pathogenesis remains elusive. Such human pathogens as the Venezuelan (16) and eastern (48) encephalitis viruses, which cause severe disease, contain an opal codon at this position, indicating its selective advantage and stability. In vitro, using SIN RNA, readthrough of the opal codon has been estimated to occur with about 20% of the efficiency of that of a sense mutant (7), and in another study, mutants with sense replacements showed a delay in the appearance of nsP3 and reduced levels of this protein (23). Also, in the same report, in cell cultures SIN RNA synthesis was rather reduced in mutants with in vitro replacement of the opal with different sense codons. This was ascribed to the increased turnover rate of the nsP4 protein.
The present study shows that in SFV, the opal site in nsp3 affects virus pathogenesis in mice. The introduction of an opal codon dramatically attenuated SFV4, making it nearly avirulent for adult mice, while replacement of the opal codon in rA774 with an arginine clearly increased pathogenicity even if it was not sufficient to restore full virulence. While SFV4, rA774-V4nstr, and rA774-V4nsp3 all caused neurological symptoms within a short period before the mice succumbed, the rA774-arg mutant, which killed only one mouse, nevertheless frequently caused transient paralysis and limb weakness, thus exhibiting a medium degree of virulence not observed for rA774.
Generally, the viral RNA synthesis rate correlated well with the degree of virulence, but the increased pathogenicity may not always necessitate an increase in RNA synthesis, as indicated by the fact that the viral RNA levels in rA774-V4nsp3-infected neurons were much lower than those obtained with mutants of similar virulence. Also, the levels of RNA in rA774-arg-infected hamster cells were comparable to those found in asymptomatic A7(74) infection, but much lower than those produced by the completely avirulent rA774-V4str chimera. It is interesting that in neurons, the natural host cells of SFV, rA774-V4str produced much lower amounts of RNA than it did in hamster kidney cells, indicating cell type-specific regulation and control as well.
Virulence correlated poorly with the levels of the viral structural proteins in neuronal cell cultures, as of the pathogenic clones, only SFV4 and rA774-V4nstr produced remarkably increased amounts, while the lethal rA774-V4nsp3 showed glycoprotein levels similar to those with A7(74) (Fig. 6B). In contrast, expression of nsP4 in hamster cells was in accordance with the degree of pathogenicity, with levels clearly elevated for the lethal mutants and significantly increased for rA774-arg, which had moderate virulence (Fig. 6C).
As demonstrated by the pathogenicity tests using the rA774-V4nsp34 and rA774-V4nsp3 chimeras, nsP3 of SFV4 alone was sufficient to mediate lethal neurovirulence, showing that the two amino acid differences found in nsP4 do not significantly alter virus phenotype. However, it cannot be excluded that if used to replace the corresponding rA774 gene, nsp4 of SFV4 could alter virulence. Like the nsP4 of SFV4 and other alphaviruses, the nsP4 of rA774 displayed an N-terminal tyrosine, the only amino acid residue at this site that allows full replication efficiency (39). The significantly increased virulence observed for the rA774-V4nsp12 chimera compared with parental rA774 indicated that the SFV4 nsp12 locus partially compensates for the attenuation deriving from nsp3. Although the contribution to pathogenicity of the nsp1 gene was not separately tested, it probably is involved, because in a reciprocal analysis, an SFV4 derivative expressing nsp1 of rA774 displayed a remarkably attenuated phenotype in adult mice (unpublished data). This does not, however, exclude the involvement of nsp2 as well, because a few amino acid mutations were found in this region (Table 1). These results fit into the present view of alphavirus replication that nsP1 participates in forming a vacuole-docking unit of the replicase complex together with nsP3 (30), and for such vacuoles, the membrane affinity of the replicase multiprotein complex can be assumed to determine its surface concentration, a parameter potentially affecting virus replication efficiency.
