ABSTRACT
Introduction
The aim of the study was to evaluate the modulating effects of five commonly used sweetener (glucose, inulin, isomaltulose, tagatose, trehalose) containing mouth rinses on the oral microbiome.
Methods
A single-centre, double-blind, parallel randomized clinical trial was performed with healthy, 18–55-year-old volunteers (N = 65), who rinsed thrice-daily for two weeks with a 10% solution of one of the allocated sweeteners. Microbiota composition of supragingival dental plaque and the tongue dorsum coating was analysed by 16S RNA gene amplicon sequencing of the V4 hypervariable region (Illumina MiSeq). As secondary outcomes, dental plaque red fluorescence and salivary pH were measured.
Results
Dental plaque microbiota changed significantly for two groups: inulin (F = 2.0239, p = 0.0006 PERMANOVA, Aitchison distance) and isomaltulose (F = 0.67, p = 0.0305). For the tongue microbiota, significant changes were observed for isomaltulose (F = 0.8382, p = 0.0452) and trehalose (F = 1.0119, p = 0.0098). In plaque, 13 species changed significantly for the inulin group, while for tongue coating, three species changed for the trehalose group (ALDEx2, p < 0.1). No significant changes were observed for the secondary outcomes.
Conclusion
The effects on the oral microbiota were sweetener dependant with the most pronounced effect on plaque microbiota. Inulin exhibited the strongest microbial modulating potential of the sweeteners tested. Further full-scale clinical studies are required.
KEYWORDS: Oral microbiome, microbiome modulation, oral health, dental plaque, tongue microbiome, clinical trial, sequence analysis, sweetening agents
Introduction
Dental caries is the most widespread preventable noncommunicable disease in humans, affecting roughly two billion people worldwide [1]. A diet high in sugar-sweetened beverages has been linked with caries and type-2 diabetes [2].
Dental caries is defined as a gradual loss and breakdown (decay) of tooth hard tissues (enamel and dentine) that results when free sugars contained in food or drink are converted by bacteria into acids that destroy the tooth over time [1]. Caries is a process caused by demineralization of dental tissues at low pH primarily due to carbohydrate metabolism by oral microorganisms. It should be noted that besides aciduric and acidogenic taxa, there are arginolytic, ureolytic and proteolytic bacteria that can neutralize acidic pH [3,4]. With poor oral hygiene, biofilm can grow undisturbed, increasing in both thickness and surface area. This changes the dynamics within the oral niche, leading to changes in microbial composition, additionally, a thick biofilm also intensifies the exposure to bacterial metabolic end products and the damage these can cause [5–7]. Diet can significantly impact the abundance of both acid-producing and acid-neutralizing microorganisms in the oral microbiome as it can benefit certain microbial metabolic pathways more than others [3,8]. Predominant taxa in a healthy oral microbiome mostly consist of Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes and Fusobacteria, with a varying degree of proportions based on the intraoral niche [9]. The combination of diet and poor oral hygiene can increase the abundance of plaque forming and acid producing bacteria such as streptococci and Corynebacterium and later further increase in aciduric bacteria such as lactobacilli, commonly found in carious lesions [3].
The past decade has seen a rapid development of probiotics and prebiotics that are aimed at modulating the human microbiome and steering it into a healthier equilibrium. Modulation of the gut microbiome has shown positive effects in the treatment of diseases such as depression and obesity [10–12]. However, there is limited research on the oral microbiome [10,12]. The nutrients carrying the highest risk for caries are carbohydrates, especially sucrose [13,14]. Sucrose is extremely appealing to the human sense of taste, while it is also responsible for the changes in microbiome that promote selection of more acid-producing and acid resistant bacteria that can increase the risk for caries [15,16]. One could argue that it would be of great benefit if sucrose could be replaced by alternative sweeteners that have prebiotic properties and also a more oral-health positive effect on the oral microbiome [17]. There have been studies that have made an effort to evaluate the effects of common alternative sweeteners such as isomaltulose, trehalose or inulin but the vast majority of these focus on interactions with gut microbiome or analyse the effects in vitro [18–21]. Only a very limited number of clinical studies have directly compared microbiome-modulating properties of carbohydrate alternatives and the effects on oral microbiome, and even more scarce are studies that use next-generation sequencing to analyse the microbial profile instead of selected microbial taxa [22–25].
The aim of this study was to analyse the effects of five different commonly used sweeteners (inulin, glucose, isomaltulose, tagatose and trehalose) on the oral microbiome.
Materials and methods
Participants
The inclusion criteria were as follows: 1) Healthy, ASA score I (American Society of Anaesthesiologists classification system) as assessed by medical questionnaire [26]; 2) Non-smokers: <1 cigarette a day for at least one year [27]; 3) Minimum of 20 natural teeth: at least 5 evaluable teeth in each quadrant; 4) 17–50% bleeding on probing.
The exclusion criteria were as follows: smokers, individuals with overt dental caries, DPSI (Dutch Periodontal Screening index) score ≥ 3+ [28], users of removable dentures or night guards, those with oral piercings, apparent oral lesions (excluding aphthous ulcers), orthodontic banding (except for lingual retention wire), recent clinical study participation, excessive gum consumption, self-reported pregnancy or breastfeeding, antibiotic use during the last 2 months, need for antibiotic prophylaxis, regular anti-inflammatory drug use, evidence of systemic disease or compromised health, adverse medical history, long-term prescribed medication (excluding contraceptives), or allergies to soy, milk, eggs, gluten or lupin.
The study was reviewed and approved by the Medical Research Ethics Committee (Dutch CCMO research protocol nr: NL68654.100.19; registration in the Netherlands Trial Register (NTR-new): NL7525). The participants provided their written informed consent to participate in this study.
Intervention and measurements
This single-centre, double-blind parallel randomized clinical trial studied the effects of five mouth rinses (inulin, glucose, isomaltulose, tagatose and trehalose) for their microbiome-modulating effects. The study consisted of a screening visit, two baseline visits and four study visits (Figure 1). During the two baseline visits following the screening for eligibility, the following information was collected: general information (age, sex, tooth and tongue brushing frequency), red fluorescence photos, samples from tongue and dental plaque for microbiota analysis. The subjects were randomly allocated to one of the five test groups. The oral microbiome was challenged with a mouth rinse consisting of one of the test sweetener solutions (10%) in water. Five carbohydrates (sweeteners) were selected (tagatose, trehalose, inulin, isomaltulose, glucose), based on previous rinse-studies showing oral ecosystem modulating effects [17,24,25,29,30]. Before each study visit, subjects were instructed not to eat or drink (except water) 2 h before the appointment and to refrain from oral hygiene for 24 h.
Figure 1.
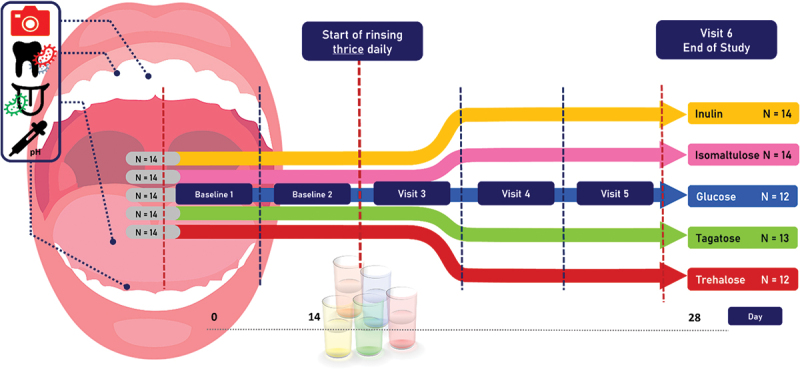
Study outline. After the screening, standard toothpaste was provided once the informed consent forms were signed. Mouth rinse period started from day 14 and lasted two weeks..
After the screening visit, and once the informed consent was signed, the subjects received commercially available toothpaste (1450 ppm F, Cool Mint, Prodent, Unilever) and were asked to use it until the end of the study. At visit 2, all subjects received 16 vials, each containing eight grams of the carbohydrate. Subjects were instructed to take one vial a day and fill with handwarm tap water until the 80-mL indicator on the side of the vial. Thereafter, they had to close the vial with a screw cap and briefly shake the vial until a clear solution was obtained. Subjects were instructed to draw 10 mL of solution from the vial with the provided plastic syringe and to dispense the aliquot into their mouth, followed by rinsing for 30 seconds. After rinsing, subjects spat out the solution and repeated the procedure with an additional 10 mL of the solution from the vial. Subjects were instructed to refrain from oral hygiene, food, and drinks for 30 minutes after rinsing. The subjects were asked to rinse three times daily with the solution. From the second baseline visit, the subjects were seen at the clinic weekly for 2 weeks (Figure 1).
