Abstract
Lasso peptides are a class of ribosomally synthesized and post-translationally modified peptides (RiPPs) defined by a macrolactam linkage between the N-terminus and the side chain of an internal Asp or Glu residue. Instead of adopting a branched-cyclic conformation, lasso peptides are “threaded”, with the C-terminal tail passing through the macrocycle to present a kinetically trapped rotaxane conformation. The availability of enhanced bioinformatics methods has led to a significant increase in the number of secondary modifications found on lasso peptides. To uncover new ancillary modifications in a targeted manner, a bioinformatic strategy was developed to discover lasso peptides with modifications to tryptophan. This effort identified numerous putative lasso peptide biosynthetic gene clusters with core regions of the precursor peptides enriched in tryptophan. Parsing of these Trp-rich biosynthetic gene clusters uncovered several putative ancillary modifying enzymes, including halogenases and dimethylallyltransferases expected to act upon Trp. Characterization of two gene products yielded a lasso peptide with two 5-Cl-Trp modifications (chlorolassin) and another bearing 5-dimethylallyl-Trp and 2,3-didehydro-Tyr modifications (wygwalassin). Bioinformatic analysis of the requisite halogenase and dimethylallyltransferase revealed numerous other putative Trp-modified lasso peptides that remain uncharacterized. We anticipate the Trp-centric strategy reported herein may be useful in discovering ancillary modifications for other RiPP classes and, more generally, guide the functional prediction of enzymes that act on specific amino acids.
Graphical Abstract

Introduction
Lasso peptides are a class of natural product defined by a unique rotaxane (lariat) structure, which is formed by a macrolactam between the N-terminal amine and the side-chain carboxylate of a downstream aspartic/glutamic acid, with the remaining C-terminal tail threaded through the macrocycle.1 Lasso peptides have a wide variety of reported biological activities, and their lariat conformation imparts traits unusual for polypeptides, including enhanced stability towards heat and proteolytic degradation.2,3 Biosynthetic studies of lasso peptides have identified the core genes required for assembly: the precursor peptide, which is comprised of leader and core regions; a RiPP recognition element (RRE) responsible for leader-peptide mediated substrate recognition; a leader peptidase; and an ATP-dependent lasso cyclase that catalyzes macrolactam and lariat formation.1,2,4,5 Beyond the class-defining threaded macrolactam modification, a variety of secondary modifications have been reported in lasso peptides, extending the chemical diversity beyond the common proteinogenic amino acids.6
Like other natural products, the earliest known lasso peptides were discovered serendipitously through phenotypic screens; however, the increase in publicly available genome sequences and increasingly sophisticated bioinformatic tools have enhanced prediction and targeted discovery of new natural products.7–9 Lasso peptides, and prokaryotic RiPPs more generally, are attractive genome mining targets due to the genomically encoded nature of the precursor peptides, rendering gene-to-molecule prediction more straightforward for RiPPs compared to other natural products.10–12 Automated tools, such as Rapid ORF Description and Evaluation Online (RODEO), have enabled high-throughput and high-confidence prediction of lasso peptides and other RiPP classes.13–17 Although most secondary modifications of lasso peptides were discovered serendipitously, recent examples have been discovered bioinformatically, such as Arg deimination (citrulassin),18 aspartimide formation (cellulonodin-2/lihuanodin),19 as well as biaryl crosslinking and Met S-methylation (nocapeptin A/longipeptin A).20
Recent bioinformatic surveys to catalog lasso peptides from sequenced genomes have revealed thousands of uncharacterized lasso peptide biosynthetic gene clusters (BGCs).4,21,22 With this wealth of untapped biosynthetic potential, the characterization of new compounds remains rate-limiting, necessitating prioritization of which BGCs are worthy of experimental investigation. We reasoned that lasso peptides with new types of secondary modifications could justify an isolation campaign, especially if the discovery method could be more broadly applied. In devising a method for discovering novel ancillary modifications, we were drawn to the amino acid tryptophan. Despite being one of the rarest amino acids in proteins (~1.4%),21,23 the repertoire of Trp modifications found in RiPP and non-RiPP natural products is rich, spanning amino acid crosslinks, C-C and C-heteroatom linkages, hydroxylation, epimerization, halogenation, and prenylation (Figure S1).1 Comparatively few Trp modifications have been characterized in lasso peptides, which currently include RES-701–4 (C-terminal 7-hydroxytryptophan),24,25 MS-271 (C-terminal D-Trp),26,27 and the nocapeptins/longipeptins (C-N biaryl linkage of Trp with another Trp/Tyr).20 Given this gap, we reasoned that a substrate-informed search based on Trp would be a fruitful strategy for finding new tailoring chemistry for lasso peptides. As a proof of concept, we bioinformatically collected a set of lasso peptides enriched in Trp and queried the dataset for genes with predicted Trp-modifying activity. Using this method, we discovered and characterized two new lasso peptides endowed with three new ancillary tailorings unprecedented in lasso peptide biosynthesis.
Materials and Methods
Genome Mining for Lasso Peptides with Trp-Rich Core Sequences.
Confirmed lasso leader peptidase sequences (protein family PF13471) from a diverse selection of bacterial phyla were selected as input for Position-Specific Iterated (PSI)-BLAST queries, according to Table S1. Results from all PSI-BLAST28 searches were combined and redundant sequences were removed for a total of 11,781 putative lasso peptidases (as of August 2023). All sequences were used as input for Rapid ORF Detection and Evaluation Online (RODEO, https://webtool.ripp.rodeo) ,29 with the default settings for open-reading frame (ORF) retrieval (±5 ORFs of the query), RiPP precursor Recognition Element (RRE) Finder scoring,30 and lasso peptide precursor scoring. The resulting RODEO co-occurrence output (Supplemental Dataset 1) was used to identify complete lasso peptide biosynthetic gene clusters (BGCs), which minimally require a locally encoded lasso cyclase (members of protein family PF00733)31 and a lasso precursor peptide. Loci that excluded either the lasso cyclase or a high-confidence precursor peptide (≥12 RODEO precursor score) were removed from the dataset. The resulting set of 4,797 leader peptidase sequences was used as input to generate a sequence similarity network (SSN) using the Enzyme Function Initiative-Enzyme Similarity Tool (https://efi.igb.illinois.edu/efi-est/).32 SSNs were visualized at RepNode100, meaning proteins sharing 100% sequence identity were conflated into a single node, and at an alignment score of 25 using Cytoscape, corresponding to clusters of proteins sharing ~40% identity or higher.33 RODEO output was used to annotate SSNs with co-occurring protein domains and attributes related to predicted precursor peptides. The custom node attribute “percent Trp in core” was calculated as the number of Trp in the core region divided by the number of total core residues x 100. The list of leader peptidases used as RODEO input are provided (Supplemental Dataset 2).
Growth and Extraction of Bacterial Cultures.
Actinomycete starter cultures (5 mL) were inoculated in liquid ATCC 172 growth medium (20 g/L soluble starch, 10 g/L glucose, 5 g/L yeast extract, 5 g/L N-Z amine, 1 g/L calcium carbonate) at 30 °C on a tube roller for 2–4 d. If necessary, glass beads were used to disrupt mycelium aggregates before the inoculation of plates. Approximately 500 μL of the starter culture was used to inoculate 10 cm agar plates (25 mL solid medium per plate) of ISP2 (4 g/L yeast extract, 10 g/L malt extract, 4 g/L glucose, 15 g/L agar, pH 7.2), GUBC (6.25 g/L glycerol, 10 g/L sucrose, 10 g/L beef extract, 5 g/L casamino acids [Bacto], 20 mM Na2HPO4-KH2PO4, and 10 mL of a solution of Balch’s vitamins [described below], 15 g/L agar, pH 7.2), CFood (25 g/L glucose, 15 g/L fish meal [Dr. Earth], 2 g/L yeast extract, 4 g/L calcium carbonate, 15 g/L agar, pH 7.2), V8 (200 mL/L V8 vegetable juice, 3 g/L calcium carbonate, 15 g/L agar, pH 7.2), MS (20 g/L mannitol, 20 g/L soya flour [Kinako], 10 mM magnesium chloride, 15 g/L agar, pH 7.2) and ISP4 (10 g/L soluble starch, 1 g/L potassium phosphate dibasic, 1 g/L magnesium sulfate heptahydrate, 1 g/L sodium chloride, 2 g/L ammonium sulfate, 2 g/L calcium carbonate, 1 mg iron (II) sulfate heptahydrate, 1 mg zinc sulfate heptahydrate, 1 mg manganese (II) chloride, 15 g/L agar, pH 7.2) media. To the media specified was added Balch’s vitamins34, which consists of 2 mg biotin, 2 mg folic acid, 10 mg pyridoxine hydrochloride, 5 mg thiamine hydrochloride, 5 mg riboflavin, 5 mg nicotinic acid, and 5 mg DL-calcium pantothenate in 100 mL deionized water. Agar plates were grown at 30 °C for 10 d. Cell mass and the top layer of agar were scraped from the plate with a sterile razor blade and placed into a 1.5 mL microfuge tube. The cell mass was extracted with HPLC-grade methanol at 25 °C for 2 h with occasional shaking. Solid material was removed by centrifugation (17,000 × g, 30 min) followed by the removal of the liquid extract. Extracts were typically screened from recently harvested cultures (<24 h) and without concentration. In cases where the signal intensity from matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was low, extracts were concentrated under reduced pressure, reconstituted in water, and desalted using a C18 ZipTip (EMD Millipore) according to manufacturer specifications, and eluted into 80% aq. acetonitrile. As needed, extracts were stored at −20 °C as a lyophilized powder.
MALDI-TOF-MS and High-Resolution Mass Spectrometry.
Bacterial extracts were subjected to MALDI-TOF-MS (Bruker Daltonics UltrafleXtreme MALDI TOF) in reflector positive using α-cyano-4-hydroxycinnamic acid (CHCA) as a matrix. Suspected ions corresponding to new lasso peptides of interest were verified with a second biological replicate to assure reproducibility. The cultures/fractions containing the lasso peptide features of interest were then desalted using a C18 ZipTip following the manufacturer’s instructions (EMD Millipore). The peptide fragmentation by high-resolution-tandem mass spectrometry (HRMS/MS) was achieved by the direct infusion onto a ThermoFisher Orbitrap Electrospray ionization mass spectrometer (ESI-MS) using an Advion TriVersa NanoMate. The MS was calibrated weekly using a calibration mixture, following manufacturer instructions (ThermoFisher), and tuned daily with Pierce LTQ Velos ESI Positive Ion Calibration Solution. Spectra were collected in profile mode with a resolution of 100,000. Ions were selected for fragmentation in the Ultra-High-Field Orbitrap Mass Analyzer using an isolation width of 5 m/z, a normalized collision energy of 35, an activation q value of 0.4, and an activation time of 30 ms. Data analysis was performed using Thermo Xcalibur software.
Native Production and Isolation of Chlorolassin.