Previous analyses of the structural genome of the SFV derivatives rA774 (34, 35) and CA7 (9) suggested that the two strains are nearly identical, as only four amino acid differences were detected. The present analysis of the nonstructural genome, however, clearly established that these are two essentially different SFV strains, although they both derive from the original Bradish isolate (4). The nsp regions of these two strains showed major differences in the sequence per se; also, whereas in rA774 the avirulence was primarily mediated by nsp3, with less pronounced contributions by nsp1 or nsp2 and with the structural genome being fully functional, in CA7 the attenuating domains were found distributed in both structural and nonstructural parts of the genome (46). However, in contrast to rA774, the nsp3-nsp4 region of CA7 was not found to contain major attenuating determinants. Hence, these strains should not be considered equivalent, as this might produce contradictory results because the causative mutations reside in structures with fundamentally different functions in viral replication.
Our analysis has shown that the genetic loci responsible for the natural avirulence of SFV A7(74) reside entirely in the nonstructural genome and that they mainly derive from changes in the nsp3 gene, with a significant contribution by the opal mutation, not found in SFV4, whereas the 21-nt deletion at the nonconserved carboxy terminus does not affect virus phenotype. Further analyses are required to assess the role of the single nsP3 amino acid mutations in the regulation of SFV replication.
ACKNOWLEDGMENTS
This work was supported by the Sigrid Juselius Foundation, the Finnish MS-Foundation, and the Åbo Akademi University.
We thank Michael Courtney for help with the rat cerebellar granule neurons and for revising the English and Petri Auvinen for critical reading of the manuscript. We also thank Seija Lindqvist for excellent animal care, Marja Aaltonen for help in plaque titration, Hanna Laurén for prA774-V4nsp34 construction, and Jaakko Liippo for experienced preparation of the photographs. We are grateful to Leevi Kääriäinen for providing the nsP antibodies. Thanks are also due to Gregory Atkins and Harry Kujari for helpful discussions.
REFERENCES
- 1.Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor S, Scallan M F, Morris M M, Dyson H, Fazakerley J K. Role of immune responses in protection and pathogenesis during Semliki Forest virus encephalitis. J Gen Virol. 1996;77:281–291. doi: 10.1099/0022-1317-77-2-281. [DOI] [PubMed] [Google Scholar]
- 3.Atkins G J, Sheahan B J, Mooney D A. Pathogenicity of Semliki Forest virus for the rat central nervous system and primary rat neural cell cultures: possible implications for the pathogenesis of multiple sclerosis. Neuropathol Appl Neurobiol. 1990;16:57–68. doi: 10.1111/j.1365-2990.1990.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradish C J, Allaner K, Maber H B. The virulence of original and derived strains of Semliki Forest virus for mice, guinea-pigs and rabbits. J Gen Virol. 1971;12:141–162. doi: 10.1099/0022-1317-12-2-141. [DOI] [PubMed] [Google Scholar]
- 5.Courtney M J, Lambert J J, Nicholls D G. The interactions between plasma membrane depolarization and glutamate receptor activation in the regulation of cytoplasmic free calcium in cultured cerebellar granule cells. J Neurosci. 1990;10:3873–3879. doi: 10.1523/JNEUROSCI.10-12-03873.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis N L, Powell N, Greenwald G F, Willis L V, Johnson B J B, Smith J F, Johnston R E. Attenuating mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus: construction of single and multiple mutants in a full-length cDNA clone. Virology. 1991;183:20–31. doi: 10.1016/0042-6822(91)90114-q. [DOI] [PubMed] [Google Scholar]
- 7.de Groot R J, Hardy W R, Shirako Y, Strauss J H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase: evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fazakerley J K, Pathak S, Scallan M, Amor S, Dyson H. Replication of the A7(74) strain of Semliki Forest virus is restricted in neurons. Virology. 1993;195:627–637. doi: 10.1006/viro.1993.1414. [DOI] [PubMed] [Google Scholar]
