Abstract
Although the full sequence of the human immunodeficiency virus type 1 (HIV-1) genome has been known for more than a decade, effective genetic antivirals have yet to be developed. Here we show that, of 22 regions examined, one highly conserved sequence (ACTCTTTGGCAACGA) near the 3′ end of the HIV-1 gag-pol transframe region, encoding viral protease residues 4 to 8 and a C-terminal Vpr-binding motif of p6Gag protein in two different reading frames, can be successfully targeted by an antisense peptide nucleic acid oligomer named PNAPR2. A disrupted translation of gag-pol mRNA induced at the PNAPR2-annealing site resulted in a decreased synthesis of Pr160Gag-Pol polyprotein, hence the viral protease, a predominant expression of Pr55Gag devoid of a fully functional p6Gag protein, and the excessive intracellular cleavage of Gag precursor proteins, hindering the processes of virion assembly. Treatment with PNAPR2 abolished virion production by up to 99% in chronically HIV-1-infected H9 cells and in peripheral blood mononuclear cells infected with clinical HIV-1 isolates with the multidrug-resistant phenotype. This particular segment of the gag-pol transframe gene appears to offer a distinctive advantage over other regions in invading viral structural genes and restraining HIV-1 replication in infected cells and may potentially be exploited as a novel antiviral genetic target.
The persistence of integrated proviruses in host cells presents formidable challenges to the treatment of human immunodeficiency virus type 1 (HIV-1) infection. Replication-competent HIV-1 can be recovered from resting CD4+ T lymphocytes even from individuals receiving potent combination antiretroviral therapy, whose plasma virus levels have remained below the detection limit for a prolonged period (19, 33, 36, 147, 153). The discovery of a long-lived viral reservoir, which is established early in the infection (18, 33, 147), has suggested that HIV-1 cannot be easily eradicated from infected individuals with the current treatments. Another impediment to controlling HIV-1 infection is the emergence of drug-resistant viral strains (21, 47, 50, 61, 62, 67, 78, 90, 107, 110, 111, 130). HIV-1 protease, in particular, seemingly tolerates extensive sequence variations (8, 76, 79), contributing to a rapid emergence of protease inhibitor-resistant strains, which can be cross-resistant to multiple protease inhibitors (21, 47, 90, 107, 110, 111). Indeed, multidrug-resistant (MDR) HIV-1 strains are isolated increasingly from patients who have been extensively treated with various antiretroviral agents of similar classes (151), and the spread of these MDR strains may become a serious threat to the containment of the AIDS epidemic in the future. The identification of novel viral targets is clearly needed to empower anti-HIV-1 therapeutic strategies. In recent years, viral coreceptors (25, 26, 39, 99, 105, 132), integrase (7, 29, 31, 93, 101, 115, 154, 155), and the viral nucleocapsid protein zinc finger motif (94, 117–120, 137) have emerged as novel antiviral targets. However, substantial progress has yet to be made before any of the candidate compounds can be brought to practical applications.
Another possible antiviral target pursued over the years is the HIV-1 genome itself. Antisense reagents, in particular, have been extensively investigated primarily in the form of nuclease-resistant phosphorothioate oligodeoxynucleotides (PsODN). However, these previous attempts targeting various regions of the HIV-1 genome with antisense PsODN have produced inconsistent results in chronically infected cells (2, 4, 5, 64, 66, 83–86, 92, 141, 148). The reasons for a lack of consistency may vary. Selected target sequences may have been less critical for virion production in chronically HIV-1-infected cells or less accessible to the PsODN molecules because of the RNA-binding proteins or intracellular folding of the target RNA. It is also possible that antisense PsODN molecules were simply ineffective in abating ribosome elongation (9, 14, 139) or that available antisense molecule numbers were insufficient to overcome the enormous amount of viral transcripts expressed in the chronically infected cells (140).
Selection of optimal genetic targets and the use of potent gene-intervening reagents are equally critical elements of a successful antigene or antisense strategy. Peptide nucleic acid (PNA) (27, 45, 103, 145), initially developed as a reagent for strand invasion of the duplex DNA, is a DNA mimic, consisting of a peptide backbone of N-(2-aminoethyl)glycine units in place of a deoxyribose backbone. Although unmodified PNA has a relatively poor cellular uptake compared to that of ODN (104, 144), it has unique molecular characteristics which may enhance its utility as a genome-intervening tool, such as resistance to nucleases and proteases (24) and sequence-specific hybridization to DNA or RNA targets using Watson-Crick base pair formation (27) with much higher thermal stability than that of ODN (27, 28, 51).
Using a PNA oligomer as a prototype molecular tool, we explored viral sequences that were susceptible to PNA-mediated inhibition of gene expression and asked whether such sequences might be considered potential anti-HIV-1 genetic targets in the current study. Target sequences were selected from the previously less explored gag-pol gene, with a particular focus on the pol gene, as few studies have tried to directly block the expression of viral enzymes. In particular, we were interested in the protease-encoding sequence that begins upstream of the 3′ end of the gag-pol transframe gene. Despite its extensive sequence variations, a short segment of sequence toward the 5′ end of the viral protease-encoding gene, ACTCTTTGGCAACGA, which also encodes the C-terminal Vpr-binding motif of p6Gag protein, LXXLFG, by a different reading frame (17, 68, 88), is highly conserved among various HIV-1 subtypes (Fig. 1). We hypothesized that nucleotide sequences of certain segments of the transframe domain must be invariably conserved if a single transcript has to encode critical amino acid sequences of two different proteins by a ribosomal frameshift, thus reducing the probability of escape mutants, and considered this particular sequence as one of the prime targets.
FIG. 1.
Schematic representation of the gag and pol regions of HIV-1 (top) and the amino acid sequences of portions of p6Gag protein and viral protease encoded by two different reading frames (bottom). The amino acid residues conserved in >98% of the majority HIV-1 substrains (group M), which include subtypes A, B, C, D, F, G, H, and J and circulating recombinant forms (AE, AG, AGI, and AB) (HIV Sequence Database [http://hiv-web.lanl.gov]), are shown in large capital letters. The B subtype consensus sequences are shown in small capital letters. The nucleotide sequence of our interest is shown in open letters. PR, protease; RH, RNase H; INT, integrase.
MATERIALS AND METHODS
PNA oligomers.
Because of the propensity of PNA molecules to be confined within the cytoplasm (104) (also see below), we decided to focus on the antisense intervention, primarily targeting the translation of viral RNA, and designed 22 PNA oligomers, 14- to 15-mers in length, complementary to the site of our interest within the gag-pol transframe domain or other highly conserved regions within the gag and pol genes for the initial screening (Table 1). PNA oligomers were synthesized on the Expedite Nucleic Acid Synthesis System (PerSeptive Biosystems, Inc., Framingham, Mass.) by Research Genetics, Inc., Huntsville, Ala. Although all these PNA oligomers were easily dissolved in water, some oligomers, including PNAGAG1, PNAGAG2, PNAPR1, PNAPR2, PNART1, PNART5, PNART8, and PNAINT3, formed fine precipitates when added to the culture medium at higher concentrations, as has previously been reported (104). There was no discernible relationship between the precipitate formation and effects on cell growth or HIV-1 production (see below).
TABLE 1.
Sequences of PNA oligomers and locations of targeted genes relative to the HXB2 numbering system
| PNA oligomer | Sequence (5′-3′) | Position in HIV-1 genome |
|---|---|---|
| PNAGag1 | CGCTCTCGCACCCAT | 790–804 |
| PNAGag2 | TCCCTGCTTGCCCAT | 894–908 |
| PNAGag3 | TCATCATTTCTTCT | 1818–1831 |
| PNAPR1 | CCAAAGAGTGATTTT | 2256–2270 |
| PNAPR2 | TCGTTGCCAAAGAGT | 2262–2276 |
| PNAPR3 | ATCATCTGCTCCTGT | 2328–2342 |
| PNAPR4 | CCCCCTATCATTTTT | 2384–2398 |
| PNART1 | TTTTACTGGTACAGT | 2568–2582 |
| PNART2 | CTGTCAATGGCCATT | 2617–2631 |
| PNART3 | ATTTTCAGGCCCAA | 2698–2711 |
| PNART4 | CTGTCTTTTTTCTTT | 2738–2752 |
| PNART5 | ATTGTACTGATATCT | 2976–2990 |
| PNART6 | AATCATCCATGTATT | 3094–3108 |
| PNART7 | TGCTGCCCTATTTCT | 3128–3142 |
| PNART8 | ATTGACAGTCCAGCT | 3300–3314 |
| PNARH1 | TGTGCTGGTACCCAT | 4151–4165 |
| PNAINT1 | TAGCCATTGCTCTCC | 4285–4299 |
| PNAINT2 | TCTACTTGTCCATGC | 4379–4393 |
| PNAINT3 | CTTCCTGCTAATTTT | 4535–4549 |
| PNAINT4 | CCCCCAATCCCCCCT | 4793–4807 |
| PNAINT5 | AATTTTGAATTTTT | 4883–4896 |
| PNAINT6 | TTGCTGGTCCTTTCC | 4933–4947 |
Evaluation of cellular uptake of PNA oligomers.
H9 cells chronically infected with HIV-1LAI (H9LAI) were incubated with fluorescein-tagged PNAPR2, PNAPR4, or PNART2 at 10, 30, and 100 μM in fresh RPMI 1640 complete medium supplemented with 13% fetal bovine serum (HyClone, Logan, Utah), 4 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml at 37°C in 5% CO2-containing humidified air overnight. The cells were fixed with 2% paraformaldehyde and examined by fluorescence microscopy or by fluorescence-activated cell sorter analysis.
Assessment of effects of PNA oligomers on HIV-1 infection in various cell culture systems.
