Observational studies and clinical trials have investigated the effect of lipid-lowering drug targets (LLDT) on the risk of stroke. Understanding the potential impact of LLDT perturbation on stroke risk in different ethnic populations is crucial for personalized medicine and targeted therapies. Research comparing the efficacy of LLDTs for ischemic stroke in European and African ancestries can shed light on potential ethnic ancestry-specific effects.
We, therefore, undertook a drug target Mendelian randomization (MR) analysis to examine the relationship between genetic proxies for the inhibition of (PCSK9, LDLR, and APOB) on ischemic stroke risk in European and African ethnic ancestry groups.
We used data from the Global Lipid Genetics Consortium to generate genetic instruments to proxy LLDT perturbation. Specifically, we used ancestry-specific summary data from the study by Graham et al, which included analyses in 1 751 523 individuals of the European ancestry and 739 703 of the African ancestry.1 Ischemic stroke summary statistics were extracted from genome-wide association studies of the GIGASTROKE consortium.2 This comprises Africans (ncases=3961; ncontrols=20 030), and Europeans (ncases=73 552; ncontrols=1 234 808).
To proxy LLDT perturbation, we identified cis variants within ±200 kb of each gene location, associated with low-density lipoprotein cholesterol at a genome-wide association study threshold of P=5×10−8 for the European ancestry and P=5×10−6 for the African ancestry. P=5×10−6 was used for the African ancestry due to the smaller sample size and lower statistical power. The variants that survived this threshold were then clumped at linkage disequilibrium correlation threshold R2=0.01 and 0.1 for African and European data, respectively, using ancestry-specific reference panels. MR was performed using the random-effects inverse variance–weighted method for associations with multiple variants. The Cochran Q test was conducted to test heterogeneity and any form of pleiotropy among the instrumental variables, with a P value of <0.05 for ascertaining statistical significance.3 MR-Egger regression was used to further test for pleiotropy.4 The strength of the instrumental variables was evaluated using F statistics. The analyses were performed with the aid of TwoSampleMR package.5
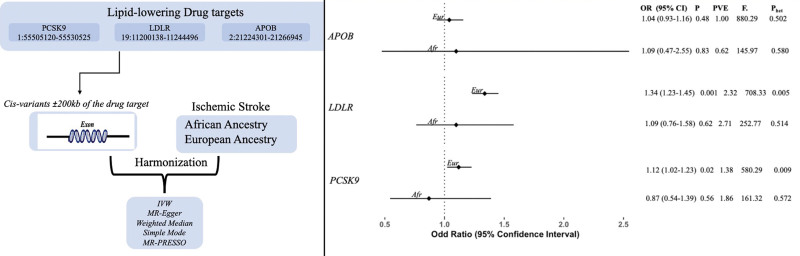
Genetically proxied PCSK9 perturbation was associated with a higher risk of ischemic stroke in European ancestry individuals (odds ratio, 1.118 [95% CI, 1.021–1.226]; P=0.016) but not in African ancestry individuals (odds ratio, 0.869 [95% CI, 0.543–1.389]; P=0.558). We found significant association in LDLR (odds ratio, 1.336 [95% CI, 1.229–1.1452]; P=0.001) inhibition with a higher risk of ischemic stroke in the European ancestry (Figure).
Figure.
Forest plot highlighting the association of genetically proxied lipid-lowering drug targets and ischemic stroke across the European (Eur) and African (Afr) ancestry populations. No significant association was found in Africans; however, same direction of effect was observed (odds ratio [OR], 1.10 [95% CI, 0.763–1.600]; P=0.616). APOB perturbation showed no significant association with risk of ischemic stroke in both European (OR, 1.039 [95% CI, 0.934–1.157]; P=0.477) and African ancestries (OR, 1.097 [95% CI, 0.473–2.547]; P=0.828). The findings of the MR-Egger, weighted median, and weighted mode sensitivity analyses revealed consistency in the effect estimates for all 3 techniques. We did not detect any evidence of heterogeneity in the 3 genes in African ancestry individuals, but some heterogeneity was detected in LDLR (low-density lipoprotein receptor) and PCSK9 (proprotein convertase subtilisin/kexin type 9) in the European ancestry. F indicates F statistics; IVW, inverse variance weighted; MR, Mendelian randomization; MR-PRESSO, Mendelian Randomization Pleiotropy Residual Sum and Outlier; Phet, P value for heterogeneity; and PVE, percentage variance explained.
The limitations of this study include potential bias from pleiotropic effects of the variants used as instruments through pathways unrelated to drug target perturbation. There may be additional differences within these populations examined that could impact the results, such as variations in lifestyle, diet, or other risk factors. Likewise, the data used in the study, especially for Africans, may not be representative of the whole diverse population and may limit the generalizability of the results. Finally, as some of the individuals in the studies were on lipid-lowering drugs, this may have also introduced a degree of bias into the results.
In conclusion, because the CIs for both ancestry groups overlap, it is possible that the risk of ischemic stroke is not statistically different for both groups, although the evidence is not conclusive and more research is needed. The findings may help prioritize further research in this area.
ARTICLE INFORMATION
Sources of Funding
Dr Soremekun is supported by the Africa Research Excellence Fund (AREF-325-SORE-F-C0904). Dr Fatumo is supported by the Wellcome Trust grant (220740/Z/20/Z) at MRC/UVRI and LSHTM. Dr Chikowore is an international training fellow supported by the Wellcome Trust grant (214205/Z/18/Z). Dr Gill is supported by the British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College.
Disclosures
Dr Gill is employed part-time by Novo Nordisk, outside this work. The other authors report no conflicts.
Footnotes
S. Fatumo and D. Gill contributed equally.
For Sources of Funding and Disclosures, see page e185.
Contributor Information
Gloria Kirabo, Email: gloriakirabo18@gmail.com.
Tinashe Chikowore, Email: tinashe.chikowore1@wits.ac.za.
Dipender Gill, Email: dipender.gill@imperial.ac.uk.
REFERENCES
- 1.Graham SE, Clarke SL, Wu KHH, Kanoni S, Zajac GJM, Ramdas S, Surakka I, Ntalla I, Vedantam S, Winkler TW, et al. ; VA Million Veteran Program. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra A, Malik R, Hachiya T, Jürgenson T, Namba S, Posner DC, Kamanu FK, Koido M, Le Grand Q, Shi M, et al. ; COMPASS Consortium. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature. 2022;611:115–123. doi: 10.1038/s41586-022-05165-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemani G, Bowden J, Smith GD. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–R208. doi: 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]