Abstract
Transcriptional transactivation of the human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR) promoter element by the essential viral Tat protein requires recruitment of positive transcription elongation factor b (P-TEFb) to the viral TAR RNA target. The recruitment of P-TEFb, which has been proposed to be necessary and sufficient for activation of viral gene expression, is mediated by the highly cooperative interaction of Tat and cyclin T1, an essential component of P-TEFb, with the HIV-1 TAR element. Species, such as rodents, that encode cyclin T1 variants that are unable to support TAR binding by the Tat-cyclin T1 heterodimer are also unable to support HIV-1 Tat function. In contrast, we here demonstrate that the bovine immunodeficiency virus (BIV) Tat protein is fully able to bind to BIV TAR both in vivo and in vitro in the absence of any cellular cofactor. Nevertheless, BIV Tat can specifically recruit cyclin T1 to the BIV TAR element, and this recruitment is as essential for BIV Tat function as it is for HIV-1 Tat activity. However, because the cyclin T1 protein does not contribute to TAR binding, BIV Tat is able to function effectively in cells from several species that do not support HIV-1 Tat function. Thus, BIV Tat, while apparently dependent on the same cellular cofactor as the Tat proteins encoded by other lentiviruses, is nevertheless unique in terms of the mechanism used to recruit the BIV Tat-cyclin T1 complex to the viral LTR promoter.
Lentiviruses can be divided into two subgroups based on whether they express an RNA sequence-dependent transcriptional transactivator functionally equivalent to the human immunodeficiency virus type 1 (HIV-1) Tat protein (reviewed in references 10 and 35). All primate lentiviruses, as well as equine infectious anemia virus (EIAV) and bovine immunodeficiency virus (BIV), encode an HIV-1 Tat homolog (4–7, 13, 16, 25, 37). In contrast, feline immunodeficiency virus and the ovine and caprine lentiviruses lack an equivalent RNA sequence-dependent transcriptional activator (11, 29).
In addition to its unique RNA sequence dependence, HIV-1 Tat (hTat) is also unusual in that it acts mainly at the level of transcription elongation rather than initiation (14, 22). hTat activity requires the recruitment of both hTat and a cellular cofactor, termed cyclin T1 (CycT1), to the HIV-1 TAR (hTAR) RNA stem-loop structure (2, 18, 38, 42). CycT1, together with cdk9, forms part of positive transcription elongation factor b (P-TEFb) (27, 38, 39). Recruitment of P-TEFb to TAR has been proposed to be both necessary and sufficient for activation of transcription elongation from the HIV-1 long terminal repeat (LTR) promoter (3).
Binding of hTat and CycT1 to hTAR is highly cooperative. Thus, human CycT1 (hCycT1) is unable to bind hTAR in the absence of hTat, and hTat binding to hTAR, while detectable, is very inefficient in the absence of hCycT1 (2, 4, 20, 26, 38). Recruitment of the hTat-hCycT1 heterodimer to hTAR involves a direct interaction between hTat and a U-rich RNA bulge, while hCycT1 is believed to bind the TAR terminal loop (12, 26, 32, 38, 41). Interestingly, the ability of CycT1 to bind TAR is not evolutionarily conserved, so that the murine CycT1 (mCycT1) protein, for example, can bind to hTat but is unable to mediate the recruitment of the hTat-mCycT1 heterodimer to hTAR (2, 18). This deficiency, which results from a single amino acid difference between mCycT1 and hCycT1 (2, 17, 18, 24), renders hTat inactive in murine cells and can explain the observed species tropism of hTat (1, 26).
Analysis of Tat function in HIV-2, in the simian immunodeficiency viruses (SIVs), and in the distantly related EIAV has demonstrated that these Tat proteins also recruit CycT1 to their cognate TAR elements and, in particular, has revealed that TAR binding by the relevant Tat-CycT1 heterodimer is again highly cooperative (4, 5, 38). Further, HIV-2, SIV, and EIAV Tat all show species tropisms, and this is again due to the inability of the CycT1 proteins present in certain species to contribute to TAR binding (1, 4, 5, 26). Thus, both hCycT1 and equine CycT1 bind EIAV Tat, but the former differs from the latter in being unable to mediate binding of the resultant heterodimer to EIAV TAR (4).