H9LAI cells or H9 cells chronically infected with another HIV-1 strain, HIV-1RF (H9RF), were extensively washed to remove previously produced virions and incubated in a 96-well culture plate at 104 cells/well in 200 μl of RPMI 1640 complete medium in the absence or presence of PNA oligomers at various concentrations. After 4 days in culture, supernatant was collected from each well and the level of p24 Gag antigen was determined by radioimmunoassay (RIA) (HIV-1 p24 RIA kit; NEN Life Science Products, Boston, Mass.). To compare the antiviral effects of PNAPR2 and PNA oligomers targeting the sequences proximate to the PNAPR2-annealing site, H9LAI cells were cultured as described above in the absence or presence of 100 μM PNAPR2 or other PNA oligomers tested. The relative antiviral effects were computed from the following formula: (% p24 antigen suppression by a given PNA oligomer)/(% p24 antigen suppression by PNAPR2). Each PNA oligomer was tested in triplicate. H9LAI and H9RF cells were also cultured in a six-well culture plate at 105 cells/ml in the absence or presence of 30 to 100 μM PNAPR2. After 4 days, cells were counted by the trypan blue dye exclusion method and lysed at 106 cells/ml in phosphate-buffered saline (PBS) containing 0.5% Triton X-100. The amounts of p24 antigen in the culture supernatant and cell lysate were measured by RIA. The experiments were repeated at least three times.
Phytohemagglutinin (PHA)-activated peripheral blood mononuclear cells (PBMC) obtained from healthy blood bank donors (2 × 106 cells/ml) were infected with seven MDR HIV-1 clinical isolates (151) in RPMI 1640 complete medium supplemented with 2.5 ng of recombinant human interleukin-2 (R&D Systems, Minneapolis, Minn.) per ml and cultured for at least 2 weeks or until the p24 antigen levels in the culture medium persistently exceeded 30 ng/ml. The PNAPR2-targeting nucleotide sequence of the transframe domain of each isolate was consistent with the consensus B sequence. Freshly PHA-stimulated PBMC from healthy donors (2 × 106 cells/ml) were added to each culture flask every 3 to 4 days. PBMC (106 cells/ml) infected with each MDR isolate were vigorously washed and cocultured with PHA-stimulated PBMC (106 cells/ml) in 200 μl of recombinant human interleukin-2-containing complete medium in the absence or presence of 10 to 60 μM PNAPR2. The number of infectious virions released after 7 days in culture was determined by an assay with multinuclear activation of a galactosidase indicator (MAGI assay) (see below) using an indicator cell line, MAGI-CCR5 cells (16) (AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, contributed by Julie Overbaugh). All assays were performed in triplicate.
MT2 cells (104 cells/ml) were exposed to 64 50% tissue culture infectious doses of HIV-1NL4-3 at 37°C for 1 h. After a vigorous virus washout, the cells were cultured in 200 μl of RPMI 1640 complete medium at 37°C in 5% CO2-containing humidified air in the absence or presence of various PNA oligomers at 100 μM. The amount of p24 antigen produced by the MT2 cells was determined by RIA on day 7. All assays were performed in triplicate.
COS-7 cells were transfected with 1 μg of pNL4-3 by FuGENE 6 transfection reagent (Boehringer Mannheim, Indianapolis, Ind.) in a six-well culture plate (35-mm diameter) for 5 h, immediately followed by incubation in Dulbecco's modified Eagle medium (DMEM) supplemented with 13% fetal bovine serum (HyClone), 4 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml in the absence or presence of 100 μM PNAPR2. After 48 h, cells and supernatants were harvested from each sample and evaluated for viral protein expression and virion production. HLtat cells (AIDS Research and Reference Reagent Program; contributed by Barbara K. Felber and George Pavlakis) were transfected with 1 μg of p55M1-10 (kindly provided by George Pavlakis, National Cancer Institute-Frederick Cancer Research and Development Center) by FuGENE 6 transfection reagent (Boehringer Mannheim) for 5 h, immediately followed by incubation in complete DMEM in the absence or presence of 100 μM PNAPR2. After 24 h, HLtat cells were lysed for Western blot analysis.
MAGI assay.
The MAGI assay was employed to determine the number of newly produced virion particles in the culture supernatant of HIV-1-infected cells as described previously (65). Briefly, the HeLa–CD4–long terminal repeat–β-galactosidase indicator cells were plated in a 96-well tissue culture plate at 104 cells per well, each well containing 125 μl of complete DMEM, 24 h prior to the assay. On the following day, the cells were generally 20 to 30% confluent. The cells were washed with 200 μl of Opti-MEM (Life Technologies, Inc., Rockville, Md.) twice and then exposed to serially diluted infectious culture supernatants in a total volume of 30 μl per well in the presence of 20 μg of DEAE-dextran (Sigma, St. Louis, Mo.) per ml. The infectious titers of the supernatants from PHA-PBMC infected with MDR isolates were examined with the MAGI-CCR5 indicator cell line (16), using 30 or 60 μl of inoculum. After the plates were incubated at 37°C in 5% CO2-containing humidified air for 2 h, 140 μl of complete DMEM was added to each well. The plates were incubated for another 46 h, followed by fixation at room temperature with 1% formaldehyde and 0.2% glutaraldehyde in PBS for 5 min. The cells were then washed with PBS and incubated in 100 μl (per well) of staining solution containing 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 0.4 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per ml. The blue cells were counted under a microscope.
Electron microscopy.
Electron microscopic examination was performed as previously described (38). Briefly, the harvested cells were centrifuged at 1,500 × g for 5 min. The cell pellets were fixed in 1.25% glutaraldehyde and then in 1% osmium, dehydrated in graded alcohol, and embedded in pure epoxy resins. Thin sections (60 nm) were stained with uranyl acetate and lead citrate and stabilized by carbon evaporation for an examination.
RNA analysis.
The culture supernatant was subjected to microcentrifugation at 32,800 × g for 2 h to pellet virions (35, 142). Pelleted virion particles were subjected to RNA extraction as previously described (6), followed by reverse transcriptase PCR (RT-PCR) with a primer pair, SK38-SK39 (128, 129), to estimate the amount of virion-derived RNA. The harvested cells were subjected to RNA extraction as previously described (129) followed by RT-PCR using two primer pairs, SK38-SK39 (128, 129) and BSS-KPNA (102, 124), in order to evaluate the levels of unspliced and singly spliced HIV-1 RNA, respectively.
Western blot analysis.
The virion particles pelleted from the culture supernatant (see above) were lysed in viral lysis buffer (10 mM Tris [pH 7.4], 1 μM EDTA, 0.02% NP-40). The harvested cells were washed in PBS and lysed in cell lysis buffer (10 mM Tris [pH 7.4], 50 mM NaCl, 100 mM KCl, 1 mM EDTA, 1% NP-40, 1 mM phenylmethanesulfonyl fluoride) at 2 × 107 cells/ml at 4°C for 30 min, followed by centrifugation at 13,800 × g to remove cell debris. Protein concentrations of the cell lysates were determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). Virion-associated protein derived from 100 μl of supernatant and 2 to 25 μg of cellular protein were resolved by electrophoresis on sodium dodecyl sulfate–4 to 12% polyacrylamide gradient gels (Novex, San Diego, Calif.) under reducing conditions, followed by electroblotting onto a polyvinylidene difluoride membrane (Novex). The HIV-1 Gag proteins were visualized by a chemiluminescence detection system (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) using anti-p24Gag antiserum (Advanced Biotechnologies, Inc., Columbia, Md.) (2 μg of cellular protein per lane loaded for this antibody), anti-p17Gag monoclonal antibody (Advanced Biotechnologies, Inc.) (2 μg of protein per lane), and anti-p6Gag antiserum (kindly provided by Louis E. Henderson, National Cancer Institute-Frederick Cancer Research and Development Center) (5 μg of protein per lane). For the evaluations of pol gene products, antiprotease antiserum (kindly provided by Louis E. Henderson), (25 μg of protein loaded per lane) and anti-p66RT monoclonal antibody (AIDS Research and Reference Reagent Program; contributed by Paul Yoshihara) (15 μg of protein per lane) were employed. For a resolution of anti-p66RT antibody-reactive proteins, sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis was used instead of a 4 to 12% gradient gel. The anti-p66RT antibody is reactive with the p66RT but not the p51RT band by Western blot analysis (NIH AIDS Research and Reference Reagent Program Catalog). Anti-gp120 antiserum (AIDS Research and Reference Reagent Program; contributed by Margarita Quiroga) (10 μg of protein loaded per lane) was used to evaluate the amount of env gene product expressed in HIV-1-infected cells.
Peptide ELISA.
The affinity of rabbit anti-p6Gag antiserum (kindly provided by Louis E. Henderson) for a series of HIV-1 HXB2 (subtype B) Gag synthetic peptides, 441–458 (YKGRPGNFLQSRPEPTAP), 451–470 (SRPEPTAPPEESFRSGVETT), 459–479 (PEESFRSGVETTTPPQKQEPI), 470–489 (TTPPQKQEPIDKELYPLTSL), and 480–500 (DKELYPLTSLRSLFGNDPSSQ) (obtained through AIDS Research and Reference Reagent Program), which encompassed the entire p6Gag domain, was determined by enzyme-linked immunosorbent assay (peptide ELISA). Briefly, 96-well plates were coated with serially diluted Gag peptides (9.8 to 2,500 ng/well) in triplicate. Anti-p6Gag antiserum diluted 1:500 was incubated in the peptide-coated plates for 90 min at room temperature. After washing, horseradish peroxidase-labeled anti-rabbit secondary antibody was added to each well, followed by the peroxidase substrate. The antibody affinity for each peptide was determined by the optical density (OD) at 405 nm.
RESULTS
Cellular uptake of PNA and its intracellular localization.
First, in order to determine concentrations of PNAs required to achieve a sufficient cellular delivery in tissue culture, H9LAI cells were incubated with fluorescein-tagged PNA at 10 to 100 μM. We elected to deliver PNA by simply adding it to the culture medium, thus relying on endocytosis (42, 104, 131) rather than the microinjection technique commonly adopted in the previous studies (10, 45, 104). After an overnight incubation, H9LAI cells were examined under a fluorescence microscope. A clear fluorescent signal was demonstrated in the majority of cells incubated with PNA at ≥30 μM compared to untreated H9LAI cells (Fig. 2A to C), albeit the signal was virtually confined to the cytoplasm, as has previously been reported (104) (Fig. 2D). The fluorescence intensity increased in a dose-dependent manner as demonstrated by fluorescence-activated cell sorter analysis regardless of the sequences tested (Fig. 2E).
FIG. 2.