Outcomes
Main study parameters
The main study outcome was the shift in microbial composition in dental plaque and tongue biofilm.
Secondary study parameters
The secondary study outcomes were the proportion of red fluorescent plaque (RFP) and unstimulated salivary pH before and after the 2-week challenge.
Sample size
A recommendation of minimum group size of 12 individuals in pilot studies was used and adjusted for the drop-out rate [31]. Drop-out rate was estimated from a recent clinical study at ACTA to be 6% (this number was low due to the reminders that were sent via the WhatsApp Messenger application a day before the visit) [32]. We doubled this and 14 subjects were included per group.
Randomisation, blinding, and treatment allocation
The investigators and the subjects were blinded to the investigational products. Experienced laboratory technician allocated the five sweeteners to a number 1–5 in a randomization key, which was kept in a sealed file with the sponsor of the study. A list of random numbers was generated with the online application www.random.org, from which a number was assigned to each participant. Using computer-generated stratified block randomization, each subject number was randomly assigned to one of the five groups. The sweeteners were only de-blinded after the data of the primary outcome had been analysed.
Procedures at each study visit
All of the study visits were located in the university clinic (UC) of the Academic Centre for Dentistry Amsterdam (ACTA). At each study visit, first, three fluorescence and three white-light photographs were taken to assess the red fluorescence of plaque (RFP) [32,33]. This was done on the vestibular aspect of the front and lateral teeth using cheek retractors and Canon 450-D SLR camera equipped with a Biluminator tube (QLF-D system, Inspektor Research Systems BV, Amsterdam, the Netherlands), via image capture software and a computer as described previously [32]. The average fluorescence of the three images was calculated.
After the photos were taken, a supragingival plaque sample and a tongue coating sample were collected using a sterile plastic spatula and a sterile microbrush, respectively, as described previously [9]. In brief, a supra-gingival plaque sample was collected from the buccal surface of first and second molars in the 1st quadrant (or 2nd quadrant if one of the teeth was missing). A tongue dorsum was brushed by four strikes with a sterile microbrush (Microbrush International, Grafton, USA) in a longitudinal direction. After sample collection, tubes with plaque samples were centrifuged for 1 min at 14,000 rpm, immediately put on ice, and transferred to the laboratory within 2 hours and stored at − 80◦C until further processing for 16S rRNA gene amplicon sequencing.
At each study visit, the pH of the saliva was measured by collecting two drops of saliva with a disposable pipette from the floor of the mouth and wetting the sensor of the pH-meter (Sentron, Leek, the Netherlands) with the saliva. Calibration of the pH-meter was done before each measurement day. At visit 5, the caries experience was assessed by recording decayed, missing, or filled surfaces (DMFS), according to the WHO criteria on all teeth except third molars [34]. At the last visit (visit 6), a compliance was measured based on the returned used and unused vials: compliance rate = used vials/total vials given * 100%.
Microbiological analysis
16S rRNA gene amplicon sequencing of the V4 hypervariable region was performed as previously described [35,36]. After the addition of lysis buffer and homogenization by bead beating, samples were split into two halves (by volume), one transferred to DNA extraction, one kept as a back-up. DNA extraction was performed using DNeasy 96 PowerSoil Pro QIAcube HT Kit (Qiagen, Carlsbad, CA), following manufacturer’s instructions. Bacterial DNA was quantified by quantitative polymerase chain reaction (qPCR) using universal 16S rRNA gene primers and probe [36]. After DNA quantification, each sample was PCR amplified using 1 ng of template DNA as described by Kozich et al. 2013 [37] with primers F515/R806, targeting the V4 hypervariable region of the 16S ribosomal gene. In each batch of samples, a mix of pure culture isolates (mock), DNA extraction blank, non-template PCR control and pooled plaque-tongue controls were included. The amount of DNA per sample was quantified using the Quant-iT™ PicoGreen® dsDNA Assay Kit (Thermo Fisher Scientific). After pooling in equimolar amounts, the amplicons were purified using AMPureTM XP (Agencourt, Beckmann Coulter, USA) and Gel Band Purification Kit (GE Healthcare, Eindhoven, The Netherlands). Amplicon quality and size were analysed on the Fragment Analyzer (Advanced Analytical). Paired-end sequencing was conducted in five separate runs using the Illumina MiSeq platform (Illumina, Eindhoven, The Netherlands). Procedures regarding drop-out samples: 1) In instances of low DNA yield (qPCR <1000 pg/µl), the sample was excluded followed by repeated extraction using the back-up. The sample was completely excluded after the second failure. 2) In instances when the DNA yield (qPCR) was sufficient, but the amplicon PCR resulted in a low yield, the amplicon PCR was repeated following sensitive sample protocol, the sample was completely excluded after the second failure. 3) In instances of low number of raw sequences (reads <2000), the sample was included in a subsequent run and the sample was completely excluded after the second failure.
Sequencing data processing
The sequencing reads were merged, quality-filtered, and denoised after which the merged reads were mapped to the zero-radius operational taxonomic units (ZOTUs) [38], using USEARCH [39] as described previously [40]. Taxonomy was assigned for each ZOTU using the human oral microbiome database (HOMD v. 14.51) [41] and the RDP classifier [42]. For a part of the analysis, the final ZOTU table was subsampled at equal depth (9800 reads/sample).
Statistical analysis
The Shapiro-Wilk test (with p > 0.05 the data considered normal) was done to test for normality, while Levene’s test of variance was used to test for equal homogeneity of variances for primary and secondary study outcomes as well as for the general information of each group. With normal data distribution and equal homogeneity of variances, the one-way ANOVA was used, otherwise the Kruskal-Wallis test was used. Categorical variables (e.g. gender, tongue brushing habit) were compared among the groups using the Chi-square test.
Next, changes over time were assessed. The second baseline visit was used as a reference to compare the changes at each following visit, as the subjects had been using the same toothpaste for two weeks. With parametric data, a one-way repeated measures ANOVA was used together with paired t-test. With non-parametric data, the Friedman’s test followed by the Wilcoxon signed-rank test was used. The Benjamin-Hochberg post-hoc correction was used to adjust for multiple comparisons. All these tests were performed using IBM SPSS Statistics version 28.
Principal component analysis (PCA) was performed on centered log-ratio (CLR) transformed raw data (pseudo count 0.5, microbiome package ver. 1.18.0) [43]. The Aitchison distance was used to assess the dissimilarity in microbial profiles (R version 4.3.1) [44], as a method that does take into account compositionality of microbiome data [45,46]. Within group, changes in dissimilarity were analysed by Friedman’s test followed by Wilcoxon signed-rank test to compare changes between two timepoints (IBM SPSS Statistics 28). Both alpha and beta diversities were calculated using the vegan package in R (vegan 2.6–4) [47–49]. The Alpha diversity was assessed using the Shannon Diversity Index (the data was first subsampled to an even depth of 9800 and then transformed with a pseudo count of 0.5). The changes in beta-diversity between baseline and further visits were determined with Permutational Multivariate Analysis of Variance (PERMANOVA 99,999 permutations) with permutations restricted by the individual subjects and performed using Aitchison distance. PERMANOVA tests were performed using the adonis2 function from the vegan package (version 2.6–4). The p values were corrected for multiple testing (Bonferroni correction), with an alpha level of 0.05. When PERMANOVA was significant for the within-group analyses, ZOTUs were aggregated to species level (and to genus level) using the ‘tax_glom’ function from phyloseq package (version 1.44.0) [50]. After aggregation to species level, the differential (relative) abundance and effect size was analysed using ALDEx2 (version 1.31.0) package in R [51–54]. Note: for Aldex2 analysis default alpha level is set to 0.1 not 0.05, authors of ALDEX do emphasize the use of effect size over exclusively p-values [53].