- 9.Glasgow G M, Killen H M, Liljeström P, Sheahan B J, Atkins G J. A single amino acid change in the E2 spike protein of a virulent strain of Semliki Forest virus attenuates pathogenicity. J Gen Virol. 1994;75:663–668. doi: 10.1099/0022-1317-75-3-663. [DOI] [PubMed] [Google Scholar]
- 10.Glasgow G M, McGee M M, Sheahan B J, Atkins G J. Death mechanisms in cultured cells infected by Semliki Forest virus. J Gen Virol. 1997;78:1559–1563. doi: 10.1099/0022-1317-78-7-1559. [DOI] [PubMed] [Google Scholar]
- 11.Glasgow G M, Sheahan B J, Atkins G J, Wahlberg J M, Salminen A, Liljeström P. Two mutations in the envelope glycoprotein E2 of Semliki Forest virus affecting the maturation and entry patterns of the virus alter pathogenicity for mice. Virology. 1991;185:741–748. doi: 10.1016/0042-6822(91)90545-m. [DOI] [PubMed] [Google Scholar]
- 12.Grieder F B, Davis N L, Aronson J F, Charles P C, Sellon D C, Suzuki K, Johnston R E. Specific restrictions in the progression of Venezuelan equine encephalitis virus-induced disease resulting from single amino acid changes in the glycoproteins. Virology. 1995;206:994–1006. doi: 10.1006/viro.1995.1022. [DOI] [PubMed] [Google Scholar]
- 13.Haseloff J, Goelet P, Zimmern D, Ahlquist R, Dasgupta R, Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci USA. 1984;81:4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kääriäinen L, Söderlund H. Structure and replication of alphaviruses. Methods Enzymol. 1978;82:15–69. doi: 10.1007/978-3-642-46388-4_2. [DOI] [PubMed] [Google Scholar]
- 15.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinney R M, Johnson B J, Welch J B, Tsuchiya K R, Trent D W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170:19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 18.Laakkonen P, Hyvönen M, Peränen J, Kääriäinen L. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J Virol. 1994;68:7418–7425. doi: 10.1128/jvi.68.11.7418-7425.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanciotti R S, Ludwig M L, Rwaguma E B, Lutwama J J, Kram T M, Karabatsos N, Cropp B C, Miller B R. Emergence of epidemic O'nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus. Virology. 1998;252:258–268. doi: 10.1006/viro.1998.9437. [DOI] [PubMed] [Google Scholar]
- 20.LaStarza M W, Grakoui A, Rice C M. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsp3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology. 1994;202:224–232. doi: 10.1006/viro.1994.1338. [DOI] [PubMed] [Google Scholar]
- 21.LaStarza M W, Lemm J A, Rice C M. Genetic analysis of the nsp3 region of Sindbis virus: evidence for roles in minus-strand and subgenomic RNA synthesis. J Virol. 1994;68:5781–5791. doi: 10.1128/jvi.68.9.5781-5791.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemm J A, Durbin R K, Stollar V, Rice C M. Mutations which alter the level or structure of nsp4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64:3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G P, Rice C M. Mutagenesis of the in-frame opal termination codon preceding nsp4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez S, Bell J R, Strauss E G, Strauss J H. The nonstructural proteins of Sindbis virus as studied with an antibody specific for the C terminus of the nonstructural readthrough polyprotein. Virology. 1985;141:235–247. doi: 10.1016/0042-6822(85)90254-5. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh B M, Brookworth C, Kokernot R H. Isolation of Semliki Forest virus from Aedes (Aedimorphus) argenteopunctatus (Theobald) collected in Portuguese East Africa. Trans R Soc Trop Med Hyg. 1961;55:192–198. doi: 10.1016/0035-9203(61)90025-6. [DOI] [PubMed] [Google Scholar]
- 27.Oliver K R, Scallan M F, Dyson H, Fazakerley J K. Susceptibility to a neurotropic virus and its changing distribution in the developing brain is a function of CNS maturity. J Neurovirol. 1997;3:38–48. doi: 10.3109/13550289709015791. [DOI] [PubMed] [Google Scholar]
- 28.Peränen J. Localization and phosphorylation of Semliki Forest virus non-structural protein nsp3 expressed in COS cells from a cloned cDNA. J Gen Virol. 1991;72:195–199. doi: 10.1099/0022-1317-72-1-195. [DOI] [PubMed] [Google Scholar]