Cellular uptake of PNA. (A to C) Chronically HIV-1-infected H9LAI cells were incubated with fluorescein-tagged PNA at various concentrations overnight, fixed with 3.7% formaldehyde for 20 min, washed and resuspended in PBS, and examined by fluorescence microscopy. Shown are the cells incubated with 0 (A), 30 (B), and 60 (C) μM fluorescein-tagged PNAPR2. Note that the cells treated with fluorescein-tagged PNAPR2 were resuspended in a slightly larger volume of PBS than was the untreated control so that each cell could be clearly visualized. The panel on the right-hand side is the fluorescence image, and that on the left is the phase-contrast image of the same field. The discernible fluorescent signal was seen in the majority of cells at 30 μM PNA or greater. (D) Shown at a higher magnification are the cells incubated with 30 μM PNAPR2. Note that the fluorescent signal is virtually confined to the cytoplasm. (E) H9LAI cells incubated with fluorescein-tagged PNA were also evaluated by fluorescence-activated cell sorter analysis. Curves a, b, and c represent H9LAI cells incubated with 0, 30, and 60 μM fluorescein-tagged PNAPR2, respectively. Similar results were obtained with fluorescein-tagged PNAPR4 and PNART2.
PNAPR2, targeting the 3′ end of the transframe domain, decreases extracellular virion production while increasing intracellular concentration of Gag protein in HIV-1-infected cells.
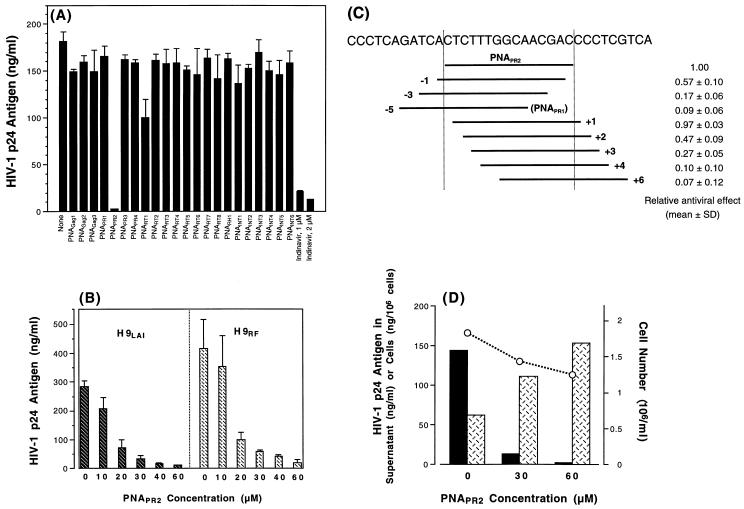
Preliminary effects of various PNA oligomers were evaluated in chronically HIV-1-infected H9LAI cells. While the majority of PNA oligomers, all tested at 100 μM, exhibited virtually no antiviral effect, PNAPR2 reduced the HIV-1 p24 antigen production in the culture supernatant by up to 98.4% (Fig. 3A). This p24-inhibitory effect of PNAPR2 appeared to be dose dependent in H9LAI as well as in H9RF cells (Fig. 3B) and specific to the PNAPR2-targeting region, as shifting the target sequence by two or more nucleotides upstream or downstream resulted in a more than 50% decrease in the inhibitory effect (Fig. 3C). Significant inhibition of p24 by PNAPR2 was also observed in MT2 cells acutely infected with HIV-1NL4-3 (inhibition of [99.6 ± 0.4] % [mean ± standard deviation (SD) of triplicate values]) compared to untreated HIV-1-infected MT2 cells. Similarly, PNAPR2 reduced infectious virion production from PBMC infected with clinical MDR isolates, originally isolated from extensively pretreated patients (151), by 97.3 to 99.4% (Table 2). These data suggested a potential antiviral effect of PNAPR2 against a broad spectrum of HIV-1 strains.
FIG. 3.
Effects of PNA oligomers on HIV-1 production in chronically HIV-1-infected cells. (A) Levels of p24 antigen in the supernatants of H9LAI cells cultured in the absence or presence of various PNA oligomers used at a 100 μM concentration. All compounds were tested in triplicate, and the values shown are means with SDs. The p24 antigen level in a supernatant of H9LAI treated with an HIV-1 protease inhibitor, indinavir, is shown as a reference. (B) Levels of p24 antigen in the supernatants of H9LAI and H9RF cells cultured in the absence or presence of various concentrations of PNAPR2. Each dose was tested in triplicate. The values shown are means with SDs. (C) The degrees of inhibition of p24 antigen production in H9LAI cells cultured with 100 μM PNAPR2 or other PNA oligomers targeting the sequences proximate to the PNAPR2-annealing site were compared. Each PNA oligomer was tested in triplicate. Each bar represents the target sequence shifted upstream (−) or downstream (+) by one to six nucleotides away from the PNAPR2-annealing site. The values shown are relative antiviral effects (means ± SDs). (D) Amounts of p24 antigen in the supernatant (solid bars) and cell lysate (crosshatched bars) of H9LAI cells cultured in the absence or presence of 30 and 60 μM PNAPR2. The cell numbers are indicated by circles. The experimental results shown are representative of three separate experiments.
TABLE 2.
HIV-1 virion production from PHA-PBMC infected with clinical MDR isolates and cultured in the absence or presence of 60 μM PNAPR2
| MDR isolate no. | Result of MAGI-CCR5 assaya
|
|
|---|---|---|
| No PNAPR2 | 60 μM PNAPR2 | |
| 1 | 467.7 ± 138.6 | 12.7 ± 4.2 (97.3) |
| 2 | 486.7 ± 112.0 | 4.7 ± 5.5 (99.0) |
| 3 | 210.0 ± 16.10 | 1.3 ± 0.6 (99.4) |
| 4 | 589.7 ± 32.10 | 7.7 ± 4.2 (98.7) |
| 5 | 214.0 ± 142.6 | 5.7 ± 1.5 (97.4) |
| 6 | 219.7 ± 25.20 | 3.7 ± 1.5 (98.3) |
| 7 | 208.3 ± 96.70 | 2.0 ± 1.0 (99.0) |
Results are shown as the mean numbers ± SDs of blue cells per well. Values in parentheses are the mean percent reductions.
In contrast to a marked decrease in the supernatant p24 antigen levels, the amount of intracellular p24 antigen was increased in H9LAI cells treated with 30 or 60 μM PNAPR2 (Fig. 3D) with moderately reduced cell numbers at the time of harvest. Comparable results were obtained in other experiments, in which PNAPR2 was tested up to 100 μM. Percent cell growth of H9LAI cells incubated with 60 to 100 μM PNAPR2 ranged from 68.3 to 78.1% (mean ± SD, [73.9 ± 5.1]%). Similar degrees of impaired cell growth were observed in H9RF as well as uninfected H9 cells incubated with up to 100 μM PNAPR2, while other PNA oligomers had no effect on cell proliferation in H9LAI, H9RF, or uninfected H9 cells (data not shown). These findings suggested that the impaired cell growth was specific to PNAPR2-targeted sequence intervention and may have resulted from an accumulation of p24 Gag protein within the cytoplasm and/or a down-modulation of certain host proteins that have yet to be identified, rather than from general toxicity of the PNA molecules.
Next, the processes of virion morphogenesis and virion release from the cell surface were examined by electron microscopy. Treatment with PNAPR2 appeared to hinder the steps of virion assembly, unlike protease inhibitors, which halt the process of virion morphological maturation. For 100 cells surveyed within one 60-nm thin section, the estimated average number of HIV-1 virions formed and released from the cells was reduced to less than two per sectioned cell (62 cells with no virion and 38 cells with less than five virions per cell) in PNAPR2-treated H9LAI cells compared to 10 to 15 virions per sectioned cell in untreated H9LAI cells, while there were no apparent changes in virion morphology (Fig. 4).
FIG. 4.
Electron microscopy of H9LAI cells cultured in the absence (A) or presence (B) of 60 μM PNAPR2. Shown are representatives of 100 cells surveyed in each sample (magnification, ×13,500). Virions are shown at a higher magnification (×54,000) in each inset on the right-hand side at the bottom.
PNAPR2 blocks production of Pr160Gag-Pol polyprotein and induces excessive intracellular cleavage of Pr55Gag.
To determine the mechanism(s) of virion assembly inhibition by PNAPR2, H9LAI cells cultured in the presence of 30 to 100 μM PNAPR2 were evaluated for viral protein expression and virion production. Consistent with the preliminary results, the level of p24 antigen production and the amount of pelletable virions were significantly reduced in the supernatant of PNAPR2-treated cells compared to the untreated control (Fig. 5A). The amounts of intracellular HIV-1 unspliced and singly spliced RNA transcripts were comparable between PNAPR2-treated and untreated cells (Fig. 5A), indicating that PNAPR2 did not interfere with the transcription of the HIV-1 genome. Strikingly, Western blot analysis with anti-p24Gag antibody demonstrated that PNAPR2-treated H9LAI cells contained predominantly p24 Gag protein with Gag precursor Pr55Gag markedly reduced and Pr160Gag-Pol virtually undetectable within the cells (Fig. 5A).
FIG. 5.
Effects of PNAPR2 on Gag-Pol expression and virion production in HIV-1-infected cells (A and B) and on Gag protein expression in HLtat cells transfected with the Gag expression plasmid p55M1-10 (C). (A) The amounts of viral RNA in the culture supernatant (pelleted virion derived) (lanes 1 to 3) and in the cells (lanes 4 to 6) were evaluated by RT-PCR (top). Arrows indicate the pertinent PCR products for each primer pair. The virion-derived Gag protein in the culture supernatant (lanes 1 to 3) and intracellular Gag protein processing (lanes 4 to 6) were evaluated by Western blot analysis using anti-p24Gag antibody (bottom). Shown are the positions of Pr160Gag-Pol, Pr55Gag, p41, and p24 Gag proteins. Lanes of each gel represent H9LAI cells cultured alone (lanes 1 and 4) or incubated with 100 μM PNAPR2 (lanes 2 and 5) and 2 μM indinavir (lanes 3 and 6). (B) Western blot analyses with anti-p24Gag antibody and anti-p17Gag antibody (top) and with anti-p6Gag antibody (bottom) of cell lysates processed from H9LAI cells cultured alone (lane 1) or in the presence of 30 μM PNAPR2 (lane 2), 60 μM PNAPR2 (lane 3), 2 μM indinavir alone (lane 4), 30 μM PNAPR2 plus 2 μM indinavir (lane 5), or 60 μM PNAPR2 plus 2 μM indinavir (lane 6). Cell lysate of uninfected H9 cells is shown in lane 7. PI − or + represents the absence or presence of a protease inhibitor, indinavir, respectively. The level of p24 antigen in each culture supernatant is indicated at the bottom. (C) Western blot analyses with anti-p24Gag antibody (left) and anti-p6Gag antibody (right) of cell lysates processed from HLtat cells transfected with pHXB2-based plasmid pSUM9 (136) (lanes 1) or with p55M1-10 and cultured alone (lanes 2) or in the presence of 100 μM PNARR2 (lanes 3). The cell lysate of mock-transfected HLtat cells is included as a reference (lanes 4). Sup, supernatant; Ag, antigen; Ab, antibody.