Results
Study participants
A total of 70 individuals were included in the study of which 64 participants were able to finish the study (Figure 2). Reasons for the drop-out were as follows: not able to attend visits (n = 3), recent antibiotic use (n = 2), cannot bear not to brush for 24 h (n = 1). Participants ranged from 18 to 54 years of age. Most of the participants (86%) were women. Among the baseline parameters, only age differed significantly among the five groups (Table 1), where further post-hoc testing revealed that glucose group on average was significantly younger than isomaltulose group (adjusted p = 0.035).
Figure 2.
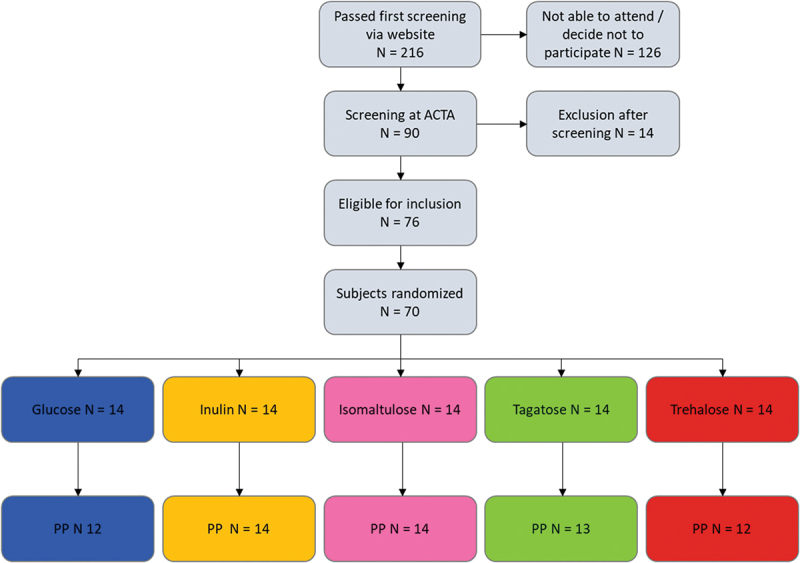
Flow chart of the study population. PP – per protocol population.
Table 1.
General information of study participants.
| Glucose (n=12) | Inulin (n=14) | Isomaltulose (n=14) | Tagatose (N=13) | Trehalose (n=12) | P-value | ||
|---|---|---|---|---|---|---|---|
| Age, years | Mean (SD) | 21.7 (2.7) | 26.6 (8.4) | 31.4 (11.0) | 28.9 (10.4) | 28.7 (11.3) | 0.022* |
| Median (Range) | 22 (18–28) | 25 (20–53) | 27.5 (21–51) | 24 (19–51) | 24.5 (20–54) | ||
| Baseline salivary pH | Mean (SD) | 6.6 (0.2) | 6.6 (0.4) | 6.6 (0.2) | 6.7 (0.3) | 6.7 (0.1) | 0.461* |
| Median (Range) | 6.6 (6.3–6.9) | 6.7 (6.1–7.2) | 6.6 (6.1–7.0) | 6.7 (6.2–7.2) | 6.7 (6.5–7.0) | ||
| 95% CI | (6.4–6.7) | (6.4–6.9) | (6.5–6.7) | (6.5–6.8) | (6.6–6.8) | ||
| DMFS | Mean (SD) | 3.3 (5.1) | 6.2 (8.9) | 6.8 (6.3) | 4 (5.4) | 8.7 (11.6) | 0.351* |
| Median (Range) | 1.5 (0–18) | 2.0 (0–26) | 6.5 (0–21) | 1.0 (0–16) | 4.0 (0–35) | ||
| 95% CI | (0.1–6.6) | (0.8–11.6) | (3.1–10.4) | (0.7–7.3) | (1.3–16.1) | ||
| Tooth brushing frequency per day | Once Twice More often |
2 (15%) 11 (85%) 0 |
1 (7%) 12 (86%) 1 (7%) |
2 (14%) 12 (86%) 0 |
0 13 (93%) 1 (7%) |
2 (14%) 10 (71%) 1 (7%) |
0.796# |
| Sex (males to females) | 2/12 | 2/12 | 1/13 | 3/11 | 1/13 | 0.775# | |
| Tongue brushing (yes/no) | 10/3 (77%) | 8/6 (57%) | 4/10 (29%) | 9/5 (64%) | 8/5 (62%) | 0.128# | |
| Study compliance (%) | 95% | 92% | 95% | 97% | 95% | 0.097* |
* Kruskal Wallis test; # Chi-square test.
Primary outcomes
The primary study outcome was a change in microbial composition. When the individual microbial profiles were ordinated by principal component analysis, plaque samples displayed a directional shift in principal components for inulin and isomaltulose, and to a lesser extent for trehalose and tagatose (Figure 3). In addition, tagatose displayed an outlier, while the shift followed the same direction as for the rest of participants within the group. All groups displayed noticeable overlap between visits, while for inulin, tagatose, and trehalose the end of the study visit clustered closer together than the baseline visit. However, when all study visits were displayed together, the progression of the shift was not uniform, yet still noticeable for the inulin group (Figure S1).
Figure 3.
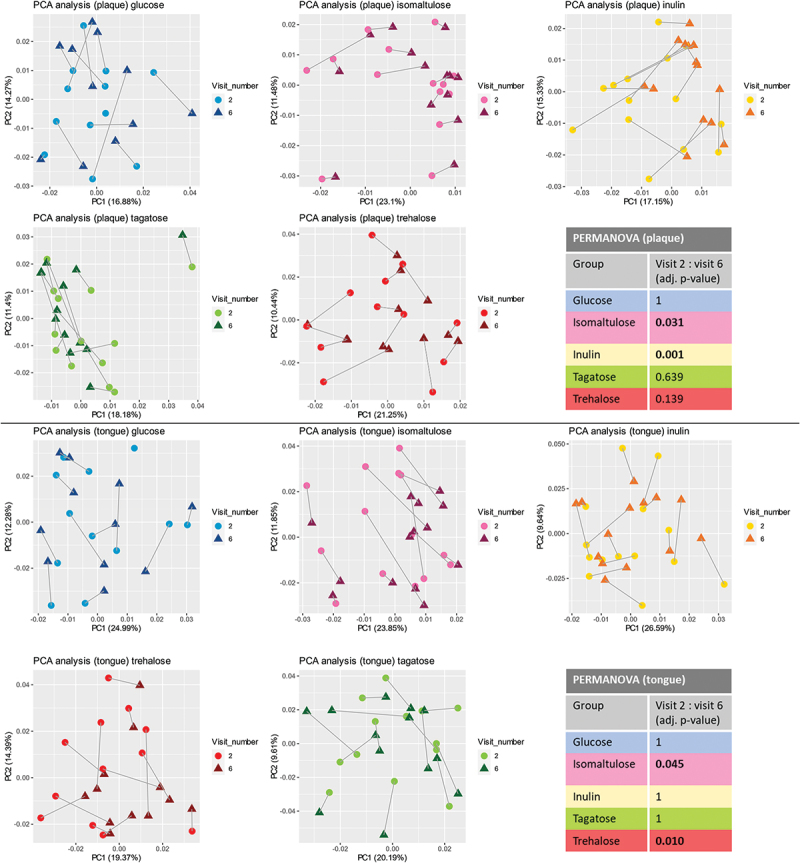
PCA analysis. Principal component analysis plots at the baseline (visit 2) and end of the study (visit 6) for plaque and tongue samples. Output of PERMANOVA analyses using Aitchison distance.
PCA for the tongue samples revealed directional shift for isomaltulose and trehalose (Figure 3, Figure S1). No directional shifts or changes in clustering were observed in the glucose group, neither for the plaque nor for the tongue.
We further quantified the differences in the microbial profiles between various time intervals using PERMANOVA. At baseline, there was no significant difference between groups. Further within-group analysis revealed that for plaque samples, two groups showed significant changes in time: inulin (F = 2.0239, p = 0.0006) and isomaltulose (F = 0.67, p = 0.0305), when comparing baseline with the end of the study. Regarding the tongue samples, significant changes were observed for isomaltulose (F = 0.8382, p = 0.0452) and trehalose (F = 1.0119, p = 0.0098).