Lentzea jiangxiensis DSM 45855 was plated from a frozen stock (25% v/v glycerol, −80 °C) onto ATCC 172 growth medium (see above) and grown at 30 °C for 2 d. Starter cultures were inoculated in liquid ATCC 172 growth medium (as above, omitting agar) and grown at 30 °C using a tube roller for 2 d. The whole starter culture was used to inoculate the larger culture of 50 mL ATCC 172 growth media in a 125 mL baffled flask and grown at 30 °C for 2 d with shaking at 220 rpm. The previous starter culture was used to inoculate the final starter culture of 250 mL ATCC172 in a 500 mL baffled flask and grown at 30 °C for 2 d with shaking at 220 rpm. Four liters of ISP2 solid medium (see above) were inoculated with the starter culture (1 mL per 15 cm plate, n = 160) and incubated for 10 d at 30 °C. The cell mass was scraped from the plates and transferred to four 50 mL falcon tubes, and each was extracted with two 30 mL portions of methanol for 1 h. The crude extract was subjected to centrifugation (4000 × g, 20 min, 4 °C), decanted, and dried to a minimal volume (~5 mL) under reduced pressure and combined with the subsequently prepared agar liquid. The agar was cubed, frozen for at least 36 h, thawed, and squeezed (filtering through cotton) to collect the bulk aqueous fraction. The squeezed agar was then extracted with 2 L of methanol for 2 h. The methanol was removed under vacuum and the remaining extract was combined with the liquid from the agar squeeze and the previously described cell extract. The combined aqueous extract was subjected to centrifugation (12,000 × g, 20 min, 4 °C) in a Sorvall RC6 Plus centrifuge with an SLA-3000 rotor and vacuum filtered through Whatman #1 filter paper using a Büchner funnel to remove dislodged solid material. The extract was loaded onto a HyperSep C18 10 g SPE column (Thermo Scientific) pre-equilibrated with 100 mL acetonitrile followed by 200 mL 50% aq. acetonitrile and finally 100 mL water before loading. After loading the unpurified extract, the column was washed with an additional 50 mL of water. The desired material was eluted with successive 50 mL portions of 10%, 20%, 30%, 40%, and 50% aq. acetonitrile, followed by 100 mL of 100% acetonitrile. Fractions were analyzed via MALDI-TOF-MS and fractions containing chlorolassin (m/z = 1817.7, [M+Na]+) were pooled and dried under reduced pressure.
Spore Preparation of Streptomyces albus.
Streptomyces albus J107435 was grown in 250 mL of ATCC 172 medium at 30 °C until high cell density was achieved (~ 3 d). Mycelia were then plated onto 10 cm petri dishes of V8 medium and allowed to grow at 30 °C for 7 d. Spores were harvested with the addition of 5 mL sterile water with 0.1% (v/v) Tween 20 and gently scraped from the cell surface with a cell spreader. The liquid-spore suspension was filtered through sterile cotton using an autoclaved syringe and harvested by centrifugation (4,000 × g, 10 min, 4 °C). After centrifugation, the supernatant was discarded, and the pellet was resuspended in 2 mL sterile water with 25% (v/v) glycerol. This mixture was then frozen in liquid nitrogen and stored at −80 °C until use.
Vector Construction for (des)-chlorolassin expression in Streptomyces albus J1074.
Integrative vector pAE4 was modified by Gibson Assembly36 to feature the strong constitutive promoter ermE*p.37 Subsequent Gibson Assembly was used to insert the chlorolassin gene cluster from the native producer (Lentzea jianxiesnsis DSM 45855) downstream of the ermE*p promoter using the primers listed in Table S2. The chlorolassin BGC was amplified from the start of the precursor (NCBI accession identifier: WP_143022795.1) to the sodium/hydrogen exchanger family protein (WP_090100301.1). The ΔchlH construct was created in the same way, but with primers designed to truncate the middle portion of the chlH gene, rendering an inactive gene product upon three component Gibson Assembly. These vectors were then transformed into conjugative Escherichia coli for insertion into S. albus spores.
E. coli - S. albus Conjugation.
E. coli conjugation strains WM602638 and WM602939 were transformed with pAE4-ermE*p constructs and selected on lysogeny broth (LB) plates containing 40 µg/mL 2,6-diaminopimelic acid (DAP) and 40 µg/mL apramycin sulfate (Apr). Colonies were selected for conjugation and inoculated in 5 mL of LB with 40 µg/mL DAP and 40 µg/mL Apr and allowed to grow at 37 °C for 16 h. Fresh LB with 40 µg/mL DAP and 40 µg/mL Apr was inoculated with 200 µL of the dense culture and incubated at 37 °C until OD600 = 0.6. Cells were then harvested by centrifugation (4,000 × g, 10 min, 4 °C) and the supernatant was discarded. The remaining cell pellet was resuspended and washed with 1 mL of 2×YT medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl). The resuspension and washing steps were repeated three additional times, after which the pellet was resuspended in 1 mL of 2×YT medium with 40 µg/mL DAP and placed on ice until used for conjugation. As the E. coli conjugation strains reached OD600 = 0.6, a 500 µL frozen suspension of S. albus J1074 spores was removed from −80 °C and heat activated at 56 °C for 15 min. Afterwards, 500 µL of 2×YT was added to the heat-activated spore stock and mixed by pipetting. Conjugation between E. coli and S. albus was initiated by aliquoting 200 µL of heat-activated S. albus J1074 spore mixture into three sterile 1.7 mL Eppendorf tubes along with 4 µL (50:1 v/v), 40 µL (5:1 v/v), and 200 µL (1:1 v/v) of E. coli. The spores and cells were mixed and then subjected to centrifugation (4,000 × g, 1 min, 25 °C) and incubated at 25 °C for 30 min. The cell pellet was then resuspended and plated on individual plates of ISP2 medium and grown at 30 °C for 16 h. After 16 h, the plates were treated with 2 mL of sterile-filtered 1 mg/mL aq. Apr solution and air-dried in a biosafety cabinet. The plates were then incubated at 30 °C until colonies appeared.
Verifying Chromosomal Insertion of Conjugative Plasmid pAE4.
Individual colonies from 1 mg/mL Apr-treated ISP2 plates were inoculated onto plates of ISP2 medium with 40 µg/mL Apr. Individual colonies were picked from the ISP2 plates and grown in 5 mL ATCC 172 medium to high density. Genomic DNA was extracted using the Qiagen DNeasy UltraClean Microbial kit and checked for insertion of ermE*p-PAD by PCR and sequencing.
Heterologous Expression and Isolation of Chlorolassin.
Streptomyces albus J1074 with the chlorolassin BGC integrated was plated from frozen stocks (25% v/v glycerol, −80 °C) onto ATCC 172 growth medium (see above) and grown at 30 °C for 2 d. Starter cultures were inoculated in liquid ATCC 172 growth medium (as above, omitting agar) and incubated at 30 °C using a tube roller for 2 d. The whole starter culture was used to inoculate the next scaleup culture of 50 mL ATCC 172 growth media in a 125 mL baffled flask and grown at 30 °C for 2 d with shaking at 220 rpm. The previous starter culture was used to inoculate the final starter culture of 250 mL ATCC172 in a 500 mL baffled flask and grown at 30 °C for 2 d with shaking at 220 rpm. Five liters of ISP2 (five 4 L baffled flasks with 1 L each) were inoculated with 250 mL starter (50 mL per baffled flask) and incubated at 30 °C for 7 d with shaking at 220 rpm. The cell mass was separated from the spent media by centrifugation (11,000 × g, 20 min, 4 °C), and decanted. The cell mass was transferred to four 50 mL falcon tubes and each was extracted with two 30 mL portions of methanol for 1 h each. The crude extract was subjected to centrifugation (4000 × g, 20 min, 4 °C), decanted, and combined with the subsequently prepared crude extract. The media was frozen and lyophilized to a crude powder using a VirTis Sentry 2.0 (~2 d). The crude powder was transferred to a 1 L flask and extracted with two 100 mL portions of methanol with agitation for 15 min for each portion. The methanol was removed from the crude powder by filtering through Whatman #1 filter paper using a Büchner funnel. The mixture was then adsorbed onto 15 g of Celite 545 under reduced pressure. The sample was then purified using a Teledyne Isco Combiflash EZprep equipped with a REdiSep Rf C18 cartridge (130 g, 60 Å pore size, 40–63 µm particle size, 230–400 mesh) with a mobile phase of 10 mM aq. NH4HCO3/acetonitrile at 75 mL/min with the following method: 1 column volume hold at 10% acetonitrile, a gradient from 10–100% acetonitrile over 12 column volumes, and a hold at 100% acetonitrile for 2 column volumes. Fractions were analyzed via MALDI-TOF-MS and fractions containing chlorolassin were pooled and dried under reduced pressure.
Purification of Chlorolassin.
Crude chlorolassin was dissolved in 2 mL 10/90 acetonitrile/NH4HCO3, the coarse sediment removed by centrifugation (17,000 x g, 15 min, 4 °C) and 0.22 µm syringe filtered (BD technologies) prior to purification using a Teledyne Isco Combiflash EZprep equipped with a RediSep Prep C18 Aq column (100 Å pore size, 40–63 µm particle size, 230–400 mesh). One mL of the chlorolassin solution was injected and purified using a flow rate of 19 mL/min, 10 mM NH4HCO3 and acetonitrile as the eluents and the following method: 10% acetonitrile (isocratic, 2.5 min), 10–50% acetonitrile (gradient, 25 min), 50% acetonitrile (isocratic, 2.5 min) 100% acetonitrile (gradient, 5 min). Absorbance of the eluate was monitored at 220, 260 and 280 nm, fractions were analyzed by MALDI-TOF-MS and those containing chlorolassin were pooled and dried under reduced pressure. The resulting semi-pure chlorolassin was dissolved in 1 mL 20/80 acetonitrile/NH4HCO3 and subjected to centrifugation (17,000 x g for 5 min) to remove particulate material. Two rounds of final purification were performed on a ThermoFisher Scientific Vanquish HPLC system equipped with an Accucore C18 column (2.4 µm, 4.6 × 150 mm) and Accucore PFP (pentafluorophenyl) column (2.4 µm, 4.6 × 150 mm), respectively. The used eluents were 0.1% aq. formic acid (A) and MeOH with 0.1% formic acid (B) with the following gradients: [30% B (isocratic, 3 min), 75% B (gradient, 5 min), 80% B (gradient, 3 min), 82% B (gradient, 4 min), 84% B (gradient, 5 min), 30% B (gradient, 2 min) and 30% (isocratic, 3 min)] and [(50% B (isocratic, 3 min), 100% B (gradient, 4 min), 100% B (isocratic, 3 min), 50% B (gradient, 1 min) and 50% B (isocratic, 2 min)], respectively. The absorbance of the eluate was monitored at 220, 260 and 280 nm, fractions were analyzed by MALDI-TOF-MS and those containing chlorolassin were pooled and dried under reduced pressure. Note: an initial titer of 100 µg/L from the native host was achieved using the same purification but with trifluoroacetic acid (TFA) adjusted agar squeeze and TFA or formic acid as the eluent buffers. The difference in yield is likely due to the poor solubility of chlorolassin under acidic conditions, which was noted with the pure compound.
Purification of des-chlorolassin.
Des-chlorolassin was isolated and partially purified in the same manner as heterologously produced chlorolassin with the following modifications: 3 L of culture was inoculated with S. albus J1074 with the chlorolassin BGC (omitting the halogenase) integrated into the genome. The final purification of des-chlorolassin was performed similarly to chlorolassin with the HPLC method modified as follows: 10% acetonitrile (isocratic, 5 min), 10–50% acetonitrile (gradient, 30 min), 95% acetonitrile (gradient, 5 min) 95% acetonitrile (isocratic, 5 min).
Proteolytic Assessment of Lasso Peptide Threadedness.
Solutions of native and heterologously produced chlorolassin (1 mg/mL) were dissolved in 20/80 acetonitrile NH4HCO3 and 50 µL (50 µg) and transferred to a PCR tube (MidSci). The relevant samples were heat-treated at 95 °C for 2 h, and both heat-treated and control samples were evaporated to dryness. The samples were then reconstituted in 50 µL of one of the three solutions: 5 ng/µL endoproteinase AspN (New England Biolabs) in 10 mM Tris-HCl, pH 8; 50 ng/µL carboxypeptidase Y (Sigma) in phosphate-buffered saline; 3.3 ng/mL (at least 0.1 U) thermolysin (Millipore Sigma) in 50 mM Tris-HCl, 100 mM NaCl, pH 8 and left to react for 18 h at room temperature for carboxypeptidase Y and 37 °C for AspN and thermolysin. The samples were then desalted with a ZipTip C18 (Sigma-Aldrich), eluted with 60% aqueous acetonitrile and analyzed by MALDI-TOF-MS as described above.
NMR Methods for Chlorolassin.