In order to further examine the expression and subsequent proteolytic processing of Gag protein in PNAPR2-treated cells, H9LAI cells were cultured with an HIV-1 protease inhibitor, indinavir, in addition to PNAPR2. The combination of indinavir and PNAPR2 resulted in a reduction of intracellular p24 and p17 Gag proteins and the reappearance of Pr55Gag protein, but not Pr160Gag-Pol (Fig. 5B). Because the PNAPR2-targeting site was near the 3′ end of the p6Gag-encoding region, the proteolytic processing of Pr55Gag, also a precursor of p6Gag protein, was examined by Western blot analysis with anti-p6Gag antibody. The peptide ELISA demonstrated that the anti-p6Gag antibody was most reactive to the peptide 480–500, which corresponds to the C-terminal domain of p6Gag protein, with the ODs linearly increasing from 0.303 ± 0.095 to 1.468 ± 0.052 (mean ± SD of triplicate values) in detecting 9.8 to 625 ng of the peptide per well, followed by the peptide 451–470, with ODs ranging from 0.212 ± 0.064 to 0.595 ± 0.067. The affinities to the other three peptides were negligible. Western blot analysis with the anti-p6Gag antibody could detect p6Gag protein in the HIV-1 virions (data not shown and reference 106) as well as its precursor (Pr55Gag) in the cell lysate of untreated H9LAI cells (Fig. 5B). This p6Gag antibody also demonstrated the p6Gag-containing intermediate (p15) in the lysate of untreated H9RF cells (data not shown). Notably, the cell lysate of H9LAI treated with 60 μM PNAPR2 contained virtually no Pr55Gag protein which could be detected by the anti-p6Gag antibody, even with the addition of a protease inhibitor, indinavir, while the anti-p24 antibody readily recognized Pr55Gag in the same lysate (Fig. 5B), indicating that the Pr55Gag expressed in PNAPR2-treated cells had insufficient binding determinants for the anti-p6Gag antibody. We then examined the effects of PNAPR2 on the expression of Pr55Gag protein in HLtat cells (32, 127) transfected with the Gag expression plasmid p55M1-10, in which Pr55Gag was not expected to be cleaved by the viral protease, as p55M1-10 contained solely the gag reading frame (126). While Western blot analysis with anti-p24Gag antibody demonstrated Pr55Gag protein in both untreated and PNAPR2-treated cells, the band intensity of anti-p6Gag antibody-reactive Pr55Gag was significantly diminished in PNAPR2-treated HLtat cells (Fig. 5C).
These data suggested that PNAPR2 treatment could induce the truncation of Pr55Gag toward the C terminus of p6Gag, presumably by disrupting a translation of the gag reading frame at its site of binding to the target mRNA in HIV-1-infected cells. It is also possible that PNAPR2 effectively blocked the translation of the gag-pol reading frame, resulting in generation of truncated Pr160Gag-Pol, namely, Pr55Gag with p6Pol which could not be detected by the anti-p6Gag antibody. Thus, the intracellular Gag precursor protein in PNAPR2-treated cells appeared to be predominantly Pr55Gag lacking the full-size p6Gag protein, which was excessively cleaved to the final products by a certain amount of viral protease.
PNAPR2 reduces synthesis of viral protease as well as triggering its premature activation.
We then asked whether preexisting viral protease at the beginning of H9LAI cell culture subsequently influenced the proteolytic processing of the viral protein. To this end, nonpermissive COS-7 cells were transfected with an infectious HIV-1 molecular clone, pNL4-3, immediately followed by incubation with 100 μM PNAPR2 in tissue culture. Virion production was significantly suppressed in PNAPR2-treated COS-7 cells as demonstrated by the reduced p24 antigen level and the decreased number of infectious virion particles in the culture supernatant determined by the MAGI assay (65). The intracellular Gag protein was predominantly p24 and p17 with significantly decreased amounts of Pr55Gag and virtually undetectable Pr160Gag-Pol as observed in chronically HIV-1-infected H9 cells (Fig. 6A). Consistent with the decreased quantity of full-size Pr160Gag-Pol, the amount of intracellular viral protease was reduced in PNAPR2-treated cells compared to untreated control (Fig. 6B). It should be noted that the antiprotease antibody that we employed in the current study detected a monomeric form of viral protease, not a homodimer, an active form of viral protease (112, 146). The latter is known to continually undergo autodegradation as it is being activated (123, 135). Thus, the amount of intracellular monomeric viral protease may not necessarily reflect the total amount of active viral protease within the cytoplasm but may simply correlate with the amount of precursor polyprotein. These results strongly suggested that the disrupted translation of gag-pol mRNA induced by PNAPR2 subsequently led to a premature activation of viral protease, albeit synthesized in probably a much smaller quantity than untreated control, resulting in an excessive intracellular cleavage of the Gag protein and an impairment of HIV-1 virion morphogenesis.
FIG. 6.
Effects of PNAPR2 on gag, pol, and env gene expression and virion production in COS-7 cells transfected with an infectious HIV-1 molecular clone, pNL4-3. Shown are results of Western blot analyses with anti-p24Gag antibody and anti-p17Gag antibody (A), antiprotease antibody and anti-p66RT antibody (B), and anti-gp120 antibody (C) of pNL4-3-transfected COS-7 cells cultured with no drug (lanes 1), 100 μM PNAPR2 (lanes 2), 1 μM saquinavir (also a protease inhibitor) (lanes 3), or 3 μM indinavir (lanes 4). The cell lysate of mock-transfected COS-7 cells is shown in lanes 5. Viral lysate from pelleted virion particles (vp) is included in the Western blot analyses for the pol gene products as a reference. The levels of p24 antigen in supernatant and the infectious titers determined by MAGI assay are shown at the bottom. Ab, antibody; Ag, antigen.
Interestingly, the amount of another pol gene product, RT, appeared to be disproportionately high compared to the low-level expression of its precursor, Pr160Gag-Pol polyprotein, in PNAPR2-treated cells. Western blot analysis using a monoclonal antibody against p66RT, which also recognizes the precursor (p160) and several intermediates, but not p51RT (NIH AIDS Research and Reference Reagent Program Catalog, 1998), clearly demonstrated p66RT in both untreated and PNAPR2-treated cells (Fig. 6B). However, the cell lysate of PNAPR2-treated COS-7 cells contained fewer precursor intermediates, smaller than 100 kDa in size (Fig. 6B). These data suggested that, in PNAPR2-treated cells, the translation of gag-pol mRNA may have been reinitiated at the appropriate AUG codon located downstream of the PNAPR2-targeting site by the mechanism described previously as ribosomal leaky scanning (71, 74, 75). A consensus sequence for the efficient initiation of translation by eukaryotic ribosomes has been reported as xxPuxxAUGG with a purine (Pu) in the −3 position (three nucleotides upstream of the AUG codon) having a dominant effect (70, 72, 73). The potential AUG codons downstream of the PNAPR2-annealing site were identified at various positions, including 2358 to 2360 (GAAAUGA), 2388 to 2390 (AAAAUGA), 2595 to 2597 (GGAAUGG), and 2670 to 2672 (GAGAUGG) of the HIV-1 HXB2 genome, preserving the amino acid sequence of the remaining Pol. If the translation is internally initiated at the sites listed above, resulting Pol proteins will be approximately 103.7, 102.5, 95.2, and 92.3 kDa, respectively. These Pol proteins may be cleaved by viral protease expressed in trans or by host proteases (34, 87). A similar pattern of p66RT expression was also observed in chronically infected H9 cells, and the addition of a viral protease inhibitor, indinavir, to PNAPR2 did not restore detection of the full-size precursor p160 (data not shown). Expression of HIV-1 Env was not affected by PNAPR2 in either COS-7 cells transfected with pNL4-3 (Fig. 6C) or chronically infected H9 cells (data not shown).
DISCUSSION
The introduction of protease inhibitors (55–57, 63, 77, 121) to conventional RT inhibitor-based antiretroviral therapy (23, 46, 95–97, 149, 150) has induced unprecedented degrees of viral load reduction and a recovery of CD4+ T-lymphocyte counts even in patients with advanced HIV-1 disease (20, 22, 43, 44, 91, 125). Substantial declines in AIDS incidence and death observed in the United States and other industrialized countries for the past 2 years have also been attributed to this improved treatment regimen (1, 15), often referred to as highly active antiretroviral therapy. While RT inhibitors, which block de novo HIV-1 infection of uninfected cells, have no control over production of virions from HIV-1-infected cells, protease inhibitors actively exert their effect in infected cells, driving them to release noninfectious immature virions and thereby preventing the spread of HIV-1. Dramatic effects of highly active antiretroviral therapy on individual cases of HIV-1 infection as well as on the global AIDS epidemic, although they may possibly be short-lived, underscore the importance of targeting the existing HIV-1 reservoir (HIV-1-infected cells) to better control HIV-1 disease, rather than simply providing protective shields for uninfected cells.