Change in microbial dissimilarity distances between two time points using Aitchison distance revealed significant changes in dissimilarity in the tongue microbiota in the isomaltulose group during the study period (Friedman’s ANOVA p = 0.0477, Figure 4), but not when comparing the baseline to the end of the study. No significant changes in dissimilarity distances were observed for the plaque samples.
Figure 4.
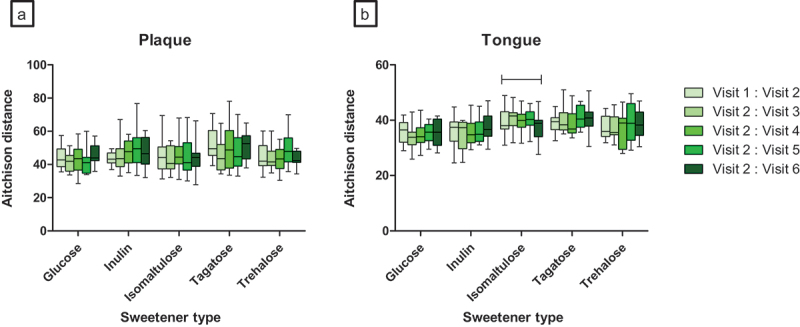
Oral microbiome beta diversity changes in plaque (A) and in tongue (B) samples across study period. First two baseline visits are compared, the second baseline visit is used as comparison for further changes. Second baseline visit is used following two-week normalization by the same toothpaste. |-| indicates significant changes when all visits are compared using Friedman’s ANOVA (p < 0.05).
Alpha diversity – the distribution of microbial species within a specific niche – the Shannon Diversity Index, changed significantly in plaque for two out of the five groups: isomaltulose and inulin (Friedman’s ANOVA, p = 0.046; p = 0.012) during the study period. However, after post-hoc testing, only the inulin group had a significant decrease in alpha diversity between baseline and the end of the study (Bonferroni adjusted, p = 0.016); (Figure S2 AB). Shannon Diversity Index changed significantly in tongue samples for tagatose group (Friedman’s ANOVA, p = 0.049), but further post-hoc testing revealed no significant changes between baseline and the end of the study.
To examine further compositional changes at the species level, we performed differential abundance analyses for the three groups which displayed significant PERMANOVA results. For the plaque samples, more than 20 different species with an effect size larger than |1| (meaning larger than 1 or less than −1) were observed for the inulin group and three species for isomaltulose (see Table 2; Figure 5, Figure S3). However, based on FDR-adjusted p-values, only the inulin group displayed significant changes (both increase and decrease in abundance) for 13 species. Noteworthy, for all groups the confidence interval of the effect size was quite large and did cross zero (Table S4); this could be narrowed if group sizes were to increase. Further looking into the species with significant changes and at the range in relative abundance in the graphs (Figure 5, Figure S3) several subjects can be seen as outliers. In some cases, the same subject was an outlier for more than one species (data not shown). When performing the differential abundance analyses at genus level (Table S3, Figure S4), only for the inulin group did several genera show significant changes: decrease in Selenomonas, Tannerella, Leptotrichia, Corynebacterium, Campylobacter, and an increase in Neisseria, Abiotrophia, Burkholderiaceae family.
Table 2.
Results from ALDEx2 analysis of plaque samples. Species with effect size larger than |1| are reported.
| Genus | Species | p-value# | Effect size* | Rel. abundance visit 2, % | Rel. abundance visit 6, % | |
|---|---|---|---|---|---|---|
| Inulin (species decreasing) | Bergeyella | oral_taxon_907 | 0.046 | −1.501 | 0.010 | 0.001 |
| Leptotrichia | (species unknown) | 0.048 | −1.089 | 0.953 | 0.126 | |
| Selenomonas | (species unknown) | 0.061 | −1.274 | 0.326 | 0.068 | |
| Fusobacterium | nucleatum_subsp._animalis | 0.066 | −1.035 | 0.330 | 0.149 | |
| Corynebacterium | matruchotii | 0.081 | −0.856 | 2.995 | 2.545 | |
| Prevotella | oral_taxon_317 | 0.090 | −1.254 | 0.092 | 0.025 | |
| Leptotrichia | wadei | 0.101 | −1.442 | 0.538 | 0.337 | |
| Selenomonas | dianae | 0.123 | −1.074 | 0.021 | 0.011 | |
| Actinomyces | georgiae/s__sp._oral_taxon_178/s__sp._oral_taxon_877 | 0.179 | −1.271 | 0.053 | 0.018 | |
| Lachnoanaerobaculum | saburreum | 0.293 | −1.497 | 0.144 | 0.022 | |
| Leptotrichia | shahii | 0.319 | −1.288 | 0.159 | 0.156 | |
| Campylobacter | concisus | 0.527 | −1.074 | 0.129 | 0.083 | |
| Inulin (species increasing) | Fusobacterium | periodonticum | 0.038 | 1.511 | 0.203 | 0.295 |
| Prevotella | melaninogenica | 0.051 | 1.097 | 0.455 | 0.453 | |
| Streptococcus | sanguinis | 0.054 | 0.970 | 3.009 | 4.715 | |
| Neisseria | (species unknown) | 0.056 | 1.000 | 9.088 | 13.561 | |
| Family Burkholderiaceae (species unknown) | 0.057 | 1.399 | 0.014 | 0.013 | ||
| Granulicatella | adiacens | 0.073 | 0.790 | 1.244 | 1.500 | |
| Abiotrophia | defectiva | 0.074 | 1.021 | 0.350 | 0.682 | |
| Prevotella | nanceiensis | 0.107 | 1.527 | 0.077 | 0.098 | |
| Veillonella | dispar | 0.114 | 1.046 | 9.701 | 10.418 | |
| Haemophilus | parainfluenzae | 0.185 | 1.975 | 6.998 | 8.515 | |
| Capnocytophaga | sputigena | 0.212 | 1.186 | 0.439 | 0.707 | |
| Isomaltulose | Selenomonas | (species unknown) | 0.563 | −1.725 | 0.974 | 0.326 |
| Actinomyces | odontolyticus/oral_taxon_180 | 0.357 | 1.236 | 0.304 | 0.558 | |
| Neisseria | (species unknown) | 0.215 | 1.454 | 9.088 | 13.561 | |
# P-values represent Wilcoxon test from ALDEx2 package, with bold font indicating p-values <0.1 (default ALDEx2 cut off).
* Effect size represents the output from ALDEX “.effect” function.
(species unknown) – represents all of the ZOTUs with unknown species level aggregated to single unknown species using ”tax_glom” function in phyloseq package in R.
Figure 5.
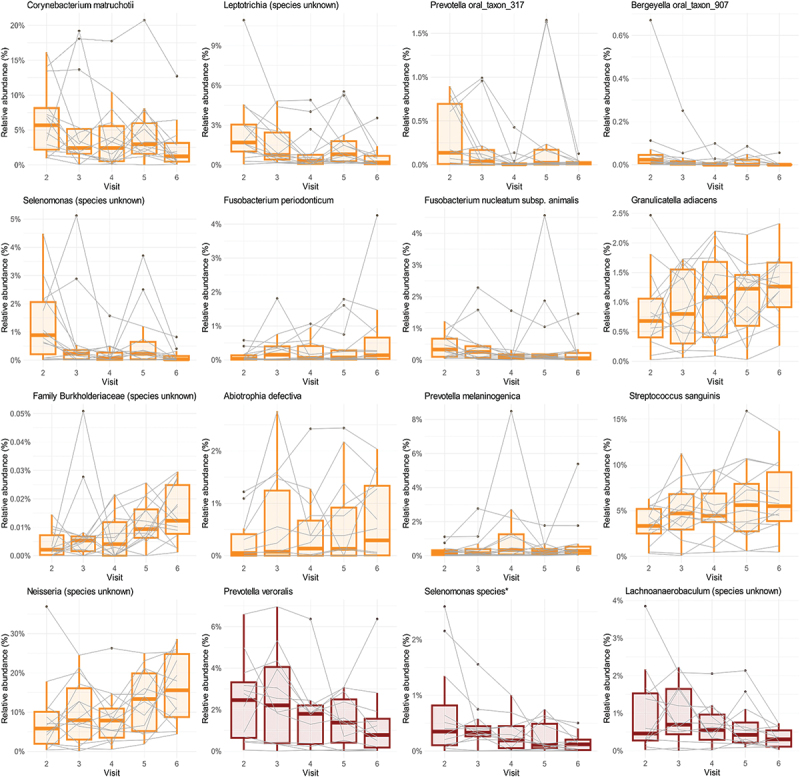
Relative abundance of bacterial species that changed significantly throughout the study. Only species with ALDEx2 adjusted p-value <0.1 are reported. Orange colour – inulin group (plaque samples), red – trehalose (tongue samples). For the full list of significantly affected species see Table 2.