While CycT1 is critical for both transcriptional activation and TAR binding by the Tat proteins enumerated above, it has been proposed that BIV Tat (bTat) is distinct in being competent for efficient BIV TAR (bTAR) binding in the absence of any cellular cofactor (7). Thus, the 17-amino-acid basic domain of bTat was shown to bind to bTAR with high affinity and specificity in vitro and could also efficiently recruit a fused heterologous effector domain to bTAR when expressed in bacteria (7, 20, 31). While these earlier experiments did not address a possible role for CycT1 in facilitating bTAR binding by bTat, the similarity in domain organization of bTat and hTat, and in particular the conservation of the cysteine-rich and core domains that mediate CycT1 binding to hTat (2, 16, 23, 25, 26, 38), suggests that CycT1 is likely to play a role in mediating bTat function.
In this report we present data strongly supporting the hypothesis that bTat, like hTat, SIV Tat, and EIAV Tat, activates viral gene expression by recruitment of the cellular P-TEFb transcription factor. However, bTat is shown to differ from these other lentiviral Tat proteins in that bTAR binding by the bTat-CycT1 heterodimer is no more efficient than binding by bTat alone.
MATERIALS AND METHODS
Construction of molecular clones.
The indicator plasmids pHIV/hTAR/CAR and pHIV/SLIIB/CAT have been previously described (4, 26, 36). Plasmid pHIV/bTAR/CAT was constructed by substituting bTAR in place of hTAR in pHIV/hTAR/CAT. Unique BglII and SacI sites allowed the excision of hTAR and the subsequent insertion of annealed oligonucleotides encoding the entire bTAR sequence.
Plasmid pcTat, encoding HIV-1 Tat, has been described elsewhere (26). The similar expression plasmid pbTat was constructed by ligating an NcoI/XhoI-digested PCR product, encoding the entire 104-amino-acid BIV Tat protein (25), into the Nco- and XhoI-cleaved expression plasmid pBC12/CMV (8). The pRev-bTat fusion protein expression plasmid was constructed by PCR amplification of the bTat cDNA, using primers that inserted flanking EcoRI sites. After cleavage with EcoRI, the resultant bTat cDNA was cloned into pcRev (26) in frame with the HIV-1 Rev protein. The expression plasmid pRev-C38S was constructed from pRev-bTat by using a Quick Change site-directed mutagenesis kit (Stratagene).
The selectable marker used in each of the following yeast expression plasmids is given in parentheses. The pGBT9/bTat yeast expression plasmid (TRP), which encodes the GAL4 DNA binding domain fused to bTat, was constructed by ligating an EcoRI restriction fragment encoding the bTat cDNA into pGBT9 (Clontech) after cleavage with EcoRI. Plasmid pGBT9/C38S (TRP) was constructed by mutation of pGBT9/bTat via Quick Change site-directed mutagenesis (Stratagene). Expression plasmids pVP16/hCycT1 and pVP16/mCycT1 (LEU), which encode the VP16 activation domain fused to hCycT1 or mCycT1, have been described elsewhere (2). The analogous yeast expression plasmid pVP16/bTat (LEU) was constructed as previously described for pVP16/hTat (2).
Plasmid pIII/MS2/bTAR (URA), encoding a hybrid MS2-bTAR RNA transcript, was constructed as previously described for the similar pIII/MS2/hTAR (2). The pPGK expression plasmid (TRP) was constructed from pGBT9 by deletion of both the adh promoter and the GAL4 DNA binding domain, followed by insertion of the pgk promoter excised from pMA91 (28). Plasmid pPGK/bTat (TRP) encodes the full-length bTat protein, attached to an amino-terminal hemagglutinin epitope tag, under the control of the pgk promoter element. The similar plasmid pPGK/C38S was derived from pPGK/bTat by site-directed mutagenesis. The expression plasmids pPGK/hCycT1 and pPGK/mCycT1 (TRP), which were constructed by insertion of the relevant full-length CycT1 cDNAs into pPGK, express nonfused hCycT1 or mCycT1 (2).
The bacterial glutathione S-transferase (GST) fusion protein expression plasmids pGEX/hCycT1 and pGEX/hTat, encoding GST-hCycT1 and GST-hTat, respectively, have been described elsewhere (4). The GST-bTat fusion protein expression plasmid pGEX/bTat was constructed by the in-frame ligation of an EcoRI fragment encoding bTat into pGEX4T-1. The in vitro transcription vector pGEM/bTAR was constructed by ligation of oligonucleotides encoding full-length bTAR into pGEM3Zf(+).
Vertebrate cell transfection assays.