- 29.Peränen J, Kääriäinen L. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J Virol. 1991;65:1623–1627. doi: 10.1128/jvi.65.3.1623-1627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peränen J, Laakkonen P, Hyvönen M, Kääriäinen L. The alphavirus replicase protein nsp1 is membrane associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 31.Peränen J, Takkinen K, Kalkkinen N, Kääriäinen L. Semliki Forest virus-specific non-structural protein nsp3 is a phosphoprotein. J Gen Virol. 1988;69:2165–2178. doi: 10.1099/0022-1317-69-9-2165. [DOI] [PubMed] [Google Scholar]
- 32.Polo J M, Johnston R E. Attenuating mutations in glycoproteins E1 and E2 of Sindbis virus produce a highly attenuated strain when combined in vitro. J Virol. 1990;64:4438–4444. doi: 10.1128/jvi.64.9.4438-4444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikkonen M, Peränen J, Kääiäinen L. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsp2. J Virol. 1994;68:5804–5810. doi: 10.1128/jvi.68.9.5804-5810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santagati M G, Itäranta P V, Koskimies P R, Määttä J A, Salmi A A, Hinkkanen A E. Multiple repeating motifs are found in the 3′-terminal nontranslated region of Semliki Forest virus A7 variant genome. J Gen Virol. 1994;75:1499–1504. doi: 10.1099/0022-1317-75-6-1499. [DOI] [PubMed] [Google Scholar]
- 35.Santagati M G, Määttä J A, Itäranta P V, Salmi A A, Hinkkanen A E. The Semliki Forest E2 gene as a virulence determinant. J Gen Virol. 1995;76:47–52. doi: 10.1099/0022-1317-76-1-47. [DOI] [PubMed] [Google Scholar]
- 36.Santagati M G, Määttä J A, Röyttä M, Salmi A A, Hinkkanen A E. The role in pathogenesis of Semliki Forest virus 3′-nontranslated region chimera and E2 envelope glycoprotein gene mutations. Virology. 1998;243:66–77. doi: 10.1006/viro.1998.9031. [DOI] [PubMed] [Google Scholar]
- 37.Scallan M F, Allsopp T E, Fazakerley J K. bcl-2 acts early to restrict Semliki Forest virus replication and delays virus-induced programmed cell death. J Virol. 1997;71:1583–1590. doi: 10.1128/jvi.71.2.1583-1590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsp4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirako Y, Strauss J H. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J Virol. 1998;72:2310–2315. doi: 10.1128/jvi.72.3.2310-2315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smithburn K C, Haddow A J. Semliki Forest virus. I. Isolation and pathogenic properties. J Immunol. 1944;49:141–157. [Google Scholar]
- 41.Smyth J M B, Sheahan B J, Atkins G J. Multiplication of virulent and demyelinating Semliki Forest virus in the mouse central nervous system: consequences in BALB/c and SJL mice. J Gen Virol. 1990;71:2575–2583. doi: 10.1099/0022-1317-71-11-2575. [DOI] [PubMed] [Google Scholar]
- 42.Strauss E G, Levinson R, Rice C M, Dalrymple J, Strauss J H. Nonstructural proteins nsp3 and nsp4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology. 1988;164:265–274. doi: 10.1016/0042-6822(88)90644-7. [DOI] [PubMed] [Google Scholar]
- 43.Strauss E, Rice C M, Strauss J H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci USA. 1983;80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986;14:5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takkinen K, Peränen J, Kääriäinen L. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein. J Gen Virol. 1991;72:1627–1633. doi: 10.1099/0022-1317-72-7-1627. [DOI] [PubMed] [Google Scholar]
- 46.Tarbatt, C. J., G. M. Glasgow, D. A. Mooney, B. J. Sheahan, and G. J. Atkins. Sequence analysis of the avirulent, demyelinating A7 strain of Semliki Forest virus. J. Gen. Virol. 78:1551–1557. [DOI] [PubMed]
- 47.Tucker P D, Griffin D. Mechanism of altered Sindbis virus neurovirulence associated with a single amino acid change in the E2 glycoprotein. J Virol. 1991;65:1551–1557. doi: 10.1128/jvi.65.3.1551-1557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver S C, Hagenbaugh A, Bellew L A, Netesov S V, Volchkov V E, Chang G J, Clarke D K, Gousset L, Scott T W, Trent D W, Holland J J. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1993;197:375–390. doi: 10.1006/viro.1993.1599. [DOI] [PubMed] [Google Scholar]