Abstract
Jaagsiekte sheep retrovirus (JSRV) is a type D retrovirus associated with a contagious lung tumor of sheep, ovine pulmonary carcinoma. Other than sheep, JSRV is known to infect goats, but there is no evidence of human infection. Until now it has not been possible to study the host range for JSRV because of the inability to grow this virus in culture. Here we show that the JSRV envelope protein (Env) can be used to pseudotype Moloney murine leukemia virus (MoMLV)-based retrovirus vectors and that such vectors can transduce human cells in culture. We constructed hybrid retrovirus packaging cells that express the JSRV Env and the MoMLV Gag-Pol proteins and can produce JSRV-pseudotype vectors at titers of up to 106 alkaline phosphatase-positive focus-forming units/ml. Using this high-titer virus, we have studied the host range for JSRV, which includes sheep, human, monkey, bovine, dog, and rabbit cells but not mouse, rat, or hamster cells. Considering the inability of the JSRV-pseudotype vector to transduce hamster cells, we used the hamster cell line-based Stanford G3 panel of whole human genome radiation hybrids to phenotypically map the JSRV receptor (JVR) gene within the p21.3 region of human chromosome 3. JVR is likely a new retrovirus receptor, as none of the previously identified retrovirus receptors localizes to the same position. Several chemokine receptors that have been shown to serve as coreceptors for lentivirus infection are clustered in the same region of chromosome 3; however, careful examination shows that the JSRV receptor does not colocalize with any of these genes.
Jaagsiekte sheep retrovirus (JSRV) is the causative agent of a contagious lung cancer of sheep known as ovine pulmonary carcinoma (OPC), also known as sheep pulmonary adenomatosis or jaagsiekte. OPC is a veterinary problem with significant economic impact in several countries. In addition, OPC shares characteristics with human bronchioalveolar carcinoma (BAC) (12, 32, 49), and BAC represents about 25% of human lung cancer cases (6). Lung cancer being the most common fatal form of cancer in humans (10), recent interest in JSRV stems from the hypothesis that OPC could be useful as a naturally occurring animal model for understanding the mechanism of pulmonary carcinogenesis (26, 49).
JSRV has been classified as a type D retrovirus, based on genomic organization, but has a type B-like Env protein (61). The sheep genome carries multiple copies of JSRV-like endogenous sheep retrovirus (ESRV) sequences (25, 27, 61), but subsequent studies have shown that JSRV is an exogenous virus distinct from ESRV sequences (4, 5, 44) and is specifically associated with OPC. Recent studies by Palmarini et al. (48) using an infectious molecular clone of JSRV have confirmed that JSRV is the causative agent of OPC. The mechanism of oncogenesis by JSRV is not known. JSRV has the genomic organization of a simple replication-competent retrovirus with no known oncogenes. The incubation period in naturally acquired OPC seems to range from months to years, suggesting insertional mutagenesis. However, OPC can be induced experimentally in 3 to 4 weeks, suggesting a mechanism of action more similar to that of a transforming retrovirus.
The main sites of JSRV replication and assembly are the transformed epithelial cells of the lung, especially the alveolar type II cells (45). The lung fluid and tumor extracts of infected sheep can be used for virus isolation and also for experimentally transmitting the disease to lambs by intratracheal inoculation (14, 37, 59), suggesting that the virus is stable in lung fluid. The stability of the virus in lung fluid as well as the ability to infect the epithelial cells of the lung indicate that JSRV vectors may be useful for gene transfer into the lung for human diseases such as cystic fibrosis. Even if the vectors were unable to transduce human airway epithelial cells, characterization of this virus would provide insights into developing improved vectors for transduction of the lung.
To study JSRV as a prospective retroviral vector, we first wanted to test whether the JSRV Env protein could be used to pseudotype a commonly used Moloney murine leukemia virus (MoMLV)-based retrovirus vector. Once this was established by transient transfection and the JSRV Env was shown to promote entry into human cells, we generated a packaging cell line for producing high-titer JSRV-pseudotype retrovirus vector which allowed us to investigate the host range for JSRV Env. Study of the JSRV host range has been hindered so far by the lack of an in vitro culture system for growing the virus, and only recently has an infectious molecular clone of the virus been developed (48). Subsequently, we used a human-hamster whole genome radiation hybrid (RH) panel to precisely localize the JSRV receptor (JVR) within human chromosome 3p21.3.
MATERIALS AND METHODS
Cell culture.