Human 293T cells were transfected by the calcium phosphate method (9) with 500 ng of reporter plasmid (pHIV/bTAR/CAT, pHIV/hTAR/CAT, or pHIV/SLIIB/CAT) and 200 ng of effector plasmid (pbTat, pRev-bTat, pRev-C38S, pcTat, or pBC12/CMV as a negative control). In addition, 1,000 ng of pBC12/CMV and 50 ng of the pBC12/CMV/β-gal internal control plasmid were added to each transfection. Mouse L cells and quail QCl-3 cells were transfected using DEAE-dextran (8, 9). At 48 h after transfection, induced levels of chloramphenicol acetyltransferase (CAT) and β-galactosidase (β-Gal) activity were determined as previously described (2, 26).
Protein-protein and RNA-protein interactions in yeast.
For two-hybrid assays (15), Saccharomyces cerevisiae Y190 cells were transformed with pGBT9, pGBT9/bTat, or pGBT9/C38S and either pVP16, pVP16/hCycT1, or pVP16/mCycT1. For three-hybrid assays (33), L40uraMS2 yeast cells (Invitrogen) were transformed with three expression plasmids expressing a hybrid MS2-(b or h)TAR RNA, the VP16-bTat (wild type or C38S), or VP16-hTat fusion protein, along with unfused human or murine CycT1. In other experiments, L40uraMS2 yeast cells were instead transformed with a hybrid RNA expression plasmid, an expression plasmid encoding nonfusion bTat or C38S and an expression plasmid encoding either the VP16-mCycT1 or VP16-hCycT1 fusion protein. After growth on selective media, β-Gal activity in cell lysates was determined as previously described (2–5).
RNA gel shift analyses.
Recombinant GST-bTat, GST-hTat, and GST-hCycT1 were expressed in the BL21 codon plus strain of Escherichia coli and affinity purified as previously described (4). A 32P-labeled bTAR RNA probe was generated by in vitro transcription using T7 RNA polymerase and the linearized plasmid pGEM3/bTAR. RNA-protein interactions were then analyzed by electrophoretic mobility shift assay as previously described (4).
RESULTS
As noted above, the Tat proteins encoded by HIV-1 and EIAV show distinct species tropisms. As tethering to the viral LTR via a heterologous RNA binding domain fully rescues both hTat function in otherwise nonpermissive rodent cells and EIAV Tat function in nonpermissive human cells (1, 26), these tropisms must largely reflect inefficient recruitment of Tat to TAR. Therefore, if bTat binding to bTAR is indeed cofactor independent, one might predict that bTat would not show a comparable species tropism. Earlier reports have not resolved this issue, as bTat has been reported to be active in bovine, human, and murine cells but poorly active in lapine cells (16). Also, while the bTat protein has been reported to activate the HIV-1 LTR in certain tissue culture cell lines (16, 25), in vitro data indicate that bTat binding to bTAR is far more efficient than binding to hTAR (7).
To address the species tropism and RNA sequences specificity of bTat, we cotransfected human 293T cells, murine L cells, and quail QCl-3 cells with expression plasmids encoding either hTat or bTat and indicator constructs consisting of the cat indicator gene linked to either the wild-type HIV-1 LTR or an HIV-1 LTR in which hTAR had been replaced by bTAR. By comparing indicator constructs in which only the TAR element was varied, confounding effects resulting from the presence or absence of transcription factors that bind the U3 regions of the HIV-1 or BIV LTR are avoided.
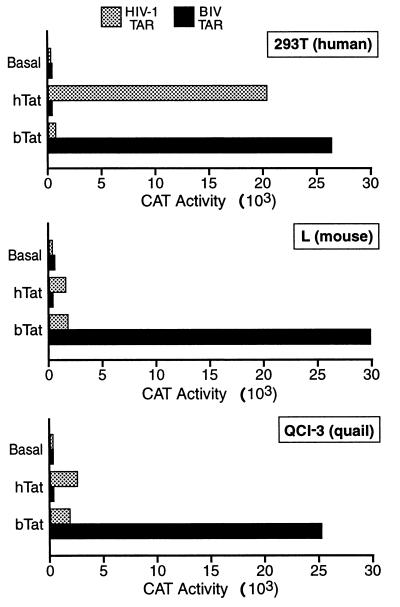
As previously reported (2, 8), hTat efficiently activated expression of a cat indicator gene linked to the wild-type HIV-1 LTR in human cells but was only weakly active in the tested murine and avian cells (Fig. 1). No hTat-induced activation of the modified HIV-1 LTR containing the bTAR RNA element was detected. In contrast, bTat potently activated cat gene expression directed by the HIV-1 LTR containing bTAR in all three cell lines tested. However, bTat had only a very weak activating effect on wild-type HIV-1 LTR-driven cat gene expression (Fig. 1). These data therefore suggest that bTat function is indeed less subject to species restriction than is hTat function and are consistent with a previous report (7) demonstrating that bTAR is a better target for bTat binding than is hTAR.