For NMR structure elucidation, ~6 mg of chlorolassin was dissolved in 0.5 mL 1:1 deionized H2O and acetonitrile, d3 (Sigma-Aldrich, 99.9% atom D). A series of 1H spectra were first collected at 25, 40 and 55 °C, then a series of 2D NMR experiments, 1H-1H COSY, 1H-1H TOCSY 1H-1H NOESY. 1H-13C HSQC and 1H-13C HMBC were acquired at 40 °C. Experiments were performed on a 750 MHz Agilent VNMRS narrow-bore NMR operating at 749.375 MHz equipped with a triple resonance 1H-13C/15N Varian probe with gradient and pulse shaping capabilities. Data was recorded on a console operating VnmrJ 3.2A. Standard Varian Biopack pulse sequences were used for all acquisitions. Mixing times for experiments were set to 30 ms for 1H-1H COSY, 70 ms for 1H-1H TOCSY and 400 ms for 1H-1H NOESY. All spectra were collected with 96 increments in the indirect dimension under WET solvent suppression. All spectra were processed in MestReNova 12.0.1 and calibrated to the residual solvent signals (δH/C 1.94/1.39). Chemical shift assignments based on all 1D and 2D data are shown in Table S3.
Marfey’s Amino Acid Analysis of Chlorolassin.
Chlorolassin (300 µg) was dissolved in 200 µL 6 M DCl in D2O (prepared as: 35% w/v DCl diluted 1:2 into D2O, obtained from Aldrich and Cambridge Isotope Laboratory, respectively). The solution was supplemented with 3% (w/v) phenol and 1% (v/v) 2-mercaptoethanol. Next, the solution was transferred to a two-necked Schlenk flask and subjected to one freeze-pump-thaw cycle and heat treatment at 95 °C for 4 h under reduced pressure using a temperature-controlled heating block (Optimag-ST, Chemglass Life Sciences). The solution was then dried using a Speedvac concentrator (Savant ISS110). The resulting hydrolysate was dissolved in 150 µL 1 M NaHCO3 and mixed with 150 µL 4 mg/mL 1-fluoro-2–4-dinitrophenyl-5-L-alanine amide (FDAA, Thermo Scientific). The reaction mixtures were heated to 60 °C for 2 h (4 mg/mL FDAA) and quenched dropwise using 30 µL 6 M HCl, and used directly in subsequent analysis. Amino acid standards were prepared by drying down 200 µL of a solution containing 0.5 µmol/mL of 4-, 5-, 6- and 7-chlorotryptophan (Biosynth) and reacted with FDAA as above. Rather than purchasing a D-amino acid standard, fluoro-2–4-dinitrophenyl-5-D-alanine amide (D-FDAA, Toronto Research Chemicals) was reacted with the L amino acid standard. The resulting adduct is the enantiomer of a D-amino acid-FDAA adduct, and thus indistinguishable via HPLC on an achiral stationary phase.40 The resulting derivatized L-/D-amino acid standards and chlorolassin hydrolysates were centrifuged to remove particulates and filtered with a 0.45 µm centrifugal filter (Thermo Scientific). LC-MS analysis was performed on a Shimadzu LC-MS 2020 with a quadrupole detector and a Macherey-Nagel Nucleodor C18 column (250 × 4.6 mm, 5 µm particle size, 100 Å pore size) The samples were analyzed in batch under the following conditions: solvent A 0.1% aq. formic acid, solvent B MeCN + 0.1% formic acid, 1 mL/min flow rate, and the following gradient: 15% B for 7 min, 15–50% B over 45 min, hold at 50% B for 5 min. 20 μL of analyte was injected and Cl-Trp-Marfey’s reagent adduct was monitored by absorbance at 340 nm and by the [M+H]+ ion of the adduct (m/z = 491)
Native Production and Isolation of Wygalassins A.
Streptomyces katrae B-16271 was plated from a frozen stock (25% v/v glycerol, −80 °C) onto ATCC 172 growth medium (see above) and grown at 30 °C for 3 d, growing as hard beige colonies. Several colonies were inoculated into 5 mL of ATCC172 in 10 mL culture tubes and grown in a tube roller at 30°C for 3 d. The cultures grew as a diffuse culture that begins to turn brown around day three. The entire 5 mL of culture was used to inoculate 400 mL of ATCC172 in a 2L baffled Erlenmeyer flask. The flask was incubated at 30 °C for 4 d at 220 rpm shaking. The starter culture was used to inoculate 3 L of ATCC172 (200 mL of culture in a 1 L baffled Erlenmeyer flask or a non-baffled Erlenmeyer flask with stainless steel coils for aeration, 20 mL inoculum per flask) and grown for 7 d at 30 °C with shaking at 220 rpm. The cells were harvested by centrifugation using a Thermo Scientific Sorval Legend XTR centrifuge (4000 × g at 4 °C for 40 min). The cell-free supernatants (SN) were extracted twice using n-BuOH (1:1) while the extraction of the cell pellet was achieved using MeOH with sonication for 30 min twice to give the methanolic extract of the cells. Under reduced pressure, the n-BuOH and MeOH extracts were evaporated to afford the crude extracts (Bu-SN and Me-cells extracts). The crude extract was resuspended in methanol followed by centrifugation to remove undissolved debris before MALDI-TOF-MS and UHPLC profiling/separation.
Purification of wygwalassin A1.
Both extracts were dissolved in 5 mL MeOH separately and 100 µL of the dissolved crudes were injected over a semi-prep C18 Hypersil GOLD column (5 µm, 10 × 250 mm) using a 0.1% aq. formic acid (A) and acetonitrile with 0.1% formic acid (B). With a 5–100% gradient of B in 35 min and a flow rate of 3 mL/min, the enrichment of wygwalassins was achieved. A further isolation step was conducted using an Accucore PFP column (2.4 µm, 4.6 × 150 mm) with the former eluents and 35–43% gradient of B in 17 min and a flow rate of 1.5 mL/min. The final purification of wygwalassin A1 was achieved using an Accucore C8 column (2.6 µm, 4.6 × 150 mm) using the formerly described eluents with 15–100% gradient of B in 40 min and a flow rate of 1.5 mL/min. The UV absorbance of the eluted fractions was monitored at 220, 280 and 320 nm and the pooling of the fractions containing wygwalassin A1 was guided by MALDI-TOF-MS measurement.
NMR Methods for Wygwalassin A1.
Wygwalassin A1 (ca. 0.12 mg) was dissolved in 300 µL of CD3OH (Sigma Aldrich, 99.9% atom D) and placed in a 5 mm MeOH-matched Shigemi tube (Millipore-Sigma). A series of 2D NMR experiments, 1H-1H COSY, 1H-1H TOCSY and 1H-1H NOESY were acquired at 23 °C. Experiments were performed on a 750 MHz Agilent VNMRS narrow-bore NMR operating at 749.375 MHz equipped with a triple resonance 1H-13C/15N Varian probe with gradient and pulse shaping capabilities. Data was recorded on a console operating VnmrJ 3.2A. Standard Varian Biopack pulse sequences were used for all acquisitions. Mixing times for experiments were set to 30 ms, 60 ms and 350 ms, respectively. All spectra were collected with 96 increments in the indirect dimension under WET solvent suppression. The 1H-13C HSQC experiment was conducted at 23 °C on a Bruker Avance NEO 600 MHz spectrometer with a 5-mm prodigy BBO probe using Topspin 4.1.4 software. All spectra were processed in MestReNova 12.0.1 and calibrated to the residual solvent signals (δH/C 3.31/49). Chemical shift assignments based on all 1D and 2D data are provided (Table S4).
Solution Structure Determination for Chlorolassin and Wygwalassin A1.
The solution structures were calculated using NIH-Xplor (version 2.51) software41,42 based on the distance restraints between protons which were extracted from NOESY spectra. Raw NMR data were processed in NMRPipe43 and analyzed in Sparky44. The cross peaks in the NOESY spectra were integrated, sorted by intensity, and exported in Xplor format from Sparky, giving strong, medium and weak NOE restraints. A total of 54 or 77 NOE restraints were used in either chlorolassin or wygwalassin A1 peptide, respectively (Table S5). Because there are a few modified amino acids in the peptide sequences, the files protein-3.2.top and protein-3.2.par in the NIH-Xplor package were edited to incorporate definitions for the modified residues, including chlorine or prenyl at the 5 position of Trp, and a dehydrated Ca-Cb bond of the Tyr amino acid. The force field parameters of the modified amino acids were generated by either Chem3D software (PerkinElmer), or ACPYPE server (https://www.bio2byte.be/acpype/).45,46 An ensemble of 20 lowest energy structures from 200 calculated is reported for each peptide. The quality of the NMR structures was evaluated using PROCHECK47,48 and validated using the Protein Data Bank (PDB) web portal (https://validate-rcsb-2.wwpdb.org/validservice/).49 The root-mean-square deviation (RMSD), Ramachandran statistics and plots are given in Tables S6, S7 and Figures S18, S23, respectively. Both structure coordinates (in PDB format) were deposited to the PDB along with NOE restraints used in the calculation, and chemical shift assignments are deposited in the Biological Magnetic Resonance Data Bank (BMRB, https://bmrb.io/).50 The accession codes are listed in Tables S8.
Growth Suppression Assays.
Growth suppression activity of chlorolassin and wygwalassin A1 were tested using the broth microdilution method against the following strains: Staphylococcus aureus USA300, Enterococcus faecalis ATCC 19433, Pseudomonas aeruginosa PA01, Klebsiella pneumoniae ATCC 27736, Acinetobacter baumannii ATCC 19606, Neisseria gonorrhoeae MS11, Bacillus anthracis Sterne, and Listeria monocytogenes 4b F2365. A cation-adjusted Mueller-Hinton Broth medium that contains casein, beef extract and starch (MHB, BBL iences) was used as the growth medium for all strains. The strains were grown overnight in 5 mL of appropriate media at 37 °C to stationary phase. Cultures were diluted 500-fold using the appropriate medium and grown to an OD600 of 0.5. Cultures were diluted further by 50,000-fold to obtain ~ 1 × 105 CFU/mL and were added to a 96-well plate. Chlorolassin and wygwalassin A1 (1–128 μg/mL) were added in successive 2-fold serial dilution to the cultures. Gentamicin (1– 128 μg/mL) was used as the positive control for all strains. Negative controls lacking chlorolassin, wygwalassin A1 and antibiotics were also used. The plates were grown overnight at 37 °C with 190 rpm shaking. The MIC was determined as the lowest compound concentration preventing visible bacterial growth.
Bioinformatics Methods for Trp Halogenases.