Regarding the tongue samples, a single species had an effect size less than −1 for the isomaltulose group, representing a decrease in an unknown Leptotrichia species, but no significant changes were observed. On the other hand, the trehalose group showed a notable decrease for six species of which three decreased significantly (see Table 3; Figure 5; Figure S3). When species were further agglomerated to genus level, only the genus Selenomonas decreased significantly for the trehalose group (Figure S4).
Table 3.
Results from ALDEx2 analysis of tongue swab samples. Species with effect size larger than |1| are reported.
| Genus | Species | p-value# | Effect size* | Rel. abundance visit 2 | Rel. abundance visit 6 | |
|---|---|---|---|---|---|---|
| Isomaltulose | Leptotrichia | (species unknown) | 0.314 | −1.779 | 2.459 | 1.718 |
| Trehalose | Prevotella | veroralis | 0.080 | −1.504 | 1.151 | 0.525 |
| Lachnoanaerobaculum | (species unknown) | 0.086 | −1.711 | 1.023 | 0.333 | |
| Selenomonas | oral_taxon_136/oral_taxon_149/oral_taxon_478 | 0.092 | −1.226 | 0.684 | 0.151 | |
| Prevotella | salivae | 0.137 | −1.069 | 0.836 | 0.437 | |
| Prevotella | oral_taxon_309 | 0.454 | −1.574 | 0.069 | 0.008 | |
| Prevotella | pallens | 0.462 | −1.440 | 0.523 | 0.273 | |
| Rothia | mucilaginosa | 0.703 | 1.011 | 1.930 | 2.578 |
# P-values represent Wilcoxon test from ALDEx2 package, with bold font indicating p-values <0.1 (default ALDEx2 cut off).
* Effect size represents the output from ALDEX “.effect” function.
(species unknown) – represents all of the ZOTUs with unknown species level aggregated to single unknown species using “tax_glom” function in phyloseq package in R.
Secondary outcomes
The secondary study outcome – intensity of red fluorescent plaque – changed significantly for the inulin group over the study period (Friedman’s ANOVA p = 0.002) (Figure S2(D), Table S1). However, when comparing the second baseline visit to the end of the study, no significant changes were observed. The isomaltulose group showed the largest range for red fluorescent plaque values among all groups (Table S1).
Ancillary analyses
Salivary pH showed significant changes only for the trehalose group (Friedman’s ANOVA p = 0.039) over the study period. However, when the baseline was compared to the end of the study, no significant changes were observed (adjusted Wilcoxon, p > 0.05). Noteworthy, there was a large variation in baseline values for all groups (Figure S2 C; Table S2).
Discussion
Main findings
In this double-blind randomized controlled pilot-study, we analysed the effects of five commonly used sweeteners – glucose, inulin, isomaltulose, tagatose and trehalose – on the oral microbiome. The primary study outcome – change in microbial composition – was the most distinct for the inulin group after the 2-week thrice-daily rinsing period, followed by isomaltulose and trehalose. Further analyses revealed a compositional shift of varying degree in plaque for four groups: inulin, isomaltulose, tagatose and trehalose. In contrast to plaque, the tongue microbiota exhibited a comparative stability with some compositional changes after the use of isomaltulose and trehalose mouthrinse.
Thrice-daily rinsing with glucose did not significantly change the salivary pH level or the proportion of the red fluorescent plaque. When glucose is available, it can be used for energy by most microorganisms [55]. Additionally, it is not used for the formation of extra polymer matrix (EPS), which is one of the main components of oral biofilm [56,57]. Insoluble extracellular polymeric matrix is largely synthesized by oral streptococci, using sucrose (not glucose) via glucosyltransferases synthesising both soluble and insoluble glucans. Furthermore, research indicates that EPS is essential for some of the known cariogenic species, such as Streptococcus mutans, to increase its acid production and acid resistance. Thus, without EPS the virulence of Streptococcus mutans is significantly reduced [57]. This may explain why the microbial composition or red fluorescent plaque did not change after rinsing with glucose. Nevertheless, rinsing with glucose for two weeks has been shown to result in subsurface demineralization of enamel, in a study by Holmen et al. (1985), where orthodontic bands were used to accumulate plaque and to induce enamel demineralization in teeth planned for orthodontic extraction [58]. Their in vivo model excluded any exposure to fluoride and precluded removal of plaque from the studied surfaces. Therefore, to lower potential demineralization risk, our study participants did not have active caries at the start of the study and they maintained their normal oral hygiene routine while using a fluoride containing toothpaste (1450 ppm F). Unchanged salivary pH and red plaque fluorescence levels measured after the intervention period confirmed that no ecological changes were introduced in our study population while rinsing with glucose for two weeks. There are three main limitations for generalizing the cariogenic effects of glucose rinse in our study set-up. Firstly, we did not test changes in total bacterial load or plaque amount. Secondly, we only included orally healthy individuals (young adults) with low caries risk. To conclude, we do not consider the effect of daily glucose rinse neither neutral nor positive on oral ecosystem, but we can conclude that the current study conditions with sufficient oral hygiene and fluoride in subjects with low caries risk did not lead to ecological changes that might increase their risk towards developing caries.
Our findings suggest that among the tested sweeteners inulin possesses the largest oral microbiome modulating property. Inulin is an insoluble dietary fibre belonging to fructans, which can be consumed by certain bacteria. Bacteria then metabolize fibres into short-chain fatty acids and other metabolites, which have a significant benefit on the human gut metabolism [59,60]. In the field of oral microbiology, Doran et al. (2007) observed an increase of oral streptococcal counts when healthy individuals performed twice-daily rinsing with inulin for 21 days [25]. These findings align with our results where Streptococcus sanguinis was one of the taxa with increased abundance in the inulin group. Noteworthy, this species is associated with good oral health and can out-compete a significantly more cariogenic (and caries associated) Streptococcus mutans [61,62]. If we further compare the outcomes at species level, we observed an increase in taxa associated with healthy oral microbiome (Haemophilus parainfluenzae, Neisseria spp., and Granulicatella adiacens) [63] and a decrease of taxa associated with adverse periodontal health (Leptotrichia spp., Selenomonas spp., Fusobacterium nucleatum subsp. animalis) [63,64], and caries (Corynebacterium matruchotii) [65]. Furthermore, the reduction of Leptotrichia, Selenomonas and Corynebacterium was observed also at the genus level.
A more recent study showed that inulin supplementation of orally derived in vitro biofilms led to opposite results – a reduced relative abundance of Streptococcus and an increased relative abundance for Lactobacillus and Bifidobacterium [19]. However, the study exclusively used anaerobic culturing techniques, which could have selectively benefited the growth of certain species and does not fully represent the dynamic conditions of the oral cavity. Furthermore, the saliva samples used for cultures came from individuals with stage III or IV chronic periodontitis, limiting further comparison to the healthy individuals used in our study. Outside the oral cavity, the effects of inulin have mostly been analysed on gastrointestinal microbiome, showing increased growth of probiotic strains of Bifidobacterium and Lactobacillus [66].
When looking at the effects of inulin at a cytokine level, in diabetic obese patients, inulin supplementation was able to reduce diastolic blood pressure and decrease of proinflammatory gene expression (TNF-α mRNA) [67]. This could mean that specific target groups might benefit more from inulin supplementation than others. If a full-scale study on the impact of inulin on the oral microbiome can show oral health benefits, like those shown by gastrointestinal research, inulin could be a prebiotic sweetener alternative. However, for now, we cannot make any recommendations regarding inulin (or any of the other tested sweetener) supplementation to improve oral health.