In the current study, we attempted to identify the viral gene sequences that were not only critical for the virion production but also accessible to gene-intervening molecules in chronically HIV-1-infected, provirus-laden cells. We showed that PNAPR2, which is complementary to a region of the viral gag-pol mRNA sequence near the 3′ end of the transframe domain, encoding protease residues 4 to 8 and the C-terminal Vpr binding motif of p6Gag protein, can effectively interrupt a translation of gag-pol mRNA. The disrupted translation of gag-pol mRNA at the PNAPR2-annealing site gave rise to Pr55Gag as a predominant Gag precursor, which lacked a full-length p6Gag protein due to a C-terminal truncation of p6Gag and/or production of Pr55Gag containing p6Pol protein. The arrest of gag-pol mRNA translation by PNAPR2 was not complete, thereby permitting a synthesis of a small quantity of full-size Pr160Gag-Pol polyprotein, which was presumably processed to viral protease.
Despite the small amount of viral protease synthesized in PNAPR2-treated cells, Gag precursors were almost entirely cleaved to the final products of p24 and p17 within the cytoplasm. These seemingly counterintuitive findings prompted us to examine the intracellular expression profiles of viral protease. The Western blot analysis of cell lysate with antiprotease antibody demonstrated a significantly decreased amount of monomeric viral protease in PNAPR2-treated COS-7 cells transfected with an HIV-1 molecular clone compared to untreated control, whereas an active form of homodimeric viral protease (112, 146) could not be visualized in either PNAPR2-treated or untreated cells, probably because the homodimer had undergone autolysis as it was being activated (123, 135). It has previously been reported that the level of viral protease activity required to properly process the precursor proteins may be reduced by 4- to 50-fold, below which a limited amount of proteolytic processing can still be demonstrated (122). Therefore, it would not have been surprising to see some degree of Gag precursor processing in PNAPR2-treated cells even if the actual quantity of active viral protease had been significantly decreased. However, the extent of intracellular Gag precursor cleavage observed in PNAPR2-treated cells notably exceeded that in the untreated control, resulting in a significant accumulation of p24 and p17 within the cells. These data strongly suggested that the rate or timing of protease activation was markedly accelerated within the cytoplasm of PNAPR2-treated cells.
The impact of PNAPR2-mediated translation arrest of gag-pol mRNA seems to be severalfold. Not only did it impede the synthesis of full-size Pr160Gag-Pol, leading to decreased production of viral protease, but it also appeared to trigger the premature activation of viral protease, resulting in excessive and untimely intracellular cleavage of Gag proteins and significantly reduced virion production. In HIV-1, approximately 5 to 10% of gag-pol mRNA translational events are mediated by −1 ribosomal frameshifting to produce Pr160Gag-Pol precursor polyprotein (52, 143), which is eventually incorporated into viral particles to provide viral protease, RT, and integrase. This delicately balanced synthesis of Pr55Gag and Pr160Gag-Pol and their intermolecular association are believed to play a critical role in coordinating sequential events of virion assembly and release. The final stages of virion morphogenesis begin with the interaction of Pr55Gag and Pr160Gag-Pol polyprotein (80, 81, 108, 133), which then attach to the plasma membrane of host cells (13, 41, 116). Viral protease embedded in Pr160Gag-Pol remains inactive until Gag and Gag-Pol precursors reach the plasma membrane, where precursor protein processing and virion assembly are initiated upon activation of the viral protease, followed by the budding and completion of virion maturation (58). Although viral protease-mediated precursor cleavage may take place within the cytoplasm (13, 41, 59), the membrane-associated precursor processing and assembly events must occur in a regulated sequence in order for the infectious virions to be released (58).
Excessive intracellular processing of Gag precursor proteins without concomitant extracellular production of mature virions as induced by PNAPR2 has also been observed in a number of other conditions, including (i) sole expression of Pr160Gag-Pol encoded in a single reading frame and thus lacking p6Gag (60, 109), (ii) p6Gag-deletion or -truncation HIV-1 molecular clones (40, 49, 152), and (iii) HIV-1 molecular clones containing p6Gag with mutated Vpr-binding motifs (49, 152). Notably, inactivation of the viral protease induced by mutational changes could restore the production of virions, albeit immature virions, in the p6Gag-deletion HIV-1 provirus-containing cells (49). These data suggest that the majority of viral protease may be prematurely released from the precursors and activated in the absence of fully functional p6Gag. How p6Gag protein exerts such a regulatory effect on viral protease has yet to be determined. It is possible that p6Gag may actively participate in the process of intermolecular association of Pr55Gag and Pr160Gag, in addition to the major homology domain of capsid protein (48, 134), such that the release of viral protease occurs in a coordinated fashion at the plasma membrane. Whether the binding of Vpr to p6Gag plays any role in regulating the processes of Gag and Gag-Pol precursor association and subsequent protease activation is unclear, since some studies have shown that a deletion of Vpr had no effect on viral infectivity or replication (3, 37).
In the current study, we could not determine whether the Pr55Gag synthesized in the PNAPR2-treated cells was the product of the gag reading frame and thus contained C-terminally truncated p6Gag or was predominantly the product of the gag-pol reading frame and had p6Pol protein. It is conceivable that the antisense PNA oligomer targeted downstream of the stem-loop structure within the gag-pol overlap region may have enhanced the ribosomal frameshifting, as has been demonstrated with the use of antisense oligonucleotides (138), resulting in increased synthesis of gag-pol gene products, most of which were nonetheless truncated toward the N terminus of the viral protease in PNAPR2-treated cells. Regardless, the lack of fully functional p6Gag protein was evidently as important a factor as the reduced synthesis of viral protease, both resulting from the PNAPR2-mediated interference with the translation of gag-pol mRNA, in blocking virion production from HIV-1-infected cells. In addition to these posttranscriptional events, it is also possible that the reverse transcription may be inhibited by the PNA oligomers tightly bound to the viral RNA target (69), thus preventing the integration of viral DNA in uninfected cells. This may potentially augment the overall antiviral effect of PNAPR2 in the setting of de novo HIV-1 infection.
One of the most challenging aspects of the development of antisense or antigene strategies is, in general, to identify the genetic targets the expression of which is crucial to the pathological process of disease. Targeting various regulatory genes of the HIV-1 genome has not always been effective against diverse HIV-1 strains (2, 64, 66, 83–85, 92, 141). The structural gag gene has also been targeted by antisense PsODN at a gag translation initiation site or various regions of capsid-encoding domain (4, 5, 66, 86, 141, 148). Such an anti-gag antisense strategy, however, exhibited only a modest, at most, antiviral effect in tissue culture. These previous studies have clearly demonstrated that a complete disruption of gag gene expression is difficult to achieve and that the moderate suppression of Gag protein production may not be substantially detrimental to HIV-1. Furthermore, even if the anti-gag antisense could block the translation of full-length Gag protein, eukaryotic ribosomes would most likely scan the downstream gag or gag-pol mRNA and identify the optimal initiation codon by leaky scanning as discussed above. The resulting truncated Gag and Gag-Pol may still be sufficient for virion particle assembly and release (11), or compensatory downstream mutations may arise to negate deletional effects and restore replication competency, as has been demonstrated for similar deletion mutants (82). The viral gene sequence identified by the current study appeared to be sufficiently accessible to the PNA oligomers within the living cells and presented a narrow window of opportunity to invade HIV-1 structural genes, as shifting the target by a few nucleotides upstream or downstream resulted in a significant loss of antiviral activity. This sequence, therefore, can be considered a vulnerable spot of the HIV-1 genome. Such genetically vulnerable spots may also be identified in other viruses that adopt a frameshift for the translation of critical proteins (12, 30, 53, 54, 89, 98, 100) and possibly exploited to develop pathogen-specific antiviral genetic intervention.
Development of optimal genome-blocking molecules with an appropriate delivery system is another critical step toward a successful antigene-antisense intervention. Antisense PsODN targeting the same sequence as PNAPR2 (PsODNPR2) did not show a substantial antiviral effect in chronically infected H9LAI or H9RF cells in our laboratory (data not shown). The antiviral effects of antisense PsODN are exhibited through several mechanisms, including sequence-specific suppression of transcription or translation and a sequence-independent inhibition of viral adsorption (83, 85, 148). Moreover, the translational suppression induced by antisense PsODN is predominantly mediated by degradation of target RNA by RNase H (9, 14, 139), unlike PNA, which blocks ribosomal elongation in an RNase H-independent manner (45). Thus, PsODN may not be the best possible agent to efficiently block genes in cells that are already infected with HIV-1, and this may explain the discrepancy between the antiviral effects of PNAPR2 and PsODNPR2 observed in our laboratory. Although PNA exhibits seemingly superior properties as a DNA-RNA-blocking tool compared with other oligonucleotide analogs, the current unmodified form of PNA cannot immediately be utilized as a therapeutic agent. Because of the poor cellular uptake of PNA, relatively higher concentrations of PNA molecules in tissue culture were required to sufficiently block the expression of the target sequence in our study. A conjugation of certain transporter molecules to the PNA may facilitate its cellular uptake (113, 114) and may prove useful if the gene-blocking ability of such a modified PNA construct is maintained. Innovative and optimal modifications to the existing PNA molecules or development of novel gene-intervening agents will clearly advance the efforts to develop potent antiviral therapeutics targeting viral genes.
ACKNOWLEDGMENTS
We thank Robert Wittes, John Erickson, and Yoshitatsu Sei for critical reviews of the manuscript; Louis E. Henderson for kindly providing anti-p6Gag and antiprotease antisera and George Pavlakis for kindly providing p55M1-10; Douglas Ferris, Eiichi Kodama, and Mutsuko Kumagai for helpful discussions; Elena Afonina for help with fluorescence microscopy; and Kathleen Noer for fluorescence-activated cell sorting analysis.
This work was supported, in part, by federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-56000.