Mammalian cells, including SSF-123 primary sheep skin fibroblasts (gift from William Osborne, University of Washington, Seattle), HT-1080 human fibrosarcoma cells (American Type Culture Collection [ATCC] cell line CCL-121), 293 human kidney epithelial cells (ATCC CRL 1573), IB3 immortalized human bronchial epithelial cells (62), HeLa cervical carcinoma cells (ATCC CCL-2), NIH 3T3 thymidine kinase-deficient mouse embryo fibroblasts (60), Mus dunni tail fibroblasts (11), D17 canine osteosarcoma cells (ATCC CRL-6248), 208F rat embryo fibroblasts (51), MDBK bovine kidney epithelial cells (ATCC CCL-22), Vero African green monkey kidney epithelial cells (ATCC CCL-81), MF-NAN primary mouse (BALB/c) fibroblasts, MF-H1 primary mouse (C57BL/6) fibroblasts, and RbTE rabbit tracheal epithelial cells (gifts from Christine Halbert, Fred Hutchinson Cancer Research Center), were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Hyclone). RbTE cells were immortalized by transduction with the human papillomavirus E6 and E7 genes in a retrovirus vector, LXSN16E6E7 (24). G355 feline embryonic brain cells (15) were grown in McCoy's medium supplemented with 15% fetal bovine serum. CHO cells (ATCC CCL-61), A23 hamster cells, and the A23-derived RH clones (57) (gifts from Davis Cox, Stanford University) were grown in minimal essential medium-alpha supplemented with 10% fetal bovine serum.
Retroviral vectors and virus titer.
The nomenclature for retroviral vectors and pseudotypes has been discussed before (41). LAPSN is an MoMLV-based vector encoding the human placental alkaline phosphatase (AP) and the neomycin phosphotransferase proteins (42). Vectors with a JSRV pseudotype were made by using the pSX2.Jenv plasmid (Fig. 1), which was constructed by inserting the 1,883-bp MslI-Ecl136 fragment of JSRV-JS7 containing the Env coding region into the BsaAI- and MscI-cut 4,239-bp backbone of the pSX2 plasmid (38) by blunt-end ligation. JSRV-JS7 is a proviral clone derived from a λ phage library of genomic DNA from the JS7 cell line that was derived from a spontaneous case of OPC (unpublished results). Retrovirus vector titers were determined as described previously, by assaying for either AP+ focus-forming units (FFU) (17) or G418-resistant colony-forming units (CFU) (40).
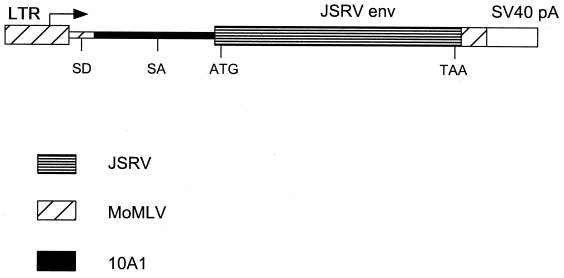
FIG. 1.
JSRV Env expression plasmid pSX2.Jenv. Abbreviations: SV40 pA, the early-region polyadenylation signal from simian virus 40; LTR, retrovirus long terminal repeat; SD and SA, splice donor and acceptor sites, respectively. The ATG start and TAA stop codons of Env are shown.
Generation of JSRV-pseudotype retrovirus packaging cells.
Stable retrovirus packaging cells expressing the JSRV Env were made using the techniques described previously for the construction of 10A1 murine leukemia virus (MLV)-pseudotype packaging cell lines (38). Briefly, NIH 3T3 cells that express the MoMLV Gag-Pol proteins (LGPS cells [39]) were cotransfected with plasmid pSX2.Jenv and a plasmid encoding hygromycin phosphotransferase (pSV2Δ13-hyg; gift from Paul Berg, Stanford University) at a 20:1 or 100:1 ratio, and 24 hygromycin-resistant clones were isolated. These clonal cell lines were tested for their ability to produce retrovirus vectors by measuring the titer of vector produced by the cells after the cells were transduced with the LAPSN vector made by using PT67 packaging cells (38). HT-1080 human cells were used as targets for measurement of the vector titer. The clonal line that produced the highest-titer vector was clone PJ14.
Marker rescue assay.