Our results also indicated minor plaque microbiota altering properties of isomaltulose, trehalose and to lower effect – tagatose. Isomaltulose also displayed slight tongue microbial modulating properties, but the effect was smaller than that on plaque. The smaller modulating effect on tongue might be affected by the low tongue brushing frequency in the isomaltulose group but to understand effects on ‘tongue brushers’ and ‘non brushers’ a larger group size would be needed. Isomaltulose (PalatinoseTM) is a popular sucrose alternative used in the food industry and can be hydrolysed into glucose and fructose albeit at a slower rate than sucrose [68]. It is worth to mention that isomaltulose induces a drop in pH in the saliva directly after use [22], however we did not observe any long-term effects on the salivary pH in our study. Since isomaltulose is absorbed more slowly and is fully metabolized, it has been widely used as a sucrose alternative and specifically recommended for diabetic patients [18]. However, it has a cariogenic potential and is around 60% less sweet than regular sugar, meaning more is needed to achieve similar sweetness, thus it should not be overlooked as a caries risk-free alternative to sucrose [68].
In addition to isomaltulose, trehalose exhibited an even stronger tongue microbial modulating potential. This was unexpected as the tongue microbial composition is regarded as being more stable than plaque [69]. Onyango et al. 2020 tested the effects of trehalose (and other sweeteners) on an in vitro oral biofilm model, using synthetic communities consisting of 19 genera and human salivary bacteria inoculum. Onyango et al. 2020 observed that cultures grown with trehalose as the only carbon source were dominated by Streptococcus at genus level with a decrease in Neisseria and Prevotella. This aligns in par with our findings, where the differential abundance of several Prevotella species decreased in tongue swab samples following the trehalose two-week rinsing period. Noteworthy, their results report similar acid production levels for trehalose and sucrose, where Lactobacillus showed metabolic preference for trehalose over sucrose, thus the effects of trehalose may be stronger for people with caries and increased abundance of lactobacilli [21,70]. Our results for trehalose group could have been further influenced by the group exhibiting the greatest DMFS score from all of the study groups and, additionally, the largest range for the scores, indicating a very diverse group of individuals.
Tagatose is a hexose monosaccharide, with 92% of the sweetness but 38% of the calories of sucrose [17]. Several studies have reported the effect of tagatose on Streptococcus mutans and other oral streptococci, reducing their activity in both acid production and EPS synthesis even in the presence of sucrose [17,23,71,72]. We, however, did not observe such effects. Most of the currently published research has been conducted by researching the growth of different microorganism cultures with supplementation of a test sweetener. The only randomized controlled trial by Nagamine et al. 2020 used tagatose chewing gum and cultured salivary samples (in both anaerobic and aerobic conditions). They showed that tagatose limits bacterial growth in a culture but has no effect on Streptococcus mutans counts when used in chewing gum [72]. Not only the dynamics of chewing are different to rinsing but also the use of sequencing may limit the comparison to our study. Furthermore, Nagamine et al. 2020 used both a higher concentration of tagatose and a longer observation period (4 weeks). Interestingly, the authors report decreased counts of Streptococcus mutans in saliva once tagatose is mixed with xylitol (sweetener, sugar alcohol) [72]. Additional non-nutrient based application of tagatose powder has been found in periodontology in air particle abrasion method to polish roots and hard to reach areas during scaling and root planning. ApaPerio prophylaxis powder (Cumdente GmbH, Germany) has been marketed as a similar product to glycine polishing powder, made of very fine particle size − 15 µm, but no research has been published regarding the possible effects on the oral microbiome. Investigating the potential use of a prebiotic for deep periodontal pocket cleaning holds considerable interest due to its dual underlying benefits working as both a cleaning agent and a microbiome modulator. By conducting further research into the effects of tagatose on subgingival microbiome composition, we may gain valuable insights into its effects outside the currently streptococci centered research.
Secondary outcomes – red fluorescent plaque and salivary pH – did not change significantly for any of the groups. Red fluorescent plaque is defined as mature plaque that exhibits porphyrin metabolism and has been suggested to be indicative of poor oral health [33,73,74]. The proportion of such plaque did not change consistently for any of the sweeteners tested. We observed what appeared to be random fluctuation between study visits with no clear effect, neither positive nor negative. This could possibly be explained by the fact that measuring red fluorescence using a QLF-camera can be very sensitive to many factors, including ambient light and operator error [33]. Interestingly, of all the sweeteners tested, the isomaltulose group displayed the most contrasting results for red fluorescent plaque, meaning that plaque of some individuals had very intense fluorescence while some – hardly any. Furthermore, this was evident already at the second baseline visit, before any intervention took place. This high inter-individual variability indicates that the group size should be larger for red fluorescent plaque analyses. To evaluate possible effects further, a larger sample size with a longer exposure to the sweetener would be needed either by increasing the rinsing time or by increasing the length of the intervention. Additionally, increasing the concentration of the sweetener in the solution might also increase its availability in the plaque after the rest of it has left the oral cavity.
Strengths and limitations
To improve the reproducibility of our findings, we chose a method of differential abundance analysis that respects compositionality of microbiome (ALDEx2) and has shown to retain low false-discovery-rate even at very low sample sizes (n = 10–25) [45,52,75]. Our study assessed the effects of sweeteners on the oral microbiota using next generation sequencing. There are very limited number of studies that have explored this effect using sequencing techniques and even fewer have compared more than two sweeteners. Sequencing improves the range of the effect measured, without targeting specific taxa of interest, thus making the conclusions more applicable to a clinical setting.
Pilot studies like ours are needed to evaluate the effect size and thus calculate the sample size required to achieve the desired statistical power. As such, this was an exploratory study not directly aimed to make any clinical recommendations. Some of the groups experienced a drop-out and this was not uniformly distributed among study groups, thus leading to possible bias in the results. Additionally, despite the randomization of subjects, the range in age for the glucose group was notably different from the other groups (mainly isomaltulose), which could have affected the observed outcomes.
Future directions: Our results indicate that inulin possesses the strongest microbial modulating properties from the tested sweeteners. The observed species level changes in oral microbiota indicates a shift towards a healthy oral microbiome. However, more research is needed to evaluate clinical effects and the full microbiome effects with improved statistical power, as well as the long-term effects. Further research is needed for the whole field of sweeteners using next-generation sequencing, enabling the assessment of the effects on the microbiome as a community. Research is needed for the already existing sweetener applications in the use of particle abrasion to aid periodontal treatment. Using a prebiotic for this function might open an unexplored dual treatment option focused not only on mechanical debridement but microbiome modulation as well.
Conclusion
The effects on the oral microbiota are sweetener dependant with the strongest effect on the plaque microbiome compared to the tongue. Inulin exhibited the strongest microbiome modulating potential of the sweeteners tested. The use of an inulin mouth rinse increased the proportion of taxa associated with oral health but was not associated with changes in salivary pH or red plaque fluorescence. A limited reduction of taxa associated with poor periodontal health and caries was also observed. Further full-scale clinical studies are required for assessment of clinical and microbiological effects.
Key messages
Different sweeteners affect the oral microbiome in diverse ways and to a different extent.
Further clinical studies are required to further evaluate the changes in microbiome and how microbiological changes may influence clinical changes.
From the five tested sweeteners inulin exhibited the strongest microbiome modulating potential.
Acknowledgments
We would like to express our sincere appreciation and gratitude to Wendy (W.E.A.J.) de Wit, Michel (M.A.) Hoogenkamp, Rob (R.A.M.) Exterkate, Mark (M.J.) Buijs, Dono (D.) Kahharova, Danuta (D.) Mazurel, Elly (E.C.) van Deutekom-Mulder, Carolien (C.J.) Bosch-Tijhof, Pauline (P.S.) Ouwerling, Maud (W.) Pullens, Sytze (S.P.) Ouwerling, Najib (N.) Sawo and Ilona (I.F.) Persoon for contributing to the successful completion of this clinical study.