FIG. 1.
Comparison of hTat and bTat function in cells from three distinct species. Human 293T cells, murine L cells, and avian QCl-3 cells were transfected with indicator constructs consisting of the wild-type HIV-1 LTR, or an HIV-1 LTR in which the hTAR element had been substituted with bTAR, linked to the cat gene. These plasmids were cotransfected with pBC12/CMV-based expression constructs expressing full-length hTat or bTat as well as a pBC12/CMV-based internal control plasmid encoding β-Gal. Plasmid pBC12/CMV also served as a negative control. Cultures were harvested at ∼48 h after transfection, and CAT and β-Gal activities were determined as described elsewhere (2–5). The indicated data are corrected for minor differences in transfection efficiency, as measured by the β-Gal internal control, and are representative of three independent transfection experiments.
The bTat protein binds CycT1 specifically.
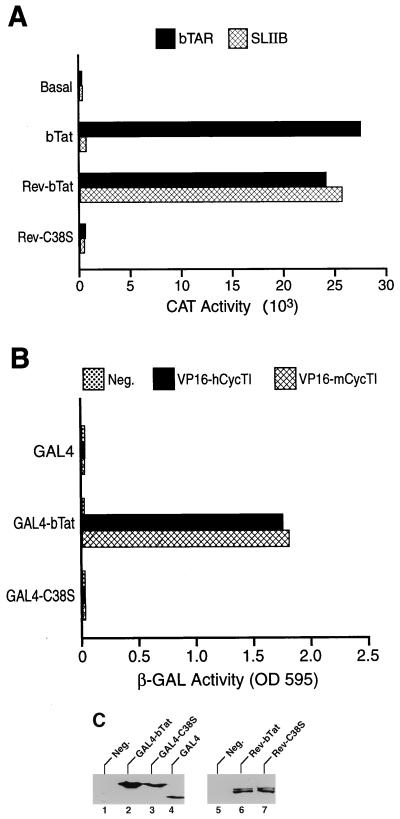
The conserved cysteine-rich domain of hTat is critical for both CycT1 binding and hTat function, and mutation of cysteine 22 in hTat to serine (C22S) therefore blocks both of these activities (2, 23, 26). Because hTat cysteine 22 is conserved in bTat (16, 23, 25), we examined whether mutation of the equivalent residue, i.e., mutation of bTat cysteine 38 to serine (C38S), would also affect bTat function and CycT1 binding. As we do not have access to a bTat-specific antiserum, we performed this mutational analysis in the context of an HIV-1 Rev-bTat fusion protein that can be readily detected using an anti-Rev antiserum. Further, in the case of hTat, we and others have previously demonstrated that an hTat-Rev fusion protein can potently activate an HIV-1 LTR in which the hTAR element has been replaced with Rev response element stem-loop IIB (SLIIB), the RNA target for HIV-1 Rev (19, 26, 34). As shown in Fig. 2A, bTat and the Rev-bTat fusion protein equivalently activate cat gene expression directed by the HIV-1 LTR containing the bTAR RNA target when tested by cotransfection into 293T cells. When the bTat and Rev-bTat proteins were instead coexpressed with an indicator construct containing the HIV-1 LTR linked to the SLIIB RNA binding site for Rev (4, 26, 36), the Rev-bTat fusion protein proved highly active whereas the bTat protein was, as expected, inactive. Thus, bTat, like hTat, can activate HIV-1 LTR-dependent gene expression when tethered to a heterologous promoter-proximal RNA target. In contrast, a fusion protein containing wild-type Rev fused to the C38S mutant of bTat was inactive on both indicator constructs (Fig. 2A), even though Rev-bTat and Rev-C38S were expressed at equivalent levels, as determined by Western analysis using an anti-Rev antiserum (Fig. 2C). As the C38S mutation inactivates Rev-bTat function via both bTAR and SLIIB (Fig. 2A), this inhibition must occur by a mechanism independent of RNA targeting, i.e., most probably by blocking cofactor recruitment.