SSF and HT-1080 cells were plated at 105 cells per 6-cm dish on day 1, transduced with LAPSN(PT67) virus at a multiplicity of infection of ∼1 in the presence of Polybrene (4 μg/ml) on day 2, and trypsinized and replated in G418 (active concentration of 250 μg/ml for SSF and 400 μg/ml for HT-1080 cells). These polyclonal populations of cells carrying the LAPSN vector were used in the marker rescue assay for helper virus as described before (38). Briefly, SSF/LAPSN or HT-1080/LAPSN cells were plated at 5 × 105 cells per 6-cm dish on day 1, infected with 0.5 ml of LAPSN(PJ) test virus (6 × 105 AP FFU/ml) per dish in the presence of Polybrene on day 2, and trypsinized and split 1:10 every 2 to 3 days for 2 weeks while being kept at high density to facilitate potential virus spread. After 2 weeks, medium harvested from confluent dishes of cells was tested for LAPSN vector rescue and transfer using SSF cells as targets. MoMLV was used as a positive control to show that helper virus could rescue the LAPSN vector in these cells. The passaged SSF/LAPSN and HT-1080/LAPSN cells were also stained for AP to ensure that they retained the LAPSN vector.
RH mapping of JVR and STRL33 genes.
The Stanford G3 panel of human whole-genome hybrid cell lines was used for phenotypic RH mapping (www-shgc.stanford.edu/Mapping/rh/) (57). For mapping JVR, the RH cell lines were plated at 5 × 104 cells per well in a 6-well plate and exposed to LAPSN(JSRV) vector the next day. AP assays were performed as explained above, and AP+ FFU were counted to measure transduction.
To map the STRL33 gene, we used the Stanford G3 panel RH DNA (Research Genetics, Huntsville, Ala.) for genotypic mapping. PCR was performed with the STRL33-specific primers 5′-GCCAGGGTTTCGAGAAGCTGCTCTGGAATT-3′ and 5′-TCATAGTCCCTGGTGCTAGTTATTCTGGAT-3′. Genomic DNA samples from the A3 hamster cell line (57) and the RM human lymphoblastoid cell line (57) were used as negative and positive controls, respectively. All PCR amplifications were performed with an initial denaturation step at 94°C for 2 min, followed by 35 cycles of amplification at 94°C for 30 s, at 62°C for 1 min, and 68°C for 4 min, with a final extension at 68°C for 10 min. The PCR products were electrophoresed on 1% agarose gels and visualized by ethidium bromide staining.
RESULTS
JSRV Env protein can pseudotype an MoMLV-based retrovirus vector and promote transduction of human cells.
We tested whether the JSRV Env could be incorporated into virions containing MoMLV Gag-Pol proteins and an MoMLV-based vector by transient transfection of the JSRV Env expression plasmid pSX2.Jenv (Fig. 1) into LGPS/LAPSN cells. These cells contain the MoMLV-based LAPSN vector and the pLGPS plasmid for expression of MoMLV Gag-Pol proteins (38). Transfection of the pSX2 plasmid DNA expressing the 10A1 MLV Env (38) was used as a positive control. Two days after transfection, the medium from these cells was harvested and assayed for vector production using SSF-123 sheep skin fibroblasts and HT-1080 human fibrosarcoma cells as targets for transduction (Table 1). Both cell types could be transduced by the JSRV-pseudotype LAPSN vector, showing the ability of JSRV Env to pseudotype MoMLV-based vectors packaged with MoMLV Gag proteins as well as the ability of JSRV Env to promote entry into human cells. The JSRV-pseudotype vector titer was ∼10-fold higher on the sheep than on the human cells. In contrast, the titers of the 10A1- and xenotropic-pseudotype LAPSN vectors were at least 10-fold higher on the human than on the sheep cells, showing the preference of the JSRV-pseudotype vector for sheep cells. These results indicated the potential to develop a JSRV-pseudotype packaging line capable of producing high-titer virus capable of transducing human cells.
TABLE 1.
Transduction of sheep and human cells by the LAPSN vector with a JSRV, 10A1, or xenotropic pseudotypea
| LAPSN pseudotype | LAPSN titer (FFU/ml) measured on:
|
|
|---|---|---|
| SSF cells | HT-1080 cells | |
| None | <1 | <1 |
| JSRV | 2 × 104 | 2 × 103 |
| 10A1 | 80 | 2 × 103 |
| Xenotropic | 1 × 103 | 1 × 104 |
JSRV and 10A1 MLV pseudotypes of the LAPSN vector were made by calcium phosphate-mediated transient transfection (40) of LGPS/LAPSN cells with the JSRV or 10A1 env construct. As a negative control, Env− LAPSN vector (no pseudotype) were generated by transient transfection of LGPS/LAPSN cells with a plasmid that did not contain an env gene. Xenotropic-pseudotype vector was made by using a packaging cell line containing the LAPSN vector and MoMLV gag-pol and xenotropic MLV env genes. Vector titers were measured on the indicated target cells, and the results are means of at least two experiments.