REFERENCES
- 1.Aalen O O, Farewell V T, De Angelis D, Day N E, Gill O N. New therapy explains the fall in AIDS incidence with a substantial rise in number of persons on treatment expected. AIDS. 1999;13:103–108. doi: 10.1097/00002030-199901140-00014. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal S, Ikeuchi T, Sun D, Sarin P S, Konopka A, Maizel J, Zamecnik P C. Inhibition of human immunodeficiency virus in early infected and chronically infected cells by antisense oligodeoxynucleotides and their phosphorothioate analogues. Proc Natl Acad Sci USA. 1989;86:7790–7794. doi: 10.1073/pnas.86.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J Virol. 1996;70:1505–1511. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anazodo M I, Duta E, Friesen A D, Wright J A. Relative levels of inhibition of p24 gene expression by different 20-mer antisense oligonucleotide sequences targeting nucleotides + 1129 to + 1268 of the HIV-1 gag genome: an analysis of mechanism. Biochem Biophys Res Commun. 1996;229:305–309. doi: 10.1006/bbrc.1996.1797. [DOI] [PubMed] [Google Scholar]
- 5.Anazodo M I, Wainberg M A, Friesen A D, Wright J A. Sequence-specific inhibition of gene expression by a novel antisense oligodeoxynucleotide phosphorothioate directed against a nonregulatory region of the human immunodeficiency virus type 1 genome. J Virol. 1995;69:1794–1801. doi: 10.1128/jvi.69.3.1794-1801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki-Sei S, Yarchoan R, Kageyama S, Hoekzema D, Pluda J M, Wyvill K M, Broder S, Mitsuya H. Plasma HIV-1 viremia in HIV-1 infected individuals assessed by polymerase chain reaction. AIDS Res Hum Retrovir. 1992;8:1263–1270. doi: 10.1089/aid.1992.8.1263. [DOI] [PubMed] [Google Scholar]
- 7.Artico M, Di Santo R, Costi R, Novellino E, Greco G, Massa S, Tramontano E, Marongiu M E, De Montis A, La Colla P. Geometrically and conformationally restrained cinnamoyl compounds as inhibitors of HIV-1 integrase: synthesis, biological evaluation, and molecular modeling. J Med Chem. 1998;41:3948–3960. doi: 10.1021/jm9707232. [DOI] [PubMed] [Google Scholar]
- 8.Barrie K A, Perez E E, Lamers S L, Farmerie W G, Dunn B M, Sleasman J W, Goodenow M M. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within gag/pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology. 1996;219:407–416. doi: 10.1006/viro.1996.0266. [DOI] [PubMed] [Google Scholar]
- 9.Boiziau C, Thuong N T, Toulme J J. Mechanisms of the inhibition of reverse transcription by antisense oligonucleotides. Proc Natl Acad Sci USA. 1992;89:768–772. doi: 10.1073/pnas.89.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonham M A, Brown S, Boyd A L, Brown P H, Bruckenstein D A, Hanvey J C, Thomson S A, Pipe A, Hassman F, Bisi J E, et al. An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borsetti A, Ohagen A, Gottlinger H G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brierley I, Boursnell M E, Binns M M, Bilimoria B, Blok V C, Brown T D, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazenave C, Stein C A, Loreau N, Thuong N T, Neckers L M, Subasinghe C, Helene C, Cohen J S, Toulme J J. Comparative inhibition of rabbit globin mRNA translation by modified antisense oligodeoxynucleotides. Nucleic Acids Res. 1989;17:4255–4273. doi: 10.1093/nar/17.11.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. HIV/AIDS surveillance report, midyear ed. vol. 10. Centers for Disease Control and Prevention, Public Health Service. Atlanta, Ga: Department of Health and Human Services; 1998. [Google Scholar]
- 16.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Checroune F, Yao X J, Gottlinger H G, Bergeron D, Cohen E A. Incorporation of Vpr into human immunodeficiency virus type 1: role of conserved regions within the P6 domain of Pr55gag. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:1–7. [PubMed] [Google Scholar]
- 18.Chun T W, Engel D, Berrey M M, Shea T, Corey L, Fauci A S. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collier A C, Coombs R W, Schoenfeld D A, Bassett R L, Timpone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichman R C, Hooper C, Corey L. Treatment of human immunodeficiency virus infection with saquinavir, zidovudine, and zalcitabine. AIDS Clinical Trials Group. N Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 21.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 22.Danner S A, Carr A, Leonard J M, Lehman L M, Gudiol F, Gonzales J, Raventos A, Rubio R, Bouza E, Pintado V, et al. A short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. European-Australian Collaborative Ritonavir Study Group. N Engl J Med. 1995;333:1528–1533. doi: 10.1056/NEJM199512073332303. [DOI] [PubMed] [Google Scholar]
- 23.De Clercq E. HIV inhibitors targeted at the reverse transcriptase. AIDS Res Hum Retrovir. 1992;8:119–134. doi: 10.1089/aid.1992.8.119. [DOI] [PubMed] [Google Scholar]
- 24.Demidov V V, Potaman V N, Frank-Kamenetskii M D, Egholm M, Buchard O, Sonnichsen S H, Nielsen P E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 25.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 26.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S H, Goetz M B, Daar E S, Doms R W, O'Brien W A. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egholm M, Buchardt O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 28.Egholm M, Buchardt O, Nielsen P E, Berg R H. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral peptide backbone. J Am Chem Soc. 1992;114:1895–1897. [Google Scholar]
- 29.Eich E, Pertz H, Kaloga M, Schulz J, Fesen M R, Mazumder A, Pommier Y. (−)-Arctigenin as a lead structure for inhibitors of human immunodeficiency virus type-1 integrase. J Med Chem. 1996;39:86–95. doi: 10.1021/jm950387u. [DOI] [PubMed] [Google Scholar]
- 30.Falk H, Mador N, Udi R, Panet A, Honigman A. Two cis-acting signals control ribosomal frameshift between human T-cell leukemia virus type II gag and pro genes. J Virol. 1993;67:6273–6277. doi: 10.1128/jvi.67.10.6273-6277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnet C M, Wang B, Hansen M, Lipford J R, Zalkow L, Robinson W E, Jr, Siegel J, Bushman F. Human immunodeficiency virus type 1 cDNA integration: new aromatic hydroxylated inhibitors and studies of the inhibition mechanism. Antimicrob Agents Chemother. 1998;42:2245–2253. doi: 10.1128/aac.42.9.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felber B K, Drysdale C M, Pavlakis G N. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 34.Flexner C, Broyles S S, Earl P, Chakrabarti S, Moss B. Characterization of human immunodeficiency virus gag/pol gene products expressed by recombinant vaccinia viruses. Virology. 1988;166:339–349. doi: 10.1016/0042-6822(88)90504-1. [DOI] [PubMed] [Google Scholar]
- 35.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retrovir. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 38.Gonda M A, Wong-Staal F, Gallo R C, Clements J E, Narayan O, Gilden R V. Sequence homology and morphologic similarity of HTLV-III and visna virus, a pathogenic lentivirus. Science. 1985;227:173–177. doi: 10.1126/science.2981428. [DOI] [PubMed] [Google Scholar]
- 39.Gong W, Howard O M, Turpin J A, Grimm M C, Ueda H, Gray P W, Raport C J, Oppenheim J J, Wang J M. Monocyte chemotactic protein-2 activates CCR5 and blocks CD4/CCR5-mediated HIV-1 entry/replication. J Biol Chem. 1998;273:4289–4292. doi: 10.1074/jbc.273.8.4289. [DOI] [PubMed] [Google Scholar]
- 40.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray G D, Basu S, Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-alkylamino oligodeoxynucleotides, 2′-O-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem Pharmacol. 1997;53:1465–1476. doi: 10.1016/s0006-2952(97)82440-9. [DOI] [PubMed] [Google Scholar]
- 43.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 44.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 45.Hanvey J C, Peffer N J, Bisi J E, Thomson S A, Cadilla R, Josey J A, Ricca D J, Hassman C F, Bonham M A, Au K G, et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 46.Hargrave K D, Proudfoot J R, Grozinger K G, Cullen E, Kapadia S R, Patel U R, Fuchs V U, Mauldin S C, Vitous J, Behnke M L, et al. Novel non-nucleoside inhibitors of HIV-1 reverse transcriptase. 1. Tricyclic pyridobenzo- and dipyridodiazepinones. J Med Chem. 1991;34:2231–2241. doi: 10.1021/jm00111a045. [DOI] [PubMed] [Google Scholar]
- 47.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang M, Martin M A. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J Virol. 1997;71:4472–4478. doi: 10.1128/jvi.71.6.4472-4478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Husson R N, Shirasaka T, Butler K M, Pizzo P A, Mitsuya H. High-level resistance to zidovudine but not to zalcitabine or didanosine in human immunodeficiency virus from children receiving antiretroviral therapy. J Pediatr. 1993;123:9–16. doi: 10.1016/s0022-3476(05)81530-6. [DOI] [PubMed] [Google Scholar]
- 51.Igloi G L. Variability in the stability of DNA-peptide nucleic acid (PNA) single-base mismatched duplexes: real-time hybridization during affinity electrophoresis in PNA-containing gels. Proc Natl Acad Sci USA. 1998;95:8562–8567. doi: 10.1073/pnas.95.15.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 53.Jacks T, Townsley K, Varmus H E, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 55.Johnson V A, Merrill D P, Chou T C, Hirsch M S. Human immunodeficiency virus type 1 (HIV-1) inhibitory interactions between protease inhibitor Ro 31-8959 and zidovudine, 2′,3′-dideoxycytidine, or recombinant interferon-alpha A against zidovudine-sensitive or -resistant HIV-1 in vitro. J Infect Dis. 1992;166:1143–1146. doi: 10.1093/infdis/166.5.1143. [DOI] [PubMed] [Google Scholar]