Funding Statement
The collaboration project between ACTA and TNO was co-funded by the PPP Allowance made available by Health~Holland, Top Sector Life Sciences & Health, to stimulate public-private partnerships. The project was further co-funded by GSK and Philips. Additional support was received from Ministry of Economic Affairs and Climate Policy (The Netherlands). DWK gratefully acknowledges the University of Amsterdam Data Science Centre for financial support provided for this project.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Davis R. Zakis: Formal Analysis, Visualization, Writing – Original Draft Preparation
Bernd W. Brandt: Formal Analysis, Visualization, Writing – Review & Editing
Suzette V. van der Waal: Conceptualization, Investigation, Writing – Review & Editing
Bart J. F. Keijser: Funding Acquisition, Conceptualization, Investigation, Writing – Review & Editing
Wim Crielaard: Funding Acquisition, Conceptualization, Investigation, Writing – Review & Editing
Derek W.K. van der Plas: Formal Analysis, Visualization, Writing – Review & Editing
Catherine M.C. Volgenant: Funding Acquisition, Conceptualization, 495 Investigation, Writing – Original Draft Preparation, Review & Editing
Egija Zaura: Funding Acquisition, Conceptualization, Investigation, Writing – Original Draft Preparation, Review & Editing
References
- [1].WHO . Global oral health status report: towards universal health coverage for oral health by 2030. Geneva: World Health Organ. 2022. doi: 10.1016/j.denabs.2011.11.005 [DOI] [Google Scholar]
- [2].Afshin A, Sur PJ, Fay KA, et al. Health effects of dietary risks in 195 countries, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet. 2019;393(10184):1958–15. doi: 10.1016/S0140-6736(19)30041-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Takahashi N. Oral microbiome metabolism: from ‘who are they?’ to ‘what are they doing? J Dent Res. 2015;94(12):1628–1637. doi: 10.1177/0022034515606045 [DOI] [PubMed] [Google Scholar]
- [4].Koopman JE, Hoogenkamp MA, Buijs MJ, et al. Changes in the oral ecosystem induced by the use of 8% arginine toothpaste. Arch Oral Biol. 2017;73:79–87. doi: 10.1016/j.archoralbio.2016.09.008 [DOI] [PubMed] [Google Scholar]
- [5].Ravald N, Birkhed D, Hamp SE. Root caries susceptibility in periodontally treated patients: results after 12 years. J Clin Periodontol. 1993;20(2):124–129. doi: 10.1111/j.1600-051X.1993.tb00326.x [DOI] [PubMed] [Google Scholar]
- [6].Teshome A, Muche A, Girma B. Prevalence of dental caries and associated factors in East Africa, 2000–2020: systematic review and meta-analysis. Front Public Health. 2021;9:1–15. doi: 10.3389/fpubh.2021.645091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu TT, Xiao J, Sohn MB, et al. Machine learning approach identified multi-platform factors for caries prediction in child-mother dyads. Front Cell Infect Microbiol. 2021;11:1–10. doi: 10.3389/fcimb.2021.727630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8(2):263–271. doi: 10.1177/08959374940080022001 [DOI] [PubMed] [Google Scholar]
- [9].Zaura E, Keijser BJ, Huse SM, et al. Defining the healthy ‘core microbiome’ of oral microbial communities. BMC Microbiol. 2009;9(1):1–12. doi: 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu RT, Walsh RFL, Sheehan AE. Prebiotics and probiotics for depression and anxiety: a systematic review and meta-analysis of controlled clinical trials. Neurosci Biobehav Rev. 2019;102:13–23. doi: 10.1016/j.neubiorev.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].DiMattia Z, Damani JJ, Van Syoc E, et al. Effect of probiotic supplementation on intestinal permeability in overweight and obesity: a systematic review of randomized controlled trials and animal studies. Adv Nutr. 2023;15(1):100162. doi: 10.1016/j.advnut.2023.100162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peckmezian T, Garcia-Larsen V, Wilkins K, et al. Microbiome-targeted therapies as an adjunct to traditional weight loss interventions: a systematic review and meta-analysis. Diabetes Metab Syndr Obes. 2022;15:3777–3798. doi: 10.2147/DMSO.S378396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ruottinen S, Karjalainen S, Pienihäkkinen K, et al. Sucrose intake since infancy and dental health in 10-year-old children. Caries Res. 2004;38(2):142–148. doi: 10.1159/000075938 [DOI] [PubMed] [Google Scholar]
- [14].Declerck D, Leroy R, Martens L, et al. Factors associated with prevalence and severity of caries experience in preschool children. Oral Epidemiol. 2008;36(2):168–178. doi: 10.1111/j.1600-0528.2007.00385.x [DOI] [PubMed] [Google Scholar]
- [15].Bowen WH. The stephan curve revisited. Odontology. 2013;101(1):2–8. doi: 10.1007/s10266-012-0092-z [DOI] [PubMed] [Google Scholar]
- [16].Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90(3):294–303. doi: 10.1177/0022034510379602 [DOI] [PubMed] [Google Scholar]
- [17].Hasibul K, Nakayama‑Imaohji H, Hashimoto M, et al. D‑Tagatose inhibits the growth and biofilm formation of streptococcus�mutans. Mol Med Rep. 2018;17:843–851. doi: 10.3892/mmr.2017.8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xie J, Li J, Qin Q, et al. Effect of isomaltulose on glycemic and insulinemic responses: a systematic review and meta-analysis of randomized controlled trials. Adv Nutr. 2022;13(5):1901–1913. doi: 10.1093/advances/nmac057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Colamarino AN, Johnson TM, Boudreaux DM, et al. Influence of Lactobacillus reuteri, Bifidobacterium animalis subsp. lactis, and prebiotic inulin on dysbiotic dental biofilm composition ex vivo. J Periodontol. 2022:1–12. doi: 10.1002/jper.22-0505 [DOI] [PubMed] [Google Scholar]
- [20].Ahmed A, Khan TA, Ramdath DD, et al. Rare sugars and their health effects in humans: a systematic review and narrative synthesis of the evidence from human trials. Nutr Rev. 2022;80(2):255–270. doi: 10.1093/nutrit/nuab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Onyango SO, De Clercq N, Beerens K, et al. Oral microbiota display profound differential metabolic kinetics and community shifts upon incubation with sucrose, trehalose, kojibiose, and xylitol. Appl Environ Microbiol. 2020;86(16):1–14. doi: 10.1128/AEM.01170-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lingström P, Lundgren F, Birkhed D, et al. Effects of frequent mouthrinses with palatinose and xylitol on dental plaque. Eur J Oral Sci. 1997;105(2):162–169. doi: 10.1111/j.1600-0722.1997.tb00195.x [DOI] [PubMed] [Google Scholar]
- [23].Mayumi S, Kuboniwa M, Sakanaka A, et al. Potential of prebiotic D-Tagatose for prevention of oral disease. Front Cell Infect Microbiol. 2021;11:1–14. doi: 10.3389/fcimb.2021.767944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Neta T, Takada K, Hirasawa M. Low-cariogenicity of trehalose as a substrate. J Dent. 2000;28(8):571–576. doi: 10.1016/S0300-5712(00)00038-5 [DOI] [PubMed] [Google Scholar]
- [25].Doran AL, Verran J. A clinical study on the effect of the prebiotic inulin in the control of oral malodour. Microb Ecol Health Dis. 2007;19(3):158–163. doi: 10.1080/08910600701521279 [DOI] [Google Scholar]
- [26].Daabiss M. American society of anaesthesiologists physical status classification. Indian J Anaesth. 2011;55(2):111–115. doi: 10.4103/0019-5049.79879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lie MA, Timmerman MF, Van der Velden U, et al. Evaluation of 2 methods to assess gingival bleeding in smokers and non-smokers in natural and experimental gingivitis. J Clin Periodontol. 1998;25(9):695–700. doi: 10.1111/j.1600-051X.1998.tb02509.x [DOI] [PubMed] [Google Scholar]
- [28].Van der Velden U. The Dutch periodontal screening index validation and its application in the Netherlands. J Clin Periodontol. 2009;36(12):1018–1024. doi: 10.1111/j.1600-051X.2009.01495.x [DOI] [PubMed] [Google Scholar]
- [29].Ooshima T, Izumitani A, Takei T, et al. Plaque formation of dietary isomaltulose in humans. Caries Res. 1990;24(1):48–51. doi: 10.1159/000261238 [DOI] [PubMed] [Google Scholar]
- [30].O’Brien-Nabors L. Alternative sweeteners, fourth edition. Boca Raton, Florida: Taylor & Francis; 2011. [Google Scholar]
- [31].Julious SA. Sample size of 12 per group rule of thumb for a pilot study. Pharm Stat. 2005;4(4):287–291. doi: 10.1002/pst.185 [DOI] [Google Scholar]
- [32].Volgenant CMC, van der Waal SV, Brandt BW, et al. The evaluation of the effects of two probiotic strains on the oral ecosystem: a randomized clinical trial. Front Oral Heal. 2022;3. doi: 10.3389/froh.2022.825017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Van Der Veen MH, Volgenant CMC, Keijser B, et al. Dynamics of red fluorescent dental plaque during experimental gingivitis - a cohort study. J Dent. 2016;48:71–76. doi: 10.1016/j.jdent.2016.02.010 [DOI] [PubMed] [Google Scholar]
- [34].WHO . Oral health surveys: basic methods. World Health Org. 1997;4(66):39–46. [Google Scholar]
- [35].Fluitman KS, van den Broek TJ, Nieuwdorp M, et al. Associations of the oral microbiota and Candida with taste, smell, appetite and undernutrition in older adults. Sci Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-02558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Keijser BJF, Agamennone V, van den Broek TJ, et al. Dose-dependent impact of oxytetracycline on the veal calf microbiome and resistome. BMC Genomics. 2019;20(1):1–14. doi: 10.1186/s12864-018-5419-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Edgar RC. UNOISE2: improved error-correction for illumina 16S and ITS amplicon sequencing. bioRxiv. 2016:081257. [Google Scholar]
- [39].Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- [40].Kahharova D, Brandt BW, Buijs MJ, et al. Maturation of the oral microbiome in Caries-free toddlers: a longitudinal study. J Dent Res. 2020;99(2):159–167. doi: 10.1177/0022034519889015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen T, Yu W-H, Izard J, et al. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010;2010:baq013. doi: 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang Q, Garrity GM, Tiedje JM, et al. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lahti L, Shetty S. microbiome R package. Bioconductor. 2017. doi: 10.18129/B9.bioc.microbiome [DOI] [Google Scholar]
- [44].R Core Team . R: a language and environment for statistical computing. 2023.