Additional chlorolassin-like BGCs were identified from the NCBI51 non-redundant protein and UniProtKB52 databases using a combination of homology-based searches, custom hidden Markov models (HMM) searches, and co-occurrence analyses, as outlined below. First, the chlorolassin halogenase (WP_090100303.1) was used as a query for the PSI-BLAST53 search tool, using the default expect threshold of 0.05 and three rounds of iterative searches. Querying the UniProtKB by BLAST search gave a similar, but non-identical, list of homologous halogenases. BLAST searching of the chlorolassin precursor peptide did not return any homologous proteins in the NCBI non-redundant database. The combined results from BLAST searches of the two databases were combined (n = 4,797), converted to NCBI accession IDs, and used as queries for the RODEO web tool with the RRE-Finder and lasso peptide precursor prediction modules enabled. The resulting list of candidate BGCs was manually curated to exclude any entries not meeting all three of the following criteria: (i) BGCs were retained if the queried halogenase co-occurs with a gene encoding a flavin reductase within 5 ORFs. (ii) BGCs were classified as “chlorolassin-like” if the queried halogenase had neighboring genes encoding a lasso peptide cyclase (PF00733), lasso leader peptidase (PF13471), and identified putative RRE domain. RREs were recognized using the custom HMMs from the RRE-Finder model, using a bit score significance threshold of 15. (iii) The lasso peptide precursor prediction module of RODEO was employed to predict likely precursor peptides, which typically contain a YxxP/WxxP recognition motif in the leader region and are typically 40–80 amino acids in length. All predicted precursor peptides were manually examined to ensure the core region contained tryptophan. As a concurrent genome mining approach, the InterPro family for flavin-dependent halogenases (IPR006905, n ~17,000) was used as the input for SSN and genome neighborhood network generation using the EFI-EST tool. Two characterized RiPP-associated Trp halogenases not contained in this family (SrpI54 and Amm355) were manually added to the networks. The SSN was visualized at an alignment score of 90, corresponding to clusters of proteins sharing ~40% identity or higher. All SSNs are shown at RepNode 60. The list of halogenases used as RODEO input is provided (Supplemental Dataset 3). SSNs were annotated and colored based on the following halogenases registered with the SwissProt database (a smaller database of proteins with known functions):52 MibH, microbisporicin/NAI-107 halogenase (E2IHC5); ChlH, chlorolassin halogenase (NCBI: WP_090100303.1); Amm3, ammosamide halogenase; SrpI, 6-brominase of a proteusin-like RiPP; RadH, radicicol halogenase (C5H881); GsfI, griseofulvin halogenase (D7PI14); PltM, phloroglucinol halogenase (Q6JHX8); OtaD, ochratoxin halogenase (A2R6G7); PtaM, pestheic acid halogenase (A0A067XMV4); RebH, rebeccamycin halogenase (Q8KHZ8); PrnA, pyrrolnitrin halogenase(P95480); SttH, Streptomyces toxytricini halogenase(E9P162); PyrH, Streptomyces rugosporus halogenase(A4D0H5); BrvH, 3-indole halogenase (B4WBL8); CmlS, chlorampheticol halogenase (Q9AL91); PltA, pyoluteorin halogenase (Q4KCZ0); MalA, malbrancheamide halogenase (L0E155); and PrnC, monodechloroaminopyrrolnitrin 3-halogenase (P95482) In cases where enzymes have known regioselectivity for tryptophan substrates, nodes were colored to indicate selectivity. Co-occurrence analysis was performed using the Genome Neighborhood Network (GNN) tool, and halogenases with co-occurring proteins belonging to known lasso peptide PFAMs (PF00733 and PF13471) were prioritized, as above. RRE-Finder is not integrated with the EFI tools, so all proteins neighboring halogenases in a ±10 ORF window were separately run through the RRE-Finder precision mode tool at a bit score threshold of 15 to assess co-occurrence of predicted RRE domains.
Bioinformatics Methods for Trp DMAT.
Additional DMAT-containing BGCs were identified in the same manner as for the Trp halogenases detailed above. The wygwalassin DMAT protein from Streptomyces katrae NRRL B-16271 (WP_030301592.1) was used as a query for PSI-BLAST, using the default expected threshold of 0.05 and three rounds of iterative searches (n = 5,000 collected as input). Querying the UniProtKB with a BLAST search gave a list of homologous DMATs (n = 337) that were also used as input. Members of the InterPro family of the aromatic prenyltransferases, DMAT-type (IPR017795, n = 3,120) were also added. A SSN was generated from these sequences using the EFI-EST and was visualized at an alignment score of 60 and RepNode 100. The list of DMATs used as RODEO input is provided (Supplemental Dataset 4). To analyze the genomic neighborhood for putative RiPP-BGCs, the non-redundant protein accessions (n = 3,440) were submitted to RODEO with all RiPP precursor prediction modules enabled, as well as RRE-Finder at a bit score significance threshold of 15. The SSN was annotated and colored based on a selected set of characterized fungal DMATs in the SwissProt database (protein name/UniProt accession): TdiB/A7XRY3, AtmD/A9JPE1, DmaW/C5FTN3, IdtF1/J7FK08, Dma1/M2Y2F4, FgaPT2/Q50EL0, DmaW2/Q6X1E1, DmaW1/Q6X2E2, PaxD/Q9C451, IfgA/W6QIM8, EchPT1/A0A017SP50, EchPT2/A0A017SPL2, AnaPT/A1DN10, RoqD/ B6HJU1; NptA/C8VEJ5, PenG/A0A1B2CTB2, PenI/A0A1B2CTB7, CriA/A0A1E3B0T7, CirF/A0A1E3B0S1. In addition, those sequences that were associated with a putative RiPP-BGC were annotated and colored.
Results
Genome Mining of Putative Trp-Modified Lasso Peptide BGCs.
To uncover new Trp-modified lasso peptides, we first set out to bioinformatically curate putative lasso peptide BGCs that encode Trp-rich core peptides. To this end, a Position-Specific Iterative (PSI)-BLAST28 search was performed using representative high-confidence lasso leader peptidases from diverse bacterial phyla as a query. RODEO analysis was performed and lasso peptide BGCs were selected for further analysis based on the presence of a peptidase (PF13471), lasso cyclase (PF00733), and a RODEO-validated lasso precursor peptide. A sequence similarity network (SSN) was then constructed32,33 from the non-redundant leader peptidases (n = 4,797) with color-coding to depict the phylogenetic origin and the percentage of Trp in the predicted core region (Figures 1, S2, Supplemental Datasets 1-2). Since Trp is enriched in lasso peptide core sequences (3–5%) compared to the overall proteome, we chose cutoffs that represented significantly higher Trp percentages than the average lasso peptide. Core regions with >10% Trp were largely concentrated in a single group comprised of Actinomycetota.
Figure 1. Genome mining for Trp-rich lasso peptides.

Non-redundant leader peptidases (PF13471) co-occurring with a lasso cyclase and predicted lasso precursor peptides were used as input (n = 4,797). SSN is visualized as RepNode 100 at alignment score 25. Coloring indicates frequency of Trp coding in the neighboring core region. The network was created using the EFI-EST32 and visualized using Cytoscape.33
Using the co-occurrence data generated by RODEO, the SSN was next annotated using the protein families of known lasso Trp-modifying enzymes (PF09587 in Trp-epimerization, PF21780 for Trp-7-hydroxylation, and PF00067 for Trp-biaryl formation/Trp-5-hydroxylation). We observed that many enzyme families involved in Trp modification colocalized with the same SSN group containing most of the Trp-rich precursor peptides (Figure S3). Thus, we closely examined this group for new Trp modifications. Manual analysis of the co-occurring gene(s) associated with these lasso peptide BGCs with ≥10% Trp-containing core regions disclosed a diversity of secondary modifying enzymes, such as oxidases/oxidoreductases belonging to several protein families, methyltransferases, and multiple hypothetical proteins (Supplemental Dataset 1). Among them, two groups of lasso peptide BGCs stood out, putatively encoding either Trp-halogenases or Trp-dimethylallyltransferases (DMATs). Since these BGCs contained genes with high-confidence predictions for Trp modification, they were prioritized for experimental characterization.
The first prioritized lasso peptide BGC, from Lentzea jiangxiensis, was designated chl (Figure 2). In addition to canonical lasso peptide biosynthetic genes (i.e., precursor peptide, chlA; leader peptidase, chlB; RRE domain, chlE; lasso cyclase, chlC) the BGC harbors a predicted Trp-halogenase (chlH, PF04820), FAD (flavin adenine dinucleotide)-reductase (chlR, PF01613), and a sodium/proton antiporter (chlF, PF00999) (Figure 2). The predicted precursor contains three Trp in the core region and the Trp-halogenase shows high sequence identity and similarity (64% / 77%, respectively) to MibH (WP_036325874.1),56 a characterized RiPP halogenase responsible for 5-Cl-Trp installation in the lanthipeptide NAI-107 (also known as microbisporicin).56 The microbisporicin BGC also contains a FAD reductase (58% identity / 65% similarity to chlR) and antiporter homolog (42% identity / 58% similarity to chlF), suggesting these genes form a common cassette for Trp halogenation.56
Figure 2.

Discovery of chlorolassin and wygwalassin. (A) BGC diagram of chlorolassin from Lentzea jiangxiensis. Precursor peptide sequence is given with Trp in green and acceptor Asp in blue. (B) MALDI-TOF-MS of L. jianxiensis extract showing natively produced chlorolassin. Inset, zoomed view to highlight the 35Cl:37Cl isotope pattern of the dichlorinated peptide, with the theoretical distribution shown below. (C) BGC diagram of wygwalassin from Streptomyces katrae. Precursor peptide is given with Trp in magenta. (D) MALDI-TOF-MS of S. katrae extract showing natively produced wygwalassin A1. Peaks labelled A1* are presumed oxidation products.
The second group of prioritized lasso peptides was a small family of nine BGCs from the genus Streptomyces, which we termed wyg (Figure 2). The BGCs contain wygB/C/E (functions assigned above), a precursor peptide (wygA) with two Trp in the core, a tryptophan dimethylallyl transferase (Trp DMAT, wygG, PF11991), a COX15-CtaA-like heme oxygenase (wygH, PF02628), and a PhyH-like oxidase (wygF, PF05721). Taken together, the presence of these enzymes suggests Trp-prenylation and additional oxidative tailoring.
Discovery of Chlorolassin and Wygwalassin.
To obtain the products of the chl and wyg BGCs, we first screened expression conditions using the respective encoding organisms. Screening of L. jiangxiensis led to the production of m/z = 1817.7, corresponding with the predicted [M+Na]+ ion of the mature lasso peptide with two chlorinations (+68 Da). Chlorine has a 35Cl:37Cl ratio of 76:24, and the isotope distribution of m/z = 1817.7 was consistent with two chlorinations (Figure 2). These observations raised confidence in the assignment of the lasso peptide, which we named chlorolassin. High-resolution mass spectrometry (HR-MS) was consistent with the expected molecular formula, and tandem mass spectrometry (MS2) localized one chlorination to Trp12 while Trp14 was unmodified (Figures 2, S4). Although the second chlorination was expected on Trp8, it could not be confirmed by the collision-induced dissociation (CID) conditions employed.2
Similarly, expression screening of two organisms harboring the wyg BGC (Streptomyces katrae NRRL B-16271 and Streptomyces iakyrus NRRL ISP-5482) yielded masses of m/z = 1709.8 and 1707.8, termed wygwalassin A1 and B1, respectively (Figure S5). The observed ions were consistent with the mature lasso peptide with one dimethylallylation (+68 Da), and an additional loss of 2 Da, possibly from a crosslink or dehydrogenation. The S. iakyrus cultures delivered an additional variant, termed wygwalassin B2, with m/z = 1775.8. This ion was speculated to contain an additional dimethylallylation (or a single geranylation) (Figure S5). HR-MS and MS2 corroborated that these masses derived from the core regions of WygA, with the daughter ion series confirming the exocyclic sequences (Figures S6-S9). The additional modifications were localized to the macrocycle region, which contains two Trp.
Production of Chlorolassin and Wygwalassin A1.
Isolation of chlorolassin from the native producer only yielded trace quantities sufficient for MS-based analysis. To obtain enough material to elucidate the structure of chlorolassin by two-dimensional (2D) NMR spectroscopy, we devised a heterologous expression strategy. The design used an integrative plasmid (pAE4) containing the chlorolassin BGC under the control of the ermE*p37 constitutive promoter. Streptomyces albus J1074 B4 was chosen as a host, since previous work removed many non-essential BGCs that provide a cleaner background.57 In addition, a second plasmid with the Trp halogenase omitted (ΔchlH) was constructed to assess the role in chlorination and possibly obtain non-chlorinated des-chlorolassin. Comparative MS screening of the chl BGC revealed a mass identical to natively produced chlorolassin while the ΔchlH strain produced m/z 1749.5, which was interpreted as [M+Na]+ for des-chlorolassin. To structurally characterize chlorolassin, we expressed 5 L and purified 20 mg (post-HPLC yield, Figure S10) from the heterologous host. HR-MS and MS2 confirmed that the two chlorination events were consistent with those from the native producer (Figures S4, S11). In addition, an overlay of the 1H-NMR spectra of chlorolassin isolated from the native and heterologous hosts was identical (Figure S12). Heterologously produced des-chlorolassin was also analyzed by HR-MS and MS2, confirming the lack of chlorination (Figure S13).