- 56.Kageyama S, Mimoto T, Murakawa Y, Nomizu M, Ford H, Jr, Shirasaka T, Gulnik S, Erickson J, Takada K, Hayashi H, Broder S, Kiso Y, Mitsuya H. In vitro anti-human immunodeficiency virus (HIV) activities of transition state mimetic HIV protease inhibitors containing allophenylnorstatine. Antimicrob Agents Chemother. 1993;37:810–817. doi: 10.1128/aac.37.4.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kageyama S, Weinstein J N, Shirasaka T, Kempf D J, Norbeck D W, Plattner J J, Erickson J, Mitsuya H. In vitro inhibition of human immunodeficiency virus (HIV) type 1 replication by C2 symmetry-based HIV protease inhibitors as single agents or in combinations. Antimicrob Agents Chemother. 1992;36:926–933. doi: 10.1128/aac.36.5.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan A H, Swanstrom R. Human immunodeficiency virus type 1 Gag proteins are processed in two cellular compartments. Proc Natl Acad Sci USA. 1991;88:4528–4532. doi: 10.1073/pnas.88.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karacostas V, Wolffe E J, Nagashima K, Gonda M A, Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 61.Kavlick M F, Shirasaka T, Kojima E, Pluda J M, Hui F, Jr, Yarchoan R, Mitsuya H. Genotypic and phenotypic characterization of HIV-1 isolated from patients receiving (−)-2′,3′-dideoxy-3′-thiacytidine. Antivir Res. 1995;28:133–146. doi: 10.1016/0166-3542(95)00044-m. [DOI] [PubMed] [Google Scholar]
- 62.Kavlick M F, Wyvill K, Yarchoan R, Mitsuya H. Emergence of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 variants, viral sequence variation, and disease progression in patients receiving antiretroviral chemotherapy. J Infect Dis. 1998;177:1506–1513. doi: 10.1086/515324. [DOI] [PubMed] [Google Scholar]
- 63.Kempf D J, Marsh K C, Denissen J F, McDonald E, Vasavanonda S, Flentge C A, Green B E, Fino L, Park C H, Kong X P, et al. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S G, Hatta T, Tsukahara S, Nakashima H, Yamamoto N, Shoji Y, Takai K, Takaku H. Antiviral effect of phosphorothioate oligodeoxyribonucleotides complementary to human immunodeficiency virus. Bioorg Med Chem. 1995;3:49–54. doi: 10.1016/0968-0896(94)00142-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinchington D, Galpin S, Jaroszewski J W, Ghosh K, Subasinghe C, Cohen J S. A comparison of gag, pol and rev antisense oligodeoxynucleotides as inhibitors of HIV-1. Antivir Res. 1992;17:53–62. doi: 10.1016/0166-3542(92)90090-r. [DOI] [PubMed] [Google Scholar]
- 67.Kojima E, Shirasaka T, Anderson B D, Chokekijchai S, Steinberg S M, Broder S, Yarchoan R, Mitsuya H. Human immunodeficiency virus type 1 (HIV-1) viremia changes and development of drug-related mutations in patients with symptomatic HIV-1 infection receiving alternating or simultaneous zidovudine and didanosine therapy. J Infect Dis. 1995;171:1152–1158. doi: 10.1093/infdis/171.5.1152. [DOI] [PubMed] [Google Scholar]
- 68.Kondo E, Gottlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koppelhus U, Zachar V, Nielsen P E, Liu X, Eugen-Olsen J, Ebbesen P. Efficient in vitro inhibition of HIV-1 gag reverse transcription by peptide nucleic acid (PNA) at minimal ratios of PNA/RNA. Nucleic Acids Res. 1997;25:2167–2173. doi: 10.1093/nar/25.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kozak M. Evaluation of the “scanning model” for initiation of protein synthesis in eucaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- 72.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 73.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5262. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 77.Lam P Y, Jadhav P K, Eyermann C J, Hodge C N, Ru Y, Bacheler L T, Meek J L, Otto M J, Rayner M M, Wong Y N, et al. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- 78.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 79.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee Y M, Tian C J, Yu X F. A bipartite membrane-binding signal in the human immunodeficiency virus type 1 matrix protein is required for the proteolytic processing of Gag precursors in a cell type-dependent manner. J Virol. 1998;72:9061–9068. doi: 10.1128/jvi.72.11.9061-9068.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 82.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lisziewicz J, Sun D, Klotman M, Agrawal S, Zamecnik P, Gallo R. Specific inhibition of human immunodeficiency virus type 1 replication by antisense oligonucleotides: an in vitro model for treatment. Proc Natl Acad Sci USA. 1992;89:11209–11213. doi: 10.1073/pnas.89.23.11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lisziewicz J, Sun D, Lisziewicz A, Gallo R C. Antitat gene therapy: a candidate for late-stage AIDS patients. Gene Ther. 1995;2:218–222. [PubMed] [Google Scholar]
- 85.Lisziewicz J, Sun D, Metelev V, Zamecnik P, Gallo R C, Agrawal S. Long-term treatment of human immunodeficiency virus-infected cells with antisense oligonucleotide phosphorothioates. Proc Natl Acad Sci USA. 1993;90:3860–3864. doi: 10.1073/pnas.90.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lisziewicz J, Sun D, Weichold F F, Thierry A R, Lusso P, Tang J, Gallo R C, Agrawal S. Antisense oligodeoxynucleotide phosphorothioate complementary to Gag mRNA blocks replication of human immunodeficiency virus type 1 in human peripheral blood cells. Proc Natl Acad Sci USA. 1994;91:7942–7946. doi: 10.1073/pnas.91.17.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lowe D M, Aitken A, Bradley C, Darby G K, Larder B A, Powell K L, Purifoy D J, Tisdale M, Stammers D K. HIV-1 reverse transcriptase: crystallization and analysis of domain structure by limited proteolysis. Biochemistry. 1988;27:8884–8889. doi: 10.1021/bi00425a002. [DOI] [PubMed] [Google Scholar]
- 88.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mador N, Panet A, Honigman A. Translation of gag, pro, and pol gene products of human T-cell leukemia virus type 2. J Virol. 1989;63:2400–2404. doi: 10.1128/jvi.63.5.2400-2404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Markowitz M, Saag M, Powderly W G, Hurley A M, Hsu A, Valdes J M, Henry D, Sattler F, La Marca A, Leonard J M, et al. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. N Engl J Med. 1995;333:1534–1539. doi: 10.1056/NEJM199512073332204. [DOI] [PubMed] [Google Scholar]
- 92.Matsukura M, Zon G, Shinozuka K, Robert-Guroff M, Shimada T, Stein C A, Mitsuya H, Wong-Staal F, Cohen J S, Broder S. Regulation of viral expression of human immunodeficiency virus in vitro by an antisense phosphorothioate oligodeoxynucleotide against rev (art/trs) in chronically infected cells. Proc Natl Acad Sci USA. 1989;86:4244–4248. doi: 10.1073/pnas.86.11.4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mazumder A, Neamati N, Ojwang J O, Sunder S, Rando R F, Pommier Y. Inhibition of the human immunodeficiency virus type 1 integrase by guanosine quartet structures. Biochemistry. 1996;35:13762–13771. doi: 10.1021/bi960541u. [DOI] [PubMed] [Google Scholar]
- 94.McDonnell N B, De Guzman R N, Rice W G, Turpin J A, Summers M F. Zinc ejection as a new rationale for the use of cystamine and related disulfide-containing antiviral agents in the treatment of AIDS. J Med Chem. 1997;40:1969–1976. doi: 10.1021/jm970147+. [DOI] [PubMed] [Google Scholar]
- 95.Mitsuya H, Broder S. Inhibition of the in vitro infectivity and cytopathic effect of human T-lymphotrophic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) by 2′,3′-dideoxynucleosides. Proc Natl Acad Sci USA. 1986;83:1911–1915. doi: 10.1073/pnas.83.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitsuya H, Jarrett R F, Matsukura M, Di Marzo Veronese F, DeVico A L, Sarngadharan M G, Johns D G, Reitz M S, Broder S. Long-term inhibition of human T-lymphotropic virus type III/lymphadenopathy-associated virus (human immunodeficiency virus) DNA synthesis and RNA expression in T cells protected by 2′,3′-dideoxynucleosides in vitro. Proc Natl Acad Sci USA. 1987;84:2033–2037. doi: 10.1073/pnas.84.7.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mitsuya H, Weinhold K J, Furman P A, St. Clair M H, Lehrman S N, Gallo R C, Bolognesi D, Barry D W, Broder S. 3′-Azido-3′-deoxythymidine (BW A509U): an antiviral agent that inhibits the infectivity and cytopathic effect of human T-lymphotropic virus type III/lymphadenopathy-associated virus in vitro. Proc Natl Acad Sci USA. 1985;82:7096–7100. doi: 10.1073/pnas.82.20.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nam S H, Copeland T D, Hatanaka M, Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J Virol. 1993;67:196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neamati N, Hong H, Owen J M, Sunder S, Winslow H E, Christensen J L, Zhao H, Burke T R, Jr, Milne G W, Pommier Y. Salicylhydrazine-containing inhibitors of HIV-1 integrase: implication for a selective chelation in the integrase active site. J Med Chem. 1998;41:3202–3209. doi: 10.1021/jm9801760. [DOI] [PubMed] [Google Scholar]
- 102.Neumann M, Harrison J, Saltarelli M, Hadziyannis E, Erfle V, Felber B K, Pavlakis G N. Splicing variability in HIV type 1 revealed by quantitative RNA polymerase chain reaction. AIDS Res Hum Retrovir. 1994;10:1531–1542. doi: 10.1089/aid.1994.10.1531. [DOI] [PubMed] [Google Scholar]
- 103.Nielsen P E, Egholm M, Berg R H, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 104.Noble S A, Bonham M A, Bisi J E, Bruckenstein D A, Brown P H, Brown S C, Cadilla R, Gaul M D, Hanvey J C, Hassman C F, Josey J A, Luzzio M J, Myers P M, Pipe A J, Ricca D J, Su C W, Stevenson C L, Thomson S A, Wiethe R W, Babiss L E. Impact of biophysical parameters on the biological assessment of peptide nucleic acids, antisense inhibitors of gene expression. Drug Dev Res. 1995;34:184–195. [Google Scholar]
- 105.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. . (Erratum, 384:288.) [DOI] [PubMed] [Google Scholar]