High-titer vector production in the absence of replication-competent virus.
Stable JSRV-pseudotype retrovirus packaging lines expressing JSRV Env and MoMLV Gag-Pol proteins were generated, and individual clones were screened for packaging function as described in Materials and Methods. Of 24 clones tested for LAPSN vector production after introduction of the vector into the clonal cell lines, 6 could produce the vector at a titer of >103 AP+ FFU/ml, 10 did not produce any vector (<10 AP+ FFU/ml), and the rest produced intermediate titers. To show that these JSRV-pseudotype packaging cells could be used to generate stable vector-producing cell lines, the clone which was able to produce the highest titer virus (PJ14) was plated at 105 cells per 6-cm dish on day 1, transduced at a multiplicity of infection of ∼1 with LAPSN(PT67) virus in the presence of Polybrene on day 2, and trypsinized and replated at a 1:100 or 1:500 dilution in medium containing G418 (500 μg/ml, active) on day 3. Individual clones were selected and screened for LAPSN vector titer using SSF cells as targets. Titers as high as 6 × 105 AP+ FFU/ml were obtained, demonstrating that the JSRV-pseudotype packaging cells can be used to generate high-titer retrovirus vectors. Testing of the LAPSN vector produced from a high-titer clone by a marker rescue assay showed that the preparations were free of replication-competent virus (<1 FFU/ml).
Host range of JSRV Env.
The JSRV-pseudotype LAPSN vector was used to measure the ability of JSRV Env to promote transduction of a variety of mammalian cells (Table 2). The JSRV-pseudotype vector transduced various human cells but at titers ∼10-fold lower than that obtained with SSF-123 cells. Although the JSRV-pseudotype vector transduced various human cell lines, including IB3 bronchial epithelial cells, HT-1080 fibrosarcoma cells, and 293 kidney epithelial cells, it could only transduce HeLa human cervical carcinoma cells at low levels. In addition to sheep and human cells, the vector transduced a wide range of mammalian cells, including monkey, bovine, dog, and rabbit cells. The vector also transduced G-355 cat cells but at very low levels. The vector was unable to transduce wild or laboratory mouse cells, rat cells, or hamster cells. All cell types that exhibited low to undetectable transduction by the JSRV-pseudotype vector (≤20 infectious units per ml) could be transduced with a 10A1 MLV-pseudotype LAPSN vector made by using PT67 retrovirus packaging cells (38) at titers of >103 G418-resistant CFU/ml (HeLa cells) or AP+ FFU/ml (all other cell types) (data not shown), indicating that the LAPSN vector and MoMLV Gag-Pol proteins are functional in these cells, as expected, and that the block to infection was at the level of virus entry mediated by the JSRV Env.
TABLE 2.
Host range of LAPSN vector produced by JSRV-pseudotype packaging cells
| Species | Target cellsa | Vector titerb (AP+ FFU/ml) |
|---|---|---|
| Sheep | Sheep skin fibroblasts | 6 × 105 |
| Human | HT-1080 | 3 × 104 |
| IB3 | 4 × 104 | |
| 293 | 4 × 104 | |
| HeLa | 10 | |
| Monkey | Vero | 1 × 105 |
| Dog | D17 | 1 × 104 |
| Bovine | MDBK | 2 × 103 |
| Rabbit | RbTE | 2 × 103 |
| Cat | G355 | 20 |
| Mouse | ||
| NIH Swiss | NIH 3T3 TK− | <1 |
| BALB/c | MF-NAN | <1 |
| C57BL/6 | MF-H1 | <1 |
| M. dunni | M. dunni tail fibroblasts | <1 |
| Rat | 208F | <1 |
| Hamster | A23 | <1 |
| CHO | <1 |
TK−, thymidine kinase deficient.
The LAPSN(PJ4) titer on HeLa cells was measured by production of G418-resistant colonies (CFU per milliliter) rather than AP+ foci because HeLa cells have high levels of endogenous heat-stable AP. Results are means of at least two experiments.
JVR maps to region p21.3 of human chromosome 3 at a site different from those of other retrovirus receptors.