- [45].Gloor GB, Macklaim JM, Pawlowsky-Glahn V, et al. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:1–6. doi: 10.3389/fmicb.2017.02224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nearing JT, Douglas GM, Hayes MG, et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat Commun. 2022;13(1):1–16. doi: 10.1038/s41467-022-28034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Oksanen J, Gavin LS, Guillaume Blanchet F, et al. Stevens and eduard szoecs and helene wagner and matt barbour and michael bedward and B. vegan. Community Ecol Package. 2022. [Google Scholar]
- [48].Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].McMurdie PJ, Holmes S, Watson M. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fernandes AD, Reid JN, Macklaim JM, et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2(1):1–13. doi: 10.1186/2049-2618-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gloor GB, Macklaim JM, Fernandes AD. Displaying variation in large datasets: plotting a visual summary of effect sizes. J Comput Graph Stat. 2016;25(3):971–979. doi: 10.1080/10618600.2015.1131161 [DOI] [Google Scholar]
- [53].Fernandes AD, Vu MTHQ, Edward L-M, et al. A reproducible effect size is more useful than an irreproducible hypothesis test to analyze high throughput sequencing datasets. arXiv. 2018;1809.02623. [Google Scholar]
- [54].Fernandes AD, Macklaim JM, Linn TG, et al. ANOVA-Like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One. 2013;8(7):e67019. doi: 10.1371/journal.pone.0067019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Utreja D, Tewari A, Chawla HS. A study of influence of sugars on the modulations of dental plaque pH in children with rampant caries, moderate caries and no caries. J Indian Soc Pedod Prev Dent. 2010;28(4):278–281. doi: 10.4103/0970-4388.76158 [DOI] [PubMed] [Google Scholar]
- [56].Jakubovics NS, Goodman SD, Mashburn-Warren L, et al. The dental plaque biofilm matrix. Periodontol 2000. 2021;86(1):32–56. doi: 10.1111/prd.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Guo L, McLean JS, Lux R, et al. The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of streptococcus mutans. Sci Rep. 2015;5(1):1–11. doi: 10.1038/srep18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Holmen L, Thylstrup A, Øgaard B, et al. A polarized light microscopic study of progressive stages of enamel caries in vivo. Caries Res. 1985;19(4):348–354. doi: 10.1159/000260866 [DOI] [PubMed] [Google Scholar]
- [59].Wiele T, Van DeBoon N, Possemiers S, et al. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J Appl Microbiol. 2007;102(2):452–460. doi: 10.1111/j.1365-2672.2006.03084.x [DOI] [PubMed] [Google Scholar]
- [60].Deleu S, Machiels K, Raes J, et al. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine. 2021;66:103293. doi: 10.1016/j.ebiom.2021.103293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: streptococcus sanguinis and streptococcus gordonii interference with streptococcus mutans. J Bacteriol. 2008;190(13):4632–4640. doi: 10.1128/JB.00276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kreth J, Giacaman RA, Raghavan R, et al. The road less traveled – defining molecular commensalism with streptococcus sanguinis. Mol Oral Microbiol. 2017;32(3):181–196. doi: 10.1111/omi.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chen WP, Chang S-H, Tang C-Y, et al. Composition analysis and feature selection of the oral microbiota associated with periodontal disease. Biomed Res Int. 2018;2018:1–14. doi: 10.1155/2018/3130607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bacteria, G. Supragingival Microbes . Atlas oral microbiol. 2015. p. 41–65. doi: 10.1016/b978-0-12-802234-4.00003-3 [DOI] [Google Scholar]
- [65].Lundtorp Olsen C, Markvart M, Vendius VFD, et al. Short-term sugar stress induces compositional changes and loss of diversity of the supragingival microbiota. J Oral Microbiol. 2023;15(1). doi: 10.1080/20002297.2023.2189770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62(8):1112–1121. doi: 10.1136/gutjnl-2012-303304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Roshanravan N, Alamdari NM, Jafarabadi MA, et al. Effects of oral butyrate and inulin supplementation on inflammation-induced pyroptosis pathway in type 2 diabetes: a randomized, double-blind, placebo-controlled trial. Cytokine. 2020;131:155101. doi: 10.1016/j.cyto.2020.155101 [DOI] [PubMed] [Google Scholar]
- [68].Matsukubo T, Takazoe I. Sucrose substitutes and their role in caries prevention. Int Dent J. 2006;56(3):119–130. doi: 10.1111/j.1875-595X.2006.tb00083.x [DOI] [PubMed] [Google Scholar]
- [69].Lloyd-Price J, Mahurkar A, Rahnavard G, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550(7674):61–66. doi: 10.1038/nature23889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Caufield PW, Schön CN, Saraithong P, et al. Oral lactobacilli and dental caries: a model for niche adaptation in humans. J Dent Res. 2015;94(9_suppl):110S–118S. doi: 10.1177/0022034515576052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sawada D, Ogawa T, Miyake M, et al. Potent inhibitory effects of D-tagatose on the acid production and water-insoluble glucan synthesis of streptococcus mutans GS5 in the presence of sucrose. Acta Med Okayama. 2015;69(2):105–111. doi: 10.18926/AMO/53339 [DOI] [PubMed] [Google Scholar]
- [72].Nagamine Y, Hasibul K, Ogawa T, et al. D-Tagatose effectively reduces the number of streptococcus mutans and oral bacteria in healthy adult subjects: a chewing gum pilot study and randomized clinical trial. Acta Med Okayama. 2020;74(4):307–317. doi: 10.18926/AMO/60369 [DOI] [PubMed] [Google Scholar]
- [73].Lee ES, Kang SM, Ko HY, et al. Association between the cariogenicity of a dental microcosm biofilm and its red fluorescence detected by quantitative light-induced fluorescence-digital (QLF-D). J Dent. 2013;41(12):1264–1270. doi: 10.1016/j.jdent.2013.08.021 [DOI] [PubMed] [Google Scholar]
- [74].Volgenant CMC, Hoogenkamp MA, Buijs MJ, et al. Red fluorescent biofilm: the thick, the old, and the cariogenic. J Oral Microbiol. 2016;8(1):30346–30349. doi: 10.3402/jom.v8.30346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cappellato M, Baruzzo G, Camillo Di B, et al. Investigating differential abundance methods in microbiome data: a benchmark study. PLoS Comput Biol. 2022;18(9):e1010467. doi: 10.1371/journal.pcbi.1010467 [DOI] [PMC free article] [PubMed] [Google Scholar]