FIG. 2.
Interaction of bTat with CycT1. (A) Mutation of the bTat cysteine motif blocks bTat function. Human 293T cells were cotransfected with either the pHIV/bTAR/CAT or pHIV/SLIIB/CAT indicator construct together with an effector plasmid encoding bTat or the Rev-bTat or Rev-C38S fusion protein. Data were derived as described in the legend to Fig. 1. (B) The bTat protein binds CycT1 specifically. The S. cerevisiae two-hybrid indicator strain Y190 was transformed with plasmids expressing the GAL4 DNA binding domain fused to wild-type or mutant bTat and with plasmids expressing the VP16 transcription activation domain in an unfused form (Neg.) or fused to wild-type hCycT1 or mCycT1. Induced β-Gal activities were determined as previously described (2–5) after selection for transformants. (C) Western analyses of GAL4 fusion protein expression levels in yeast (lanes 1 to 4) and Rev fusion expression in 293T cells (lanes 5 to 7) were performed using a commercial GAL4 DNA binding domain antibody or a rabbit polyclonal anti-Rev antiserum as previously described (3). Neg., control cells not expressing a GAL4 (lane 1) or Rev (lane 5) fusion protein. OD 595 (here and in Fig. 3 and 4), optical density at 595 nm.
Previously, we have reported that hTat, SIV Tat, and EIAV Tat all bind hCycT1 and mCycT1 specifically when analyzed in the yeast two-hybrid protein-protein interaction assay (2–5, 15). Expression of a GAL4-bTat fusion protein in the appropriate yeast indicator strain revealed a readily detectable interaction with fusion proteins consisting of the VP16 transcription activation domain linked to either hCycT1 or mCycT1 (Fig. 2B). In contrast, an equivalent fusion protein consisting of the GAL4 DNA binding domain linked to the C38S mutant of bTat failed to interact with either CycT1 protein (Fig. 2B), even though it was expressed at a comparable level (Fig. 2C). We conclude that bTat, like hTat, is able to bind to hCycT1 and mCycT1 specifically and that this interaction is, in both cases, dependent on the Tat cysteine motif.
The bTat protein binds bTAR effectively in the absence of CycT1.
We next asked whether bTat would bind bTAR cooperatively with CycT1 in vivo or whether bTat binding to bTAR would instead be unaffected by CycT1. For this purpose, we used the three-hybrid RNA-protein interaction assay in yeast (33). The tested plasmids express RNA hybrids consisting of the MS2 operator linked to either hTAR or bTAR and protein hybrids consisting of the VP16 activation domain linked to full-length hTat or bTat.
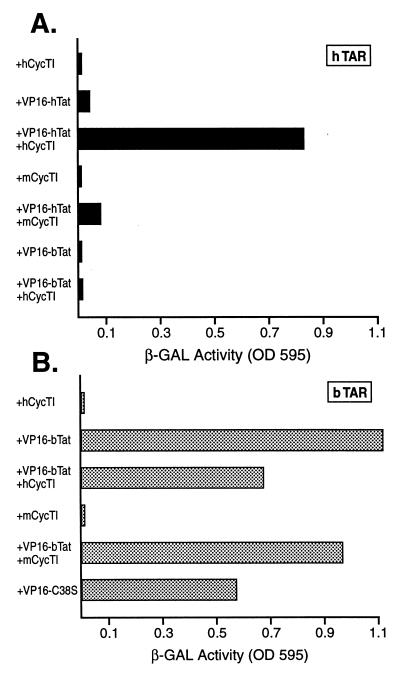
As shown in Fig. 3A, a basal level of expression of β-Gal expression was noted in yeast cells expressing the MS2-hTAR chimera and nonfused hCycT1 (all yeast cells also express a LexA-MS2 coat protein fusion protein). Coexpression of MS2-hTAR and of the VP16-hTat fusion protein gave rise to only a low induction in β-Gal expression, consistent with the previously observed poor binding of hTat to hTAR in the absence of hCycT1 (5, 20, 38). In contrast, coexpression of the MS2-hTAR RNA hybrid with both VP16-hTat and nonfused hCycT1 induced a high level of β-Gal activity, thus demonstrating that hTat recruitment to hTAR is potently activated by hCycT1 in vivo. We have previously shown that this interaction is specific, as it can be blocked by mutation of the hTAR bulge or terminal loop or by mutation of hCycT1 or hTat, e.g., the C22S mutation (2, 5). Additional evidence of specificity is provided in Fig. 3A, which shows that mCycT1 cannot substitute for hCycT1 in mediating enhanced TAR recruitment. Importantly, we were unable to detect any interaction of bTat with hTAR in either the presence or the absence of hCycT1 (Fig. 3A).