While we were interested in isolating the wygwalassin B variants due to the potential for multiple prenylations or geranylation, low yield and batch-to-batch variability in the titers of B1 and B2 were encountered in the large-scale production from S. iakyrus. To obviate this problem, large-scale cultures of S. katrae were used to isolate wygwalassin A1. UV absorbance at 280 nm was used to guide HPLC purification, (Figure S10) which yielded a low but serviceable quantity of wygwalassin A1 (120 µg purified from 3 L culture) for 2D-NMR-based structural characterization. Upon analysis of larger scale cultures, we detected a minor congener consistent with a lasso peptide lacking prenylation but displaying the −2 Da modification, which was termed wygwalassin A0 (Figure S6). While requiring further investigation, this observation indicates that the enzyme installing the −2 Da modification does not require a prenylated substrate.
NMR Characterization.
Chlorolassin.
Structure elucidation of chlorolassin was accomplished by a suite of 2D-NMR experiments, including 1H-1H COSY, TOCSY, NOESY, 1H-13C HSQC, and HMBC. A preliminary analysis of the amide region of the 1H-NMR spectrum suggested a constrained (i.e., threaded) conformation given the wide dispersion over ≈ 2 ppm (Figure S14), which was further supported by heat and protease treatment assays (Figure S15).58 Further examination of the amide region, however, revealed more signals than anticipated based on the length and amino acid composition of chlorolassin. Two possible explanations were entertained: (i) a co-eluting minor congener or (ii) conformational isomerization occurring on the NMR time scale. Owing to reports of NMR-detectable conformers on microcin J2559 and benenodin-160, we favored the latter hypothesis.
To support or refute if the additional signals in the amide region derived from interconvertible conformers, 1H-NMR spectra were collected at 25, 40, and 55 °C, and the indolic NH signals were tracked as a function of temperature (Figure S16). Because temperature changes can affect the relative distribution of conformers, a change in indolic NH integrations suggests conformational isomerism rather than contaminating congeners. Indeed, while temperature increase did not cause convergence to one set of indole signals, the relative integrations did change, strongly suggesting conformational isomerism.61
Although a high degree of spectral overlap rendered structural elucidation challenging, a full assignment of the peptide backbone was completed (Table S3). The macrolactam linkage was confirmed by strong NOE signals between Gly1-NH and both βH of Asp9 (Figures S17-S22). In addition, 1H-1H TOCSY, 1H-13C HSQC, and HMBC resonances assisted in assembling the three Trp residues with a confident assignment of Trp8 and Trp12 as 5-Cl-Trp regioisomers (Figures S20, S23-S25) To orthogonally confirm the site of Trp chlorination, chlorolassin was hydrolyzed followed by derivatization with Marfey’s reagent (1-fluoro-2–4-dinitrophenyl-5-L-alanine amide, FDAA).62 Comparative HPLC profiling of chlorolassin hydrolysate with FDAA-derivatized standards of four Cl-Trp isomers confirmed the presence of 5-Cl-L-Trp in chlorolassin (Figure S26).
Using distance constraints derived from the long-range NOE correlations (Figure S27), a solution ensemble structure was calculated for chlorolassin (Figures 3, S28).42 The solution structure formed the expected right-handed threaded confirmation with Leu13 and Trp14 assigned as the upper and lower plugs, respectively (Figure 3). To investigate the source of the observed conformational isomerism, the cis/trans isomerization of Pro7 was investigated. While the major conformer was determined to have trans-configuration at Pro7 based on NMR chemical shifts, spectral overlap precluded determination for the minor conformer (Figure S29). More chemical elucidation would be required to resolve the nature of these conformational isomers. In addition, the two Cl-Trp residues, Trp8 and Trp12, were found in close proximity such that the plane by the two indole rings creates a V shape and both pyrrole rings point towards the base of the V in most conformations. However, roughly one-third of the calculated structures have Trp12 pointing in the opposite direction (Figure S30). Interestingly, this nearly mirrors the distribution of the two most abundant conformers seen in the 20 solution structures, though further investigation is required to assess whether this explains the observed conformational isomerism.
Figure 3.

3D structure of chlorolassin. (A) Topological diagram of chlorolassin. Gray, ring region; cyan, steric-locking residues; green, 5-Cl-Trp residues. (B) NOE-based NMR ensemble of the 20 lowest energy structures of chlorolassin (only amide backbone is shown for clarity). (C) Surface-filling model of chlorolassin, with the ring and steric-locking residues tinted gray and cyan, respectively. (D) Lowest energy structure of chlorolassin. 5-Cl-Trp8 and 12 are labeled and colored green, while Leu13 and Trp14 are cyan. Chlorolassin PDB code: 8UKC.
Wygwalassin.
Structure elucidation of wygwalassin A1 was also accomplished by a series of 2D-NMR experiments. The usage of 1H-1H COSY, TOCSY, and 1H-13C HSQC enabled the assembly of nearly all the expected spin systems: 2xTrp, 3xGly, 2xAla, 2xIle, 1xAsn, 1xAsp, 1xSer, 1xThr, 1xGlu (Figures S31-S35). Sequential connectivity of the delineated residues was facilitated by 1H-1H NOESY, including the macrolactam bridge between Trp1 and Asp8 (Figures S36-S37). Homonuclear 2D correlations assigned Trp1 as unmodified while Trp4 was found to be mono-substituted at the 5-position. In addition, the couplings discerned from 1H-1H COSY, TOCSY, and NOESY uncovered the expected prenyl moiety with its characteristic triplet signal (δCH 5.42, t (7.5)/125.82) (Figure S34). The observed NOE cross-peaks between both C4H (δCH 7.28, brs/118.04) and C6H (δCH 6.92, d (8.0)/123.81) of Trp4 with C1′H (δCH2 3.46, d (6.8)/35.33) of the dimethylallyl fragment established the tailoring to be forward prenylation of C5-Trp4 (Figure S38).
Initial attempts to decipher the spin system of Tyr2 were unsuccessful. However, NOE correlations were observed between Gly3-NH, CβH of Trp1 and 2,6-CH of Tyr2 (2xδCH 7.61, d (8.3)/132.51) with a downfield unassigned NH at 9.84 ppm resonating as a broad singlet (Figure S37). While 1H-1H COSY showed no couplings with this NH, a strong correlation by TOCSY with a further singlet, δCH 7.48, s/133.63 suggested a possible long-range allylic (W) coupling, 4JNH,CβH. Tracking down this key singlet at 7.48 ppm using TOCSY and NOESY assisted in constructing the remaining Tyr residue as (Z)-α,β(2,3)-didehydro(Dh)Tyr2 (Figure S38). A solution structure of wygwalassin A1 was calculated using the observed long-range NOEs as distance constraints, confirming Ile14 as the lower steric plug (Figure 4, S39-S40). In addition, the C5-prenylated Trp4 was in close proximity to Ile14-Ile15, forming a hydrophobic patch (Figure S41). This topological vicinity resulted in a calculated distance of ≈ 3.2 Å between Ile14-CαH and the indole moiety of Trp4 in nearly all the 20 solutions NMR structures (Figure S41). As a result of having Ile14 on top of such a π-system, its αH resonated at a noticeable upfield shift (δH 3.42, m) providing further evidence of a threaded conformation.63
Figure 4.
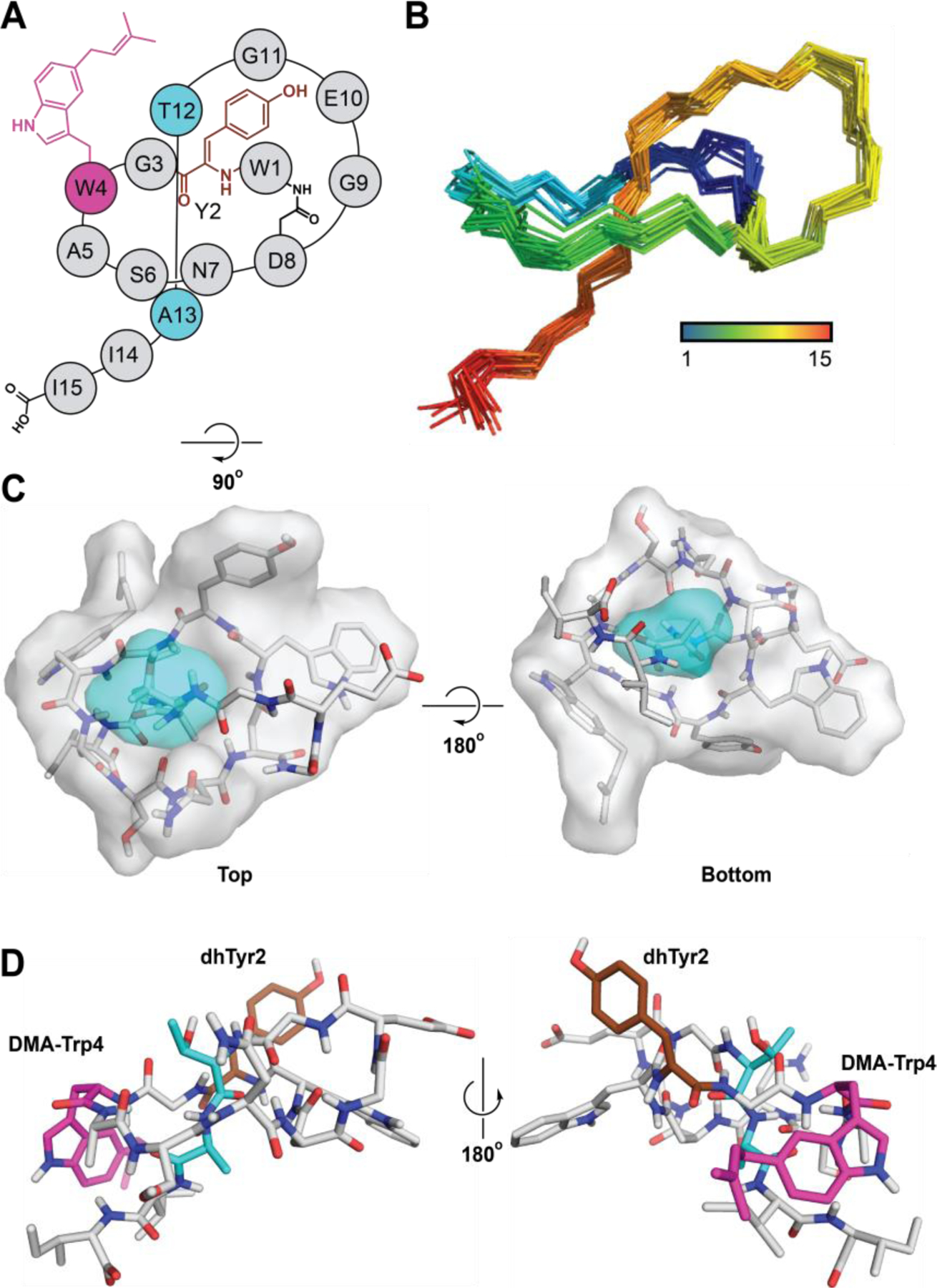
Structure of wygwalassin A1. (A) Topological diagram of wygwalassin A1. Gray, ring region; cyan, steric-locking residues; brown, (Z)-α,β-dehydro-Tyr (dhTyr); magenta, 5-dimethylallyl-Trp (DMA-Trp). (B) NOE-based NMR ensemble of the 20 lowest energy structures of wygwalassin A1 (only amide backbone is shown for clarity). (C) Surface-filling model of wygwalassin A1, with the ring and steric-locking residues tinted gray and cyan, respectively. (D) Lowest energy structure of wygwalassin A1. dhTyr2 and DMA-Trp4 are labeled and colored brown and magenta, respectively. Wygwalassin A1 PDB code: 8UKG.
Both chlorolassin and wygwalassin A1 were tested for growth inhibition in a microtiter plate assay against a diverse set of bacteria (see methods). No growth inhibition was observed for either compound up to 128 µg/mL. Future work is needed to elucidate the biological activity and role of these two compounds.
Bioinformatic Analysis.
Trp-Halogenases.