- 106.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder II R C, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Otto M J, Garber S, Winslow D L, Reid C D, Aldrich P, Jadhav P K, Patterson C E, Hodge C N, Cheng Y S. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc Natl Acad Sci USA. 1993;90:7543–7547. doi: 10.1073/pnas.90.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park J, Morrow C D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Partaledis J A, Yamaguchi K, Tisdale M, Blair E E, Falcione C, Maschera B, Myers R E, Pazhanisamy S, Futer O, Cullinan A B, Stuver C M, Byrn R A, Livingston D J. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J Virol. 1995;69:5228–5235. doi: 10.1128/jvi.69.9.5228-5235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Patick A K, Rose R, Greytok J, Bechtold C M, Hermsmeier M A, Chen P T, Barrish J C, Zahler R, Colonno R J, Lin P F. Characterization of human immunodeficiency virus type 1 variant with reduced sensitivity to an aminodiol protease inhibitor. J Virol. 1995;69:2148–2152. doi: 10.1128/jvi.69.4.2148-2152.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pearl L H, Taylor W R. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 113.Pooga M, Hallbrink M, Zorko M, Langel U. Cell penetration by transportan. FASEB J. 1998;12:67–77. doi: 10.1096/fasebj.12.1.67. [DOI] [PubMed] [Google Scholar]
- 114.Pooga M, Soomets U, Hallbrink M, Valkna A, Saar K, Rezaei K, Kahl U, Hao J X, Xu X J, Wiesenfeld-Hallin Z, Hokfelt T, Bartfai T, Langel U. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat Biotechnol. 1998;16:857–861. doi: 10.1038/nbt0998-857. [DOI] [PubMed] [Google Scholar]
- 115.Puras Lutzke R A, Eppens N A, Weber P A, Houghten R A, Plasterk R H. Identification of a hexapeptide inhibitor of the human immunodeficiency virus integrase protein by using a combinatorial chemical library. Proc Natl Acad Sci USA. 1995;92:11456–11460. doi: 10.1073/pnas.92.25.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rice W G, Baker D C, Schaeffer C A, Graham L, Bu M, Terpening S, Clanton D, Schultz R, Bader J P, Buckheit R W, Jr, Field L, Singh P K, Turpin J A. Inhibition of multiple phases of human immunodeficiency virus type 1 replication by a dithiane compound that attacks the conserved zinc fingers of retroviral nucleocapsid proteins. Antimicrob Agents Chemother. 1997;41:419–426. doi: 10.1128/aac.41.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rice W G, Schaeffer C A, Harten B, Villinger F, South T L, Summers M F, Henderson L E, Bess J W, Jr, Arthur L O, McDougal J S, et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 119.Rice W G, Supko J G, Malspeis L, Buckheit R W, Jr, Clanton D, Bu M, Graham L, Schaeffer C A, Turpin J A, Domagala J, et al. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- 120.Rice W G, Turpin J A, Schaeffer C A, Graham L, Clanton D, Buckheit R W, Jr, Zaharevitz D, Summers M F, Wallqvist A, Covell D G. Evaluation of selected chemotypes in coupled cellular and molecular target-based screens identifies novel HIV-1 zinc finger inhibitors. J Med Chem. 1996;39:3606–3616. doi: 10.1021/jm960375o. [DOI] [PubMed] [Google Scholar]
- 121.Roberts N A, Martin J A, Kinchington D, Broadhurst A V, Craig J C, Duncan I B, Galpin S A, Handa B K, Kay J, Krohn A, et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 122.Rose J R, Babe L M, Craik C S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rose J R, Salto R, Craik C S. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J Biol Chem. 1993;268:11939–11945. [PubMed] [Google Scholar]
- 124.Saltarelli M J, Hadziyannis E, Hart C E, Harrison J V, Felber B K, Spira T J, Pavlakis G N. Analysis of human immunodeficiency virus type 1 mRNA splicing patterns during disease progression in peripheral blood mononuclear cells from infected individuals. AIDS Res Hum Retrovir. 1996;12:1443–1456. doi: 10.1089/aid.1996.12.1443. [DOI] [PubMed] [Google Scholar]
- 125.Schapiro J M, Winters M A, Stewart F, Efron B, Norris J, Kozal M J, Merigan T C. The effect of high-dose saquinavir on viral load and CD4+ T-cell counts in HIV-infected patients. Ann Intern Med. 1996;124:1039–1050. doi: 10.7326/0003-4819-124-12-199606150-00003. [DOI] [PubMed] [Google Scholar]
- 126.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schwartz S, Felber B K, Benko D M, Fenyo E M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sei S, Akiyoshi H, Bernard J, Venzon D J, Fox C H, Schwartzentruber D J, Anderson B D, Kopp J B, Mueller B U, Pizzo P A. Dynamics of virus versus host interaction in children with human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:1485–1490. doi: 10.1093/infdis/173.6.1485. [DOI] [PubMed] [Google Scholar]
- 129.Sei S, Kleiner D E, Kopp J B, Chandra R, Klotman P E, Yarchoan R, Pizzo P A, Mitsuya H. Quantitative analysis of viral burden in tissues from adults and children with symptomatic human immunodeficiency virus type 1 infection assessed by polymerase chain reaction. J Infect Dis. 1994;170:325–333. doi: 10.1093/infdis/170.2.325. [DOI] [PubMed] [Google Scholar]
- 130.Shirasaka T, Kavlick M F, Ueno T, Gao W Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, et al. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shoji Y, Akhtar S, Periasamy A, Herman B, Juliano R L. Mechanism of cellular uptake of modified oligodeoxynucleotides containing methylphosphonate linkages. Nucleic Acids Res. 1991;19:5543–5550. doi: 10.1093/nar/19.20.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 133.Smith A J, Srinivasakumar N, Hammarskjold M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Srinivasakumar N, Hammarskjold M L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Strickler J E, Gorniak J, Dayton B, Meek T, Moore M, Magaard V, Malinowski J, Debouck C. Characterization and autoprocessing of precursor and mature forms of human immunodeficiency virus type 1 (HIV 1) protease purified from Escherichia coli. Proteins. 1989;6:139–154. doi: 10.1002/prot.340060205. [DOI] [PubMed] [Google Scholar]
- 136.Tanaka M, Srinivas R V, Ueno T, Kavlick M F, Hui F K, Fridland A, Driscoll J S, Mitsuya H. In vitro induction of human immunodeficiency virus type 1 variants resistant to 2′-β-fluoro-2′,3′-dideoxyadenosine. Antimicrob Agents Chemother. 1997;41:1313–1318. doi: 10.1128/aac.41.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Turpin J A, Song Y, Inman J K, Huang M, Wallqvist A, Maynard A, Covell D G, Rice W G, Appella E. Synthesis and biological properties of novel pyridinioalkanoyl thiolesters (PATE) as anti-HIV-1 agents that target the viral nucleocapsid protein zinc fingers. J Med Chem. 1999;42:67–86. doi: 10.1021/jm9802517. [DOI] [PubMed] [Google Scholar]
- 138.Vickers T A, Ecker D J. Enhancement of ribosomal frameshifting by oligonucleotides targeted to the HIV gag-pol region. Nucleic Acids Res. 1992;20:3945–3953. doi: 10.1093/nar/20.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walder R Y, Walder J A. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci USA. 1988;85:5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang S, Dolnick B J. Quantitative evaluation of intracellular sense: antisense RNA hybrid duplexes. Nucleic Acids Res. 1993;21:4383–4391. doi: 10.1093/nar/21.18.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Weichold F F, Lisziewicz J, Zeman R A, Nerurkar L S, Agrawal S, Reitz M S, Jr, Gallo R C. Antisense phosphorothioate oligodeoxynucleotides alter HIV type 1 replication in cultured human macrophages and peripheral blood mononuclear cells. AIDS Res Hum Retrovir. 1995;11:863–867. doi: 10.1089/aid.1995.11.863. [DOI] [PubMed] [Google Scholar]
- 142.Willey R L, Martin M A, Peden K W. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 144.Wittung P, Kajanus J, Edwards K, Haaima G, Nielsen P E, Norden B, Malmstrom B G. Phospholipid membrane permeability of peptide nucleic acid. FEBS Lett. 1995;365:27–29. doi: 10.1016/0014-5793(95)00409-3. . (Correction and republication, 375:27–29.) [DOI] [PubMed] [Google Scholar]
- 145.Wittung P, Nielsen P E, Buchardt O, Egholm M, Norden B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994;368:561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- 146.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana B K, Baldwin E, Weber I T, Selk L M, Clawson L, Schneider J, Kent S B. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 147.Wong J K, Hezareh M, Günthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 148.Yamaguchi K, Papp B, Zhang D, Ali A N, Agrawal S, Byrn R A. The multiple inhibitory mechanisms of GEM 91, a gag antisense phosphorothioate oligonucleotide, for human immunodeficiency virus type 1. AIDS Res Hum Retrovir. 1997;13:545–554. doi: 10.1089/aid.1997.13.545. [DOI] [PubMed] [Google Scholar]
- 149.Yarchoan R, Klecker R W, Weinhold K J, Markham P D, Lyerly H K, Durack D T, Gelmann E, Lehrman S N, Blum R M, Barry D W, et al. Administration of 3′-azido-3′-deoxythymidine, an inhibitor of HTLV-III/LAV replication, to patients with AIDS or AIDS-related complex. Lancet. 1986;i:575–580. doi: 10.1016/s0140-6736(86)92808-4. [DOI] [PubMed] [Google Scholar]
- 150.Yarchoan R, Mitsuya H, Thomas R V, Pluda J M, Hartman N R, Perno C F, Marczyk K S, Allain J P, Johns D G, Broder S. In vivo activity against HIV and favorable toxicity profile of 2′,3′-dideoxyinosine. Science. 1989;245:412–415. doi: 10.1126/science.2502840. [DOI] [PubMed] [Google Scholar]
- 151.Yoshimura K, Kato R, Yusa K, Kavlick M F, Maroun V, Nguyen A, Mimoto T, Ueno T, Shintani M, Falloon J, Masur H, Hayashi H, Erickson J, Mitsuya H. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc Natl Acad Sci USA. 1999;96:8675–8680. doi: 10.1073/pnas.96.15.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yu X F, Dawson L, Tian C J, Flexner C, Dettenhofer M. Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J Virol. 1998;72:3412–3417. doi: 10.1128/jvi.72.4.3412-3417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, Markowitz M, Ho D D. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 154.Zhao H, Neamati N, Hong H, Mazumder A, Wang S, Sunder S, Milne G W, Pommier Y, Burke T R., Jr Coumarin-based inhibitors of HIV integrase. J Med Chem. 1997;40:242–249. doi: 10.1021/jm960450v. [DOI] [PubMed] [Google Scholar]
- 155.Zhao H, Neamati N, Sunder S, Hong H, Wang S, Milne G W, Pommier Y, Burke T R., Jr Hydrazide-containing inhibitors of HIV-1 integrase. J Med Chem. 1997;40:937–941. doi: 10.1021/jm960755+. [DOI] [PubMed] [Google Scholar]