FIG. 3.
The bTat and hTat proteins differ in the ability to bind their cognate TAR elements. In these three-hybrid protein-RNA interaction assays, S. cerevisiae L40uraMS2 cells were transformed with an expression plasmid encoding the MS2 operator linked to full-length hTAR (A) or bTAR (B). In addition, these cells were transformed with a plasmid encoding the VP16 activation domain fused to hTat, bTat, or the C38S bTat mutant and finally with a plasmid encoding the nonfused form of hCycT1 or mCycT1. The appropriate empty vectors served as negative controls. Induced β-Gal activity was measured in pooled transformants as previously described (2–5).
In Fig. 3B, the ability of a VP16-bTat fusion protein to interact with an MS2-bTAR RNA target was examined using this same in vivo assay. As is readily apparent, bTat and hTat differ dramatically in the ability to bind their cognate TAR elements in vivo in the absence of CycT1. Specifically, recruitment of the VP16-bTat fusion protein to the bTAR element in the absence of CycT1 was as efficient as the recruitment of VP16-hTat to hTAR in the presence of hCycT1 (Fig. 3). Further, and in contrast to hTat, coexpression of hCycT1 or mCycT1 did not enhance binding of bTat to its cognate TAR element. This interaction is clearly specific in that, as noted above, bTat is unable to interact with hTAR (Fig. 3A). Finally, we observed that the C38S mutant of bTat retained the ability to bind to bTAR (Fig. 3B) despite lacking a functional CycT1 interaction domain (Fig. 2B). This contrasts with the equivalent C22S mutant of hTat, which lacks the ability to bind to hTAR in the presence or absence of hCycT1 (2, 26).
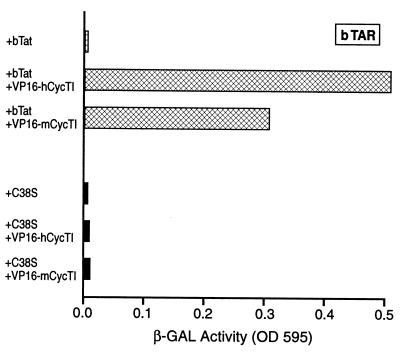
While the data presented in Fig. 3 demonstrate that bTat can bind to bTAR in the absence of CycT1, they do not reveal whether CycT1 is indeed recruited to bTAR by bTat. This question is addressed in Fig. 4, which again used the yeast three-hybrid assay. In this case, either bTat or the C38S mutant of bTat was expressed as a nonfusion protein, while hCycT1 and mCycT1 were expressed as VP16 fusions. As may be seen, bTat, but not the C38S mutant, was indeed able to recruit both hCycT1 and mCycT1 to the bTAR element. This result contrasts with our earlier work showing that hTat can recruit hCycT1, but not mCycT1, to hTAR (2) and provides an explanation for why bTat, but not hTat, is functional in murine cells (Fig. 1).
FIG. 4.
The bTat protein can recruit CycT1 to bTAR. This yeast three-hybrid assay was performed essentially as described for Fig. 5 except that the CycT1 proteins were expressed as VP16 fusions whereas bTat and the C38S bTat mutant were expressed in a nonfused form.
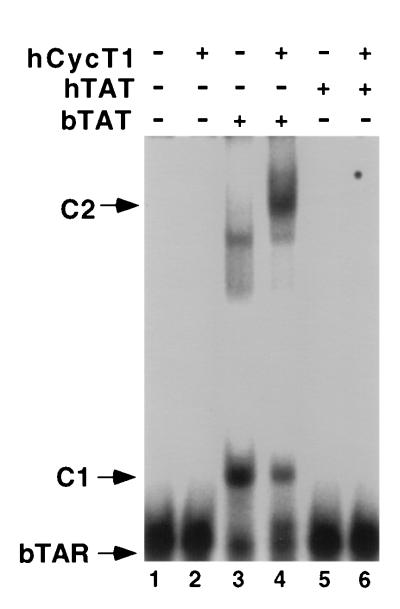
As noted above, it has previously been reported that the isolated bTat basic domain can bind to bTAR effectively in vitro (7, 20, 31). We wished to confirm this earlier result, and the in vivo data reported above, by demonstrating that full-length bTat can also bind to bTAR in vitro and that this binding is not significantly affected by hCycT1. As shown in Fig. 5, we were able to readily detect an in vitro interaction between bTAR and recombinant bTat (lane 3) but not between bTAR and hTat (lane 5). Addition of recombinant hCycT1 to the reaction resulted in the shift of the bTat-bTAR complex to a slower mobility, consistent with formation of a ternary complex, but did not result in enhanced RNA binding (lane 4). This result contrasts with our previous data that showed little or no binding of either hTat to hTAR or EIAV Tat to EIAV TAR under comparable conditions in vitro in the absence of CycT1 but high levels of RNA binding when both Tat and CycT1 were present (4).