With the Trp-halogenase (ChlH) confirmed through gene omission and its indole regioselectivity determined, additional bioinformatic analyses were conducted to contextualize ChlH in the broader flavin-dependent halogenase family (InterPro family IPR006905). A sequence similarity network (SSN) of the halogenases was generated and annotated with characterized Trp-halogenases (Figure 5, Supplemental Dataset 3). Of these, only four are known to act on RiPPs: the previously discussed MibH;56 amm3,55 a presumed 6-chlorinase critical for ammosamide biosynthesis; and SrpI,54 a 6-brominase involved in the biosynthesis of an azoline-containing proteusin-like RiPP. Unsurprisingly, ChlH groups with MibH and Amm3, while SrpI is found in a distal group.
Figure 5.

SSN of the flavin-dependent halogenases. The SSN was constructed from IPR006905 and depicts a RepNode 60 network viewed at alignment score 90. ChlH, Amm3, and SrpI were manually added given their absence in UniProt. Known proteins are denoted and colored based on halogenation regioselectivity. The network was created using the EFI-EST32 and visualized using Cytoscape.33
To determine if more halogenases are associated with putative RiPP BGCs, we employed RODEO and RRE-Finder30 to retrieve all halogenases encoded near an RRE domain. As expected, the majority of RRE-associated halogenases are gathered in this putative RiPP-modifying clade with a few scattered elsewhere in the family. Filtering further, these RRE-associated halogenases revealed several high-confidence, novel RiPP BGCs, among them lanthipeptides, lasso peptides, and unclassified examples (Figure S42). Many of these BGCs feature additional tailoring enzymes and, more importantly, their predicted precursor peptides contain multiple Trp, supporting the tentative assignment as Trp-halogenases involved in RiPP biosynthesis.
Trp-Dimethylallyltransferases (DMATs).
To glean more insight about the prevalence and context of Trp DMATs (i.e., WygG) in RiPP biosynthesis, an SSN of the aromatic prenyltransferase family (DMAT type, IPR017795) supplemented with UniProt52 and GenBank BLAST-P53 results was generated (Figure S43, Supplemental Dataset 4). Representatives associated with the wygwalassin BGCs formed a small SSN group distantly related to previously characterized DMATs.
To uncover other putative RiPP-modifying prenyltransferases, RODEO/RRE-Finder was used to analyze the family of prenyltransferases. In addition to the clade containing WygG-like enzymes, RRE-associated prenyltransferases appear in two additional SSN groups: both of which encode putative nitrile hydratase-like leader peptides (NHLPs), which is a RiPP class comprising the proteusins.1,64 These proteusins-like BGCs feature a further set of biosynthetic enzymes (Figure S44). The predicted precursor peptides also contain a conserved C-terminal Trp as a potential tailoring site for the local DMAT.
Discussion
Through a Trp-centric bioinformatic strategy, we discovered and characterized two lasso peptides bearing modifications unseen in the family to date. The first, chlorolassin, contained a core region with three Trp, two of which were modified to 5-Cl-Trp. The second, wygwalassin A1, contained two Trp, one of which was modified to 5-dimethylallyl-Trp; furthermore, this lasso peptide contained (Z)-α,β-dehydro-Tyr. The halogenase responsible for chlorolassin chlorination and DMAT responsible for wygwalassin prenylation were also bioinformatically contextualized with respect to characterized members of each enzyme family and other cases with likely involvement in RiPP biosynthesis.
Compared to the dozens of halogenases characterized in natural product biosynthesis, the subset that modify RiPPs are limited to four enzymes, of which ChlH is the newest member. The halogenase SSN indicates that three of these enzymes (ChlH, MibH, Amm3) form a RiPP-modifying group that are more distantly related to those that act on free or acyl carrier protein-bound Trp. Of the characterized RiPP-halogenases, ChlH (5-chlorinase) has the highest sequence identity with MibH (64%, 5-chlorinase), followed by Amm3 (61%, predicted 6-chlorinase). For comparison, PyrH, a free L-Trp 5-chlorinase,65 has 40% identity, confirming prior observations that the Trp substrate (peptide vs. free amino acid) is a stronger indicator of sequence similarity, as opposed to indole regioselectivity.56 Besides their indole regiospecificity and halogen selectivity, other differences in substrate preference are present in this group. MibH activity against various substrates has been reported, and this enzyme chlorinates the cyclized deschloro-NAI-107 while rejecting the linear precursor peptide MibA and mutacin 1140, a lanthipeptide with similar ring connectivity to NAI-107.56 In contrast, amm3 likely acts early in the biosynthesis of ammosamide A-C, as the omission of the amm3 gene during heterologous expression of the amm BGC does not produce the non-halogenated analog, but rather abolishes ammosamide production altogether.55 Finally, SrpI appears to have no preferred order, with the enzyme brominating the C-terminal Trp of non-cyclodehydrated, mono-, di-, and tri-cyclodehydrated precursor peptides.54
Similarly, prenylation as a RiPP modification is known for classes like cyanobactins and the quorum-sensing ComX peptides. The former contains a rich chemical diversity, with forward/reverse O/N/C-prenylation to Ser, Thr, Tyr, Arg, Trp and His residues, as well as the N/C-terminus of linear cyanobactins.1,66–70 In contrast to the variation in the prenyl acceptor residue seen in the cyanobactins, ComX peptides share a conserved forward prenylation at the C3 position of an internal Trp of ComX, with certain strains performing geranylation and others farnesylation.71 In contrast to cyanobactins and ComX pheromones, wygwalassin A1 is matured with TrpC5-forward dimethylallylation which has, to our knowledge, never been characterized in any RiPP-derived product. DMATs, particularly Trp-DMATs, are widespread in fungal biosynthetic (non-RiPP) pathways and perform similar reactions to the cyanobactin prenyltransferases.72–76 Moreover, some characterized fungal DMATs perform tandem prenylation and cyclization, further maximizing their tailoring scope.77–79 The Trp-DMATs in the wyg BGC have no significant similarity to any known RiPP-modifying enzyme, leaving their reactivity a subject for future investigation.
Besides Trp-prenylation, wygwalassin also contains (Z)-α,β-didehydro-Tyr. Dehydrated residues are present in several RiPPs classes (e.g., lanthipeptides, linaridins, linear azol(in)e-containing peptides, thiopeptides).1 Several examples of α,β-dehydro-Tyr of non-ribosomal origin have been reported in the literature, such as dityromycin,80 WS9326A,81 the thalassotalic acids (and related compounds).82,83 To the best of our knowledge, the only instance of dehydro-Tyr of ribosomal origin is green fluorescent protein,84 making wygwalassin the first known RiPP to carry this modification. Despite the bioinformatic limitation to predict the catalytic outcome of WygH/F enzymes, we propose that one or both enzymes are responsible for Tyr dehydrogenation.
WygF is functionally annotated as a PhyH-like oxidase, which is an Fe(II) 2-oxoglutarate-dependent oxygenase responsible for α-hydroxylation of phytanoyl-CoA.85 Members of this protein family are precedented in RiPP biosynthesis, for instance β-hydroxylhistidine formation in thioviridamide biosynthesis (and the related thioholgamides)86 as well as forming a C-C bond during crocagin biosynthesis.87 Additional characterized homologs of PhyH catalyze various oxidative reactions, such as hydroxylation of ectoine (ThpD),88 oxidative deamination in kanamycin B (KanJ),89 and spiro-ring formation in preaustinoid A3 biosynthesis (AusE).90 Although WygH was functionally predicted as an oxidase of the COX15-heme family, which is unprecedented in RiPP biosynthesis, a few heme-dependent oxygenases act in other natural product pathways. Two examples are TzT, which forms the N-N bond in the piperazate moiety of kutzneride 2 (and related NRPs)91 and PrnB,92 a pyrrolnitrin biosynthetic enzyme.
Conclusion
With the goal of discovering new Trp-modifications for lasso peptides, we curated a dataset of precursor peptides with cores enriched in Trp. Analyzing this dataset, we uncovered two Trp-containing lasso peptides with modifications new to the class: chlorolassin, containing two 5-Cl-Trp residues; and wygwalassin, containing a 5-DMA-Trp and (Z)-α,β-dehydro-Tyr. Analysis of the enzymes responsible for halogenation (ChlH) and prenylation (WygG) uncovered numerous other known and presumed RiPP pathways that are expected to carry similarly secondary modifications. Future work is necessary to examine chlorolassin and wygwalassin biosynthesis in more detail. While the broad functions of ChlH and WygG are clear, the role of the two oxidases, PhyH and Cox15 homologs, in the wygwalassin BGC requires clarification. Moreover, the substrate scope of these enzymes and their biocatalytic potential remain to be investigated. While the current study focused solely on Trp modifications in lasso peptides, we postulate that the bioinformatics workflow disclosed can be leveraged to discover modification chemistry for other amino acids on lasso peptides and other RiPP classes.
Supplementary Material
Acknowledgments
This work was supported in part by grant GM123998 from the NIH (to DAM). We are grateful to W. W. Metcalf for providing bacterial strains and for consultation on the conjugation experiments described in this study. Further, we thank P. A. Jordan for providing the sequence of amm3 used in our analyses, as well as G. A. Hudson, T. W. Precord, and S. Ramesh for assistance with data collection (HR MS/MS).
Footnotes
Supporting Information
Experimental methods and supporting figures (S1-S44) and tables (S1-S8) (PDF).
Supplemental Dataset 1 contains the co-occurrence analysis of lasso peptide BGCs with Trp-rich core regions in the precursor peptides (MS Excel format, XSLX).
Supplemental Dataset 2 contains a list of the leader peptidases used as RODEO input to identify Trp-rich core regions of lasso precursor peptides (MS Excel format, XSLX).
Supplemental Dataset 3 contains a list of flavin-dependent halogenases used for SSN creation (MS Excel format, XSLX).
Supplemental Dataset 4 contains a list of aromatic prenyltransferases (DMAT-type) used for SSN creation (MS Excel format, XSLX).
Accession Codes
The NMR solution structures of chlorolassin and wygwalassin A1 have been deposited to the Protein Data Bank, which have been assigned codes 8UKC and 8UKG, respectively.
Notes
The authors declare no competing financial interest.
References
- (1).Montalbán-López M; Scott TA; Ramesh S; Rahman IR; van Heel AJ; Viel JH; Bandarian V; Dittmann E; Genilloud O; Goto Y; Grande Burgos MJ; Hill C; Kim S; Koehnke J; Latham JA; Link AJ; Martínez B; Nair SK; Nicolet Y; Rebuffat S; Sahl H-G; Sareen D; Schmidt EW; Schmitt L; Severinov K; Süssmuth RD; Truman AW; Wang H; Weng J-K; van Wezel GP; Zhang Q; Zhong J; Piel J; Mitchell DA; Kuipers OP; van der Donk WA New Developments in RiPP Discovery, Enzymology and Engineering. Nat Prod Rep 2021, 38 (1), 130–239. 10.1039/D0NP00027B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hegemann JD; Zimmermann M; Xie X; Marahiel MA Lasso Peptides: An Intriguing Class of Bacterial Natural Products. Acc. Chem. Res . 2015, 48 (7), 1909–1919. 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- (3).Hegemann JD Factors Governing the Thermal Stability of Lasso Peptides. ChemBioChem 2020, 21 (1–2), 7–18. 10.1002/cbic.201900364. [DOI] [PubMed] [Google Scholar]
- (4).DiCaprio AJ; Firouzbakht A; Hudson GA; Mitchell DA Enzymatic Reconstitution and Biosynthetic Investigation of the Lasso Peptide Fusilassin. J. Am. Chem. Soc . 2019, 141 (1), 290–297. 10.1021/jacs.8b09928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Koos JD; Link AJ Heterologous and in Vitro Reconstitution of Fuscanodin, a Lasso Peptide from Thermobifida Fusca. J. Am. Chem. Soc . 2019, 141 (2), 928–935. 10.1021/jacs.8b10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Duan Y; Niu W; Pang L; Bian X; Zhang Y; Zhong G Unusual Post-Translational Modifications in the Biosynthesis of Lasso Peptides. Int. J. Mol. Sci . 2022, 23 (13), 7231. 10.3390/ijms23137231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hemmerling F; Piel J Strategies to Access Biosynthetic Novelty in Bacterial Genomes for Drug Discovery. Nat. Rev. Drug Discov . 2022, 21 (5), 359–378. 10.1038/s41573-022-00414-6. [DOI] [PubMed] [Google Scholar]
- (8).Scherlach K; Hertweck C Mining and Unearthing Hidden Biosynthetic Potential. Nat. Commun . 2021, 12 (1), 3864. 10.1038/s41467-021-24133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Medema MH; de Rond T; Moore BS Mining Genomes to Illuminate the Specialized Chemistry of Life. Nat. Rev. Genet . 2021, 22 (9), 553–571. 10.1038/s41576-021-00363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Zhong Z; He B; Li J; Li Y-X Challenges and Advances in Genome Mining of Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs). Synth. Syst. Biotechnol . 2020, 5 (3), 155–172. 10.1016/j.synbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kessler SC; Chooi Y-H Out for a RiPP: Challenges and Advances in Genome Mining of Ribosomal Peptides from Fungi. Nat. Prod. Rep . 2022, 39 (2), 222–230. 10.1039/D1NP00048A. [DOI] [PubMed] [Google Scholar]
- (12).Zdouc MM; Hooft JJJ van der; Medema, M. H. Metabolome-Guided Genome Mining of RiPP Natural Products. Trends Pharmacol. Sci . 2023, 44 (8), 532–541. 10.1016/j.tips.2023.06.004. [DOI] [PubMed] [Google Scholar]
- (13).Tietz JI; Schwalen CJ; Patel PS; Maxson T; Blair PM; Tai H-C; Zakai UI; Mitchell DA A New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol . 2017, 13 (5), 470–478. 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ramesh S; Guo X; DiCaprio AJ; De Lio AM; Harris LA; Kille BL; Pogorelov TV; Mitchell DA Bioinformatics-Guided Expansion and Discovery of Graspetides. ACS Chem. Biol . 2021, 16 (12), 2787–2797. 10.1021/acschembio.1c00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Oberg N; Precord TW; Mitchell DA; Gerlt JA RadicalSAM.Org: A Resource to Interpret Sequence-Function Space and Discover New Radical SAM Enzyme Chemistry. ACS Bio Med Chem Au 2022, 2 (1), 22–35. 10.1021/acsbiomedchemau.1c00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Georgiou MA; Dommaraju SR; Guo X; Mast DH; Mitchell DA Bioinformatic and Reactivity-Based Discovery of Linaridins. ACS Chem. Biol . 2020, 15 (11), 2976–2985. 10.1021/acschembio.0c00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Walker MC; Eslami SM; Hetrick KJ; Ackenhusen SE; Mitchell DA; van der Donk WA Precursor Peptide-Targeted Mining of More than One Hundred Thousand Genomes Expands the Lanthipeptide Natural Product Family. BMC Genomics 2020, 21 (1), 387. 10.1186/s12864-020-06785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Harris LA; Saint-Vincent PMB; Guo X; Hudson GA; DiCaprio AJ; Zhu L; Mitchell DA Reactivity-Based Screening for Citrulline-Containing Natural Products Reveals a Family of Bacterial Peptidyl Arginine Deiminases. ACS Chem. Biol . 2020, 15 (12), 3167–3175. 10.1021/acschembio.0c00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cao L; Beiser M; Koos JD; Orlova M; Elashal HE; Schröder HV; Link AJ Cellulonodin-2 and Lihuanodin: Lasso Peptides with an Aspartimide Post-Translational Modification. J. Am. Chem. Soc . 2021, 143 (30), 11690–11702. 10.1021/jacs.1c05017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Saad H; Majer T; Bhattarai K; Lampe S; Nguyen DT; Kramer M; Straetener J; Brötz-Oesterhelt H; Mitchell DA; Gross H Bioinformatics-Guided Discovery of Biaryl-Linked Lasso Peptides. Chem. Sci . 2023, 14 (45), 13176–13183. 10.1039/D3SC02380J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Si Y; Kretsch AM; Daigh LM; Burk MJ; Mitchell DA Cell-Free Biosynthesis to Evaluate Lasso Peptide Formation and Enzyme–Substrate Tolerance. J. Am. Chem. Soc . 2021, 143 (15), 5917–5927. 10.1021/jacs.1c01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kretsch AM; Gadgil MG; DiCaprio AJ; Barrett SE; Kille BL; Si Y; Zhu L; Mitchell DA Peptidase Activation by a Leader Peptide-Bound RiPP Recognition Element. Biochemistry 2023, 62 (4), 956–967. 10.1021/acs.biochem.2c00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Barik S The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. Int. J. Mol. Sci . 2020, 21 (22), 8776. 10.3390/ijms21228776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ogawa T; Ochiai K; Tanaka T; Tsukuda E; Chiba S; Yano K; Yamasaki M; Yoshida M; Matsuda Y RES-701–2, −3 and −4, Novel and Selective Endothelin Type B Receptor Antagonists Produced by Streptomyces Sp. I. Taxonomy of Producing Strains, Fermentation, Isolation, and Biochemical Properties. J. Antibiot. (Tokyo) 1995, 48 (11), 1213–1220. 10.7164/antibiotics.48.1213. [DOI] [PubMed] [Google Scholar]
- (25).Oves-Costales D; Sánchez-Hidalgo M; Martín J; Genilloud O Identification, Cloning and Heterologous Expression of the Gene Cluster Directing RES-701–3, −4 Lasso Peptides Biosynthesis from a Marine Streptomyces Strain. Mar. Drugs 2020, 18 (5), 238. 10.3390/md18050238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Feng Z; Ogasawara Y; Nomura S; Dairi T Biosynthetic Gene Cluster of a D-Tryptophan-Containing Lasso Peptide, MS-271. ChemBioChem 2018, 19 (19), 2045–2048. 10.1002/cbic.201800315. [DOI] [PubMed] [Google Scholar]
- (27).Nakashima Y; Kawakami A; Ogasawara Y; Maeki M; Tokeshi M; Dairi T; Morita H Structure of Lasso Peptide Epimerase MslH Reveals Metal-Dependent Acid/Base Catalytic Mechanism. Nat. Commun . 2023, 14 (1), 4752. 10.1038/s41467-023-40232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Altschul SF; Madden TL; Schäffer AA; Zhang J; Zhang Z; Miller W; Lipman DJ Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res . 1997, 25 (17), 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tietz JI; Schwalen CJ; Patel PS; Maxson T; Blair PM; Tai H-C; Zakai UI; Mitchell DA A New Genome-Mining Tool Redefines the Lasso Peptide Biosynthetic Landscape. Nat. Chem. Biol . 2017, 13 (5), 470–478. 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kloosterman AM; Shelton KE; van Wezel GP; Medema MH; Mitchell DA RRE-Finder: A Genome-Mining Tool for Class-Independent RiPP Discovery. mSystems 2020, 5 (5), 10.1128/msystems.00267-20. 10.1128/msystems.00267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Finn RD; Bateman A; Clements J; Coggill P; Eberhardt RY; Eddy SR; Heger A; Hetherington K; Holm L; Mistry J; Sonnhammer ELL; Tate J; Punta M Pfam: The Protein Families Database. Nucleic Acids Res . 2014, 42 (D1), D222–D230. 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Oberg N; Zallot R; Gerlt JA EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J. Mol. Biol . 2023, 435 (14), 168018. 10.1016/j.jmb.2023.168018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res . 2003, 13 (11), 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Balch WE; Fox GE; Magrum LJ; Woese CR; Wolfe RS Methanogens: Reevaluation of a Unique Biological Group. Microbiol. Rev . 1979, 43 (2), 260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Myronovskyi M; Rosenkränzer B; Nadmid S; Pujic P; Normand P; Luzhetskyy A Generation of a Cluster-Free Streptomyces Albus Chassis Strains for Improved Heterologous Expression of Secondary Metabolite Clusters. Metab. Eng . 2018, 49, 316–324. 10.1016/j.ymben.2018.09.004. [DOI] [PubMed] [Google Scholar]
- (36).Gibson DG; Young L; Chuang R-Y; Venter JC; Hutchison CA; Smith HO Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6 (5), 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- (37).Bibb MJ; Janssen GR; Ward JM Cloning and Analysis of the Promoter Region of the Erythromycin Resistance Gene (ermE) of Streptomyces Erythraeus. Gene 1985, 38 (1), 215–226. 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- (38).Blodgett JAV; Thomas PM; Li G; Velasquez JE; van der Donk WA; Kelleher NL; Metcalf WW Unusual Transformations in the Biosynthesis of the Antibiotic Phosphinothricin Tripeptide. Nat. Chem. Biol . 2007, 3 (8), 480–485. 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yu X; Price NPJ; Evans BS; Metcalf WW Purification and Characterization of Phosphonoglycans from Glycomyces Sp. Strain NRRL B-16210 and Stackebrandtia Nassauensis NRRL B-16338. J. Bacteriol . 2014, 196 (9), 1768–1779. 10.1128/jb.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bhushan R; Brückner H Marfey’s Reagent for Chiral Amino Acid Analysis: A Review. Amino Acids 2004, 27 (3), 231–247. 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- (41).Schwieters CD; Kuszewski JJ; Tjandra N; Marius Clore G The Xplor-NIH NMR Molecular Structure Determination Package. J. Magn. Reson . 2003, 160 (1), 65–73. 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- (42).Schwieters CD; Bermejo GA; Clore GM Xplor-NIH for Molecular Structure Determination from NMR and Other Data Sources. Protein Sci . 2018, 27 (1), 26–40. 10.1002/pro.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6 (3), 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (44).Lee W; Tonelli M; Markley JL NMRFAM-SPARKY: Enhanced Software for Biomolecular NMR Spectroscopy. Bioinformatics 2015, 31 (8), 1325–1327. 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sousa da Silva AW; Vranken WF ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5 (1), 367. 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kagami L; Wilter A; Diaz A; Vranken W The ACPYPE Web Server for Small-Molecule MD Topology Generation. Bioinformatics 2023, 39 (6), btad350. 10.1093/bioinformatics/btad350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Laskowski RA; MacArthur MW; Moss DS; Thornton JM PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr . 1993, 26 (2), 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- (48).Laskowski RA; Rullmann JAC; MacArthur MW; Kaptein R; Thornton JM AQUA and PROCHECK-NMR: Programs for Checking the Quality of Protein Structures Solved by NMR. J. Biomol. NMR 1996, 8 (4), 477–486. 