FIG. 5.
The bTat protein binds bTAR in vitro. In this RNA gel shift experiment, a labeled bTAR RNA probe was incubated with the indicated recombinant GST fusion proteins, and the resultant RNA-protein complexes were then resolved by nondenaturing gel electrophoresis. C1 indicates the position of the proposed bTAR-bTAT complex, while C2 indicates the position of the proposed bTAR-bTat-CycT1 ternary complex.
DISCUSSION
The mechanism of action of the essential HIV-1 Tat transcription factor now appears fairly well understood (10, 35). The Tat protein first binds to the CycT1 component of P-TEFb via the Tat cysteine and core motifs (2, 18, 38), and the resultant Tat-CycT1 heterodimer then binds the TAR element. This binding, which is highly cooperative, is mediated by direct interactions between the Tat basic domain and a TAR RNA bulge and, most probably, between CycT1 and the terminal TAR loop (12, 26, 32, 38, 41). Subsequently, the cdk9 component of P-TEFb is believed to activate efficient elongation from the HIV-1 LTR promoter by phosphorylating the carboxy-terminal domain of initiated RNA polymerase II molecules, and also possibly other substrates (3, 10, 19, 27, 35, 38, 40–42). Recent evidence demonstrating that the HIV-1 LTR can be fully activated if P-TEFb is recruited to the promoter by tethering to a heterologous RNA target (3, 19) suggests that P-TEFb recruitment, either by Tat or by some other means, is both necessary and sufficient for activation of viral transcription. Reports examining the mechanism of action of the HIV-2, SIV, and EIAV Tat proteins demonstrate that CycT1 is also critical for both TAR recruitment and activation of transcription in these other lentiviruses (4, 5).
As EIAV Tat shares far less sequence homology with hTat than does bTat (4), it was surprising that the isolated bTat basic domain had, uniquely, been reported to be fully competent for binding to bTAR (7, 20, 34). We hypothesized that an analysis of bTAR binding by full-length bTat in vivo might therefore reveal cooperative binding with CycT1, as previously reported for all other lentiviral Tat proteins (4, 5). However, as clearly demonstrated in this report, full-length bTat is in fact competent to bind to bTAR in the absence of CycT1 both in vivo (Fig. 3) and in vitro (Fig. 5), and our data therefore fully confirm the earlier work of Frankel and coworkers (7, 20, 34). Nevertheless, bTat, like all other Tat proteins, does bind to CycT1 specifically (Fig. 2B) and can recruit CycT1, and hence presumably P-TEFb, to the viral TAR element (Fig. 4 and 5). Because bTat does not depend on CycT1 for assistance in binding TAR (Fig. 3), bTat should differ from hTat in being able to recruit a wider range of CycT1 proteins to TAR, as is indeed demonstrated in Fig. 4 using mCycT1. This presumably explains the ability of bTat to function in a wider range of species than hTat (Fig. 1).