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- (49).Berman H; Henrick K; Nakamura H Announcing the Worldwide Protein Data Bank. Nat. Struct. Mol. Biol . 2003, 10 (12), 980–980. 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- (50).Hoch JC; Baskaran K; Burr H; Chin J; Eghbalnia HR; Fujiwara T; Gryk MR; Iwata T; Kojima C; Kurisu G; Maziuk D; Miyanoiri Y; Wedell JR; Wilburn C; Yao H; Yokochi M Biological Magnetic Resonance Data Bank. Nucleic Acids Res . 2023, 51 (D1), D368–D376. 10.1093/nar/gkac1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sayers EW; Bolton EE; Brister JR; Canese K; Chan J; Comeau DC; Connor R; Funk K; Kelly C; Kim S; Madej T; Marchler-Bauer A; Lanczycki C; Lathrop S; Lu Z; Thibaud-Nissen F; Murphy T; Phan L; Skripchenko Y; Tse T; Wang J; Williams R; Trawick BW; Pruitt KD; Sherry ST Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res . 2021, 50 (D1), D20–D26. 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res . 2023, 51 (D1), D523–D531. 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Altschul SF; Gish W; Miller W; Myers EW; Lipman DJ Basic Local Alignment Search Tool. J. Mol. Biol . 1990, 215 (3), 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- (54).Nguyen NA; Lin Z; Mohanty I; Garg N; Schmidt EW; Agarwal V An Obligate Peptidyl Brominase Underlies the Discovery of Highly Distributed Biosynthetic Gene Clusters in Marine Sponge Microbiomes. J. Am. Chem. Soc . 2021, 143 (27), 10221–10231. 10.1021/jacs.1c03474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Jordan PA; Moore BS Biosynthetic Pathway Connects Cryptic Ribosomally Synthesized Posttranslationally Modified Peptide Genes with Pyrroloquinoline Alkaloids. Cell Chem. Biol . 2016, 23 (12), 1504–1514. 10.1016/j.chembiol.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ortega MA; Cogan DP; Mukherjee S; Garg N; Li B; Thibodeaux GN; Maffioli SI; Donadio S; Sosio M; Escano J; Smith L; Nair SK; van der Donk WA Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis. ACS Chem. Biol . 2017, 12 (2), 548–557. 10.1021/acschembio.6b01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Myronovskyi M; Rosenkränzer B; Nadmid S; Pujic P; Normand P; Luzhetskyy A Generation of a Cluster-Free Streptomyces Albus Chassis Strains for Improved Heterologous Expression of Secondary Metabolite Clusters. Metab. Eng . 2018, 49, 316–324. 10.1016/j.ymben.2018.09.004. [DOI] [PubMed] [Google Scholar]
- (58).Xie X; Marahiel MA NMR as an Effective Tool for the Structure Determination of Lasso Peptides. ChemBioChem 2012, 13 (5), 621–625. 10.1002/cbic.201100754. [DOI] [PubMed] [Google Scholar]
- (59).Jeanne Dit Fouque K; Hegemann JD; Zirah S; Rebuffat S; Lescop E; Fernandez-Lima F Evidence of Cis/Trans-Isomerization at Pro7/Pro16 in the Lasso Peptide Microcin J25. J. Am. Soc. Mass Spectrom . 2019, 30 (6), 1038–1045. 10.1007/s13361-019-02134-5. [DOI] [PubMed] [Google Scholar]
- (60).Zong C; Wu MJ; Qin JZ; Link AJ Lasso Peptide Benenodin-1 Is a Thermally Actuated [1]Rotaxane Switch. J. Am. Chem. Soc . 2017, 139 (30), 10403–10409. 10.1021/jacs.7b04830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).de Lorimier R; Spicer LD NMR Methods Used to Derive Solution Conformations in Peptides. In Techniques in Protein Chemistry; Crabb JW, Ed.; Academic Press, 1994; Vol. 5, pp 423–430. 10.1016/B978-0-12-194710-1.50052-7. [DOI] [Google Scholar]
- (62).Bhushan R; Brückner H Marfey’s Reagent for Chiral Amino Acid Analysis: A Review. Amino Acids 2004, 27 (3), 231–247. 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- (63).Szilágyi L; Jardetzky O α-Proton Chemical Shifts and Secondary Structure in Proteins. J. Magn. Reson. 1969 1989, 83 (3), 441–449. 10.1016/0022-2364(89)90341-7. [DOI] [Google Scholar]
- (64).Haft DH; Basu MK; Mitchell DA Expansion of Ribosomally Produced Natural Products: A Nitrile Hydratase- and Nif11-Related Precursor Family. BMC Biol . 2010, 8 (1), 70. 10.1186/1741-7007-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Zehner S; Kotzsch A; Bister B; Süssmuth RD; Méndez C; Salas JA; van Pée K-H A Regioselective Tryptophan 5-Halogenase Is Involved in Pyrroindomycin Biosynthesis in Streptomyces Rugosporus LL-42D005. Chem. Biol . 2005, 12 (4), 445–452. 10.1016/j.chembiol.2005.02.005. [DOI] [PubMed] [Google Scholar]
- (66).Phan C-S; Matsuda K; Balloo N; Fujita K; Wakimoto T; Okino T Argicyclamides A–C Unveil Enzymatic Basis for Guanidine Bis-Prenylation. J. Am. Chem. Soc . 2021, 143 (27), 10083–10087. 10.1021/jacs.1c05732. [DOI] [PubMed] [Google Scholar]
- (67).Zheng Y; Cong Y; Schmidt EW; Nair SK Catalysts for the Enzymatic Lipidation of Peptides. Acc. Chem. Res . 2022, 55 (9), 1313–1323. 10.1021/acs.accounts.2c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Zhang Y; Goto Y; Suga H Discovery, Biochemical Characterization, and Bioengineering of Cyanobactin Prenyltransferases. Trends Biochem. Sci . 2023, 48 (4), 360–374. 10.1016/j.tibs.2022.11.002. [DOI] [PubMed] [Google Scholar]
- (69).Zhang Y; Hamada K; Nguyen DT; Inoue S; Satake M; Kobayashi S; Okada C; Ogata K; Okada M; Sengoku T; Goto Y; Suga H LimF Is a Versatile Prenyltransferase for Histidine-C-Geranylation on Diverse Non-Natural Substrates. Nat. Catal . 2022, 5 (8), 682–693. 10.1038/s41929-022-00822-2. [DOI] [Google Scholar]
- (70).Zhang Y; Hamada K; Satake M; Sengoku T; Goto Y; Suga H Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases. J. Am. Chem. Soc . 2023, 145 (44), 23893–23898. 10.1021/jacs.3c07373. [DOI] [PubMed] [Google Scholar]
- (71).Miyata A; Ito S; Fujinami D Structure Prediction and Genome Mining-Aided Discovery of the Bacterial C-Terminal Tryptophan Prenyltransferase PalQ. Adv. Sci n/a (n/a), 2307372. 10.1002/advs.202307372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Wang J; Machado C; Panaccione DG; Tsai H-F; Schardl CL The Determinant Step in Ergot Alkaloid Biosynthesis by an Endophyte of Perennial Ryegrass. Fungal Genet. Biol . 2004, 41 (2), 189–198. 10.1016/j.fgb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- (73).Balibar CJ; Howard-Jones AR; Walsh CT Terrequinone A Biosynthesis through L-Tryptophan Oxidation, Dimerization and Bisprenylation. Nat. Chem. Biol . 2007, 3 (9), 584–592. 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- (74).Grundmann A; Kuznetsova T; Afiyatullov S. Sh.; Li S-M. FtmPT2, an N-Prenyltransferase from Aspergillus Fumigatus, Catalyses the Last Step in the Biosynthesis of Fumitremorgin B. ChemBioChem 2008, 9 (13), 2059–2063. 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
- (75).Andersen MR; Nielsen JB; Klitgaard A; Petersen LM; Zachariasen M; Hansen TJ; Blicher LH; Gotfredsen CH; Larsen TO; Nielsen KF; Mortensen UH Accurate Prediction of Secondary Metabolite Gene Clusters in Filamentous Fungi. Proc. Natl. Acad. Sci . 2013, 110 (1), E99–E107. 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Chooi Y-H; Fang J; Liu H; Filler SG; Wang P; Tang Y Genome Mining of a Prenylated and Immunosuppressive Polyketide from Pathogenic Fungi. Org. Lett . 2013, 15 (4), 780–783. 10.1021/ol303435y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Wohlgemuth V; Kindinger F; Xie X; Wang B-G; Li S-M Two Prenyltransferases Govern a Consecutive Prenylation Cascade in the Biosynthesis of Echinulin and Neoechinulin. Org. Lett . 2017, 19 (21), 5928–5931. 10.1021/acs.orglett.7b02926. [DOI] [PubMed] [Google Scholar]
- (78).Zou Y; Zhan Z; Li D; Tang M; Cacho RA; Watanabe K; Tang Y Tandem Prenyltransferases Catalyze Isoprenoid Elongation and Complexity Generation in Biosynthesis of Quinolone Alkaloids. J. Am. Chem. Soc . 2015, 137 (15), 4980–4983. 10.1021/jacs.5b03022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Liu C; Minami A; Noike M; Toshima H; Oikawa H; Dairi T Regiospecificities and Prenylation Mode Specificities of the Fungal Indole Diterpene Prenyltransferases AtmD and PaxD. Appl. Environ. Microbiol . 2013, 79 (23), 7298–7304. 10.1128/AEM.02496-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Teshima T; Nishikawa M; Kubota I; Shiba T; Iwai Y; Ōmura S The Structure of an Antibiotic, Dityromycin. Tetrahedron Lett . 1988, 29 (16), 1963–1966. 10.1016/S0040-4039(00)82090-0. [DOI] [Google Scholar]
- (81).Yu Z; Vodanovic-Jankovic S; Kron M; Shen B New WS9326A Congeners from Streptomyces Sp. 9078 Inhibiting Brugia Malayi Asparaginyl-tRNA Synthetase. Org. Lett . 2012, 14 (18), 4946–4949. 10.1021/ol302298k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Deering RW; Chen J; Sun J; Ma H; Dubert J; Barja JL; Seeram NP; Wang H; Rowley DC N-Acyl Dehydrotyrosines, Tyrosinase Inhibitors from the Marine Bacterium Thalassotalea Sp. PP2–459. J. Nat. Prod . 2016, 79 (2), 447–450. 10.1021/acs.jnatprod.5b00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Sugumaran M; Robinson WE Bioactive Dehydrotyrosyl and Dehydrodopyl Compounds of Marine Origin. Mar. Drugs 2010, 8 (12), 2906–2935. 10.3390/md8122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Heim R; Prasher DC; Tsien RY Wavelength Mutations and Posttranslational Autoxidation of Green Fluorescent Protein. Proc. Natl. Acad. Sci . 1994, 91 (26), 12501–12504. 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Jansen GA; Hogenhout EM; Ferdinandusse S; Waterham HR; Ofman R; Jakobs C; Skjeldal OH; Wanders RJA Human Phytanoyl-CoA Hydroxylase: Resolution of the Gene Structure and the Molecular Basis of Refsum’s Disease. Hum. Mol. Genet . 2000, 9 (8), 1195–1200. 10.1093/hmg/9.8.1195. [DOI] [PubMed] [Google Scholar]
- (86).Sikandar A; Lopatniuk M; Luzhetskyy A; Koehnke J Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide β-Hydroxylation. ACS Chem. Biol . 2020, 15 (10), 2815–2819. 10.1021/acschembio.0c00637. [DOI] [PubMed] [Google Scholar]
- (87).Adam S; Zheng D; Klein A; Volz C; Mullen W; Shirran SL; Smith BO; Kalinina OV; Müller R; Koehnke J Unusual Peptide-Binding Proteins Guide Pyrroloindoline Alkaloid Formation in Crocagin Biosynthesis. Nat. Chem . 2023, 15 (4), 560–568. 10.1038/s41557-023-01153-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Prabhu J; Schauwecker F; Grammel N; Keller U; Bernhard M Functional Expression of the Ectoine Hydroxylase Gene (thpD) from Streptomyces Chrysomallus in Halomonas Elongata. Appl. Environ. Microbiol . 2004, 70 (5), 3130–3132. 10.1128/AEM.70.5.3130-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Sucipto H; Kudo F; Eguchi T The Last Step of Kanamycin Biosynthesis: Unique Deamination Reaction Catalyzed by the α-Ketoglutarate-Dependent Nonheme Iron Dioxygenase KanJ and the NADPH-Dependent Reductase KanK. Angew. Chem. Int. Ed . 2012, 51 (14), 3428–3431. 10.1002/anie.201108122. [DOI] [PubMed] [Google Scholar]
- (90).Matsuda Y; Awakawa T; Wakimoto T; Abe I Spiro-Ring Formation Is Catalyzed by a Multifunctional Dioxygenase in Austinol Biosynthesis. J. Am. Chem. Soc . 2013, 135 (30), 10962–10965. 10.1021/ja405518u. [DOI] [PubMed] [Google Scholar]
- (91).Du Y-L; He H-Y; Higgins MA; Ryan KS A Heme-Dependent Enzyme Forms the Nitrogen–Nitrogen Bond in Piperazate. Nat. Chem. Biol . 2017, 13 (8), 836–838. 10.1038/nchembio.2411. [DOI] [PubMed] [Google Scholar]
- (92).De Laurentis W; Khim L; Anderson JLR; Adam A; Phillips RS; Chapman SK; van Pee K-H; Naismith JH The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway Is Related to the Heme-Dependent Dioxygenase Superfamily. Biochemistry 2007, 46 (43), 12393–12404. 10.1021/bi7012189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