It is of interest to speculate as to why bTat has evolved a functionally autonomous RNA binding domain while all other Tat proteins can bind to TAR only as part of a CycT1-Tat heterodimer. One advantage of this latter strategy is that free Tat would not be able to compete with CycT1-Tat complexes for TAR binding, thus preventing any inhibition of transactivation by free Tat when Tat expression is saturating. Possible advantages of the noncooperative RNA binding strategy exhibited by bTat include the expanded potential species host range mentioned above and, perhaps, the ability to recruit a form of P-TEFb lacking CycT1 to TAR, i.e., a P-TEFb variant that contains one of the two known isoforms of CycT2 (30). We and others have, in fact, previously shown that a CycT2 mutant, differing by only one residue from wild-type CycT2, can fully support HIV-1 Tat function in vivo, although wild-type CycT2 is normally unable to bind to hTat (2, 5, 24). In contrast, we have observed that bTat can specifically bind to both isoforms of wild-type CycT2 in vivo and that CycT2 can, in fact, be recruited to bTAR as efficiently as CycT1 in the yeast three-hybrid assay (data not shown). As bTat is active in all cells tested, it has not been possible to confirm that this is a functionally relevant interaction by, for example, rescuing bTat function by expression of human CycT2 in trans. Nevertheless, this observation does raise the possibility that bTat, unlike hTat, may be able to utilize all forms of P-TEFb, as opposed to only the dominant CycT1 variant, for activation of viral gene expression. CycT1 is known to be expressed at low levels in, for example, resting T cells (21, 39), and the ability of bTat to recruit forms of P-TEFb lacking CycT1 to bTAR could therefore enhance bTat-dependent BIV gene expression, and hence replication, in certain contexts.
REFERENCES
- 1.Alonso A, Derse D, Peterlin B M. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J Virol. 1992;66:4617–4621. doi: 10.1128/jvi.66.7.4617-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci USA. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol Cell Biol. 1999;19:4592–4599. doi: 10.1128/mcb.19.7.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Analysis of the effect of natural sequence variation in Tat and in cyclin T on the formation and RNA binding properties of Tat-cyclin T complexes. J Virol. 1999;73:5777–5786. doi: 10.1128/jvi.73.7.5777-5786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho M, Derse D. Mutational analysis of the equine infectious anemia virus Tat-responsive element. J Virol. 1991;65:3468–3474. doi: 10.1128/jvi.65.7.3468-3474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Frankel A D. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 9.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 10.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 11.de Parseval A, Elder J H. Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial Rev activity. J Virol. 1999;73:608–617. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 Tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emerman M, Guyader M, Montagnier L, Baltimore D, Muesing M A. The specificity of the human immunodeficiency virus type 2 transactivator is different from that of human immunodeficiency virus type 1. EMBO J. 1987;6:3755–3760. doi: 10.1002/j.1460-2075.1987.tb02710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg M B, Baltimore D, Frankel A D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 16.Fong S E, Greenwood J D, Williamson J C, Derse D, Pallansch L A, Copeland T, Rasmussen L, Mentzer A, Nagashima K, Tobin G, Gonda M A. Bovine immunodeficiency virus tat gene: cloning of two distinct cDNAs and identification, characterization, and immunolocalization of the tat gene products. Virology. 1997;233:339–357. doi: 10.1006/viro.1997.8589. [DOI] [PubMed] [Google Scholar]
- 17.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada K, Martin S S, Frankel A D. Selection of RNA-binding peptides in vivo. Nature. 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann C H, Carroll R G, Wei P, Jones K A, Rice A P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 23.Kuppuswamy M, Subramanian T, Srinivasan A, Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989;17:3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z-Q, Sheridan D, Wood C. Identification and characterization of the bovine immunodeficiency-like virus tat gene. J Virol. 1992;66:5137–5140. doi: 10.1128/jvi.66.8.5137-5140.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellor J, Dobson M J, Roberts N A, Tuite M F, Emtage J S, White S, Lowe P A, Patel T, Kingsman A J, Kingsman S M. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene. 1983;24:1–14. doi: 10.1016/0378-1119(83)90126-9. [DOI] [PubMed] [Google Scholar]
- 29.Morse B A, Carruth L M, Clements J E. Targeting of the visna virus Tat protein to AP-1 sites: interactions with the bZIP domains of Fos and Jun in vitro and in vivo. J Virol. 1999;73:37–45. doi: 10.1128/jvi.73.1.37-45.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puglisi J D, Chen L, Blanchard S, Frankel A D. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 32.Roy S, Delling U, Chen C-H, Rosen C A, Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- 33.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. RNA recognition by an isolated α helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 35.Taube R, Fujinaga K, Wimmer J, Barboric M, Peterlin B M. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology. 1999;264:245–253. doi: 10.1006/viro.1999.9944. [DOI] [PubMed] [Google Scholar]
- 36.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 37.Viglianti G A, Mullins J I. Functional comparison of transactivation by simian immunodeficiency virus from rhesus macaques and human immunodeficiency virus type 1. J Virol. 1988;62:4523–4532. doi: 10.1128/jvi.62.12.4523-4532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vitro and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]