JSRV-pseudotype vectors transduce human but not hamster cells, indicating that it might be possible to map the position of the JSRV receptor by analyzing the susceptibility of human-hamster RH cell lines to transduction by a JSRV vector. We used the G3 panel of human RH cell lines from the Stanford Human Genome Center (SHGC) (57) for phenotypic mapping of JVR. The transduction result for the 83 ordered hybrid cell lines was 00000000000000000000000000000000001100001000000R000000000000001000100000000R0R00001, where 0 indicates no transduction (<10 FFU/ml), 1 indicates transduction (>100 FFU/ml), and R indicates an indeterminate result (cell clone not available for analysis). This result was submitted to the SHGC RH Server v4.0 (www-shgc.stanford.edu/RH/index.html), which mapped JVR at a distance of 18 cR10,000 (centiray distance for RH cells made using 10,000 rads of irradiation) from marker SHGC-11855 on chromosome 3 with a highly significant LOD score (log10 of the likelihood ratio) of 6.77. Subsequently, we used the multiple integrated maps at the National Center for Biotechnology Information (NCBI) Entrez Genomes site (www.ncbi.nlm.nih.gov/Entrez/Genome/org.html) to map JVR to a position within region p21.3 of chromosome 3.
The chromosomal locations of most other known retrovirus receptors have been determined (Table 3). Many of these map to chromosomes other than chromosome 3, showing that JSRV does not use these receptors for cell entry. Careful examination of the p21.3 region of human chromosome 3 showed that none of the previously mapped retroviral receptors localize to the same position. However, the lentivirus receptor STRL33 (also called Bonzo and TYMSTR) had been assigned to chromosome 3 but had not been more precisely localized (34). These studies indicated that JVR is probably a new retroviral receptor but might be the same as STRL33.
TABLE 3.
Chromosomal localization of retrovirus receptors in the human genome
| Retrovirus group | Receptor name(s) | Localization | Methoda | Reference(s)b |
|---|---|---|---|---|
| Ecotropic MLV | CAT1 (Rec1, SLC7A1) | 13q12 | In situ hybridization, RFLP | 1 |
| GALV/feline leukemia virus type B | PIT1 (GLVR1, SLC20A1) | 2q11-q14 | In situ hybridization | 33 |
| Amphotropic MLV | PIT2 (Ram1, GLVR2, SLC20A2) | 8p11-q11 | Somatic cell hybrids | 19 |
| Xenotropic and polytropic MLV | XPR1 | 1q25.1 | RH | 8 |
| RD114/simian type D retrovirus | RDR (SLC1A5) | 19q13.3 | RH | 52 |
| Feline leukemia virus type C | FLVCR | 1q32.1 | RH | A |
| T-cell leukemia viruses (human and simian T-cell lymphotropic viruses) | HTLVR | 17q23 | Somatic cell hybrids | 20, 58 |
| Lentiviruses (human, simian, and feline immunodeficiency viruses) | CD4 | 12p12-p13 | DNA sequencing | 3 |
| CCR1, -2b, -3, -5 | 3p21.3 | DNA sequencing, RH | 55 | |
| CCR4 | 3p22c | RH | 55 | |
| CCR8 | 3p22 | RH | 56 | |
| CXCR4 | 2q21 | In situ hybridization | 16 | |
| CX3CR1 (V28) | 3p22 | FISH, RH | 13, 35 | |
| GPR1 | 15q26.1 | FISH | 36 | |
| GPR15 (Bob) | 3q11.2-q13.1 | FISH | 29 | |
| ChemR23 | 12q21.2-q21.3 | RH | 54 | |
| APJ | 11q12 | FISH | 43 | |
| STRL33 (Bonzo, TYMSTR) | 3p21.3 | RH, somatic cell hybrids | 34, B | |
| JSRV | JVR | 3p21.3 | RH | B |
RFLP, restriction fragment length polymorphism; FISH, fluorescent in situ hybridization.
A, J.-L. Battini, J. E. J. Rasko, and A. D. Miller, unpublished data; B, this report.
The localization of CCR4 given by the authors (3p24) was changed to 3p22 to reflect more recent localization data for flanking markers.
STRL33 (Bonzo) maps to 3p21.3 region, ∼500 kb telomeric to the 285-kb CCR cluster.
To determine whether or not STRL33 might function as the JSRV receptor, we used the Stanford G3 panel RH DNA for mapping the STRL33 gene, as explained in Materials and Methods. The PCR results for DNA samples from the 83 ordered hybrids was 000000000000 0100R00100000000000000010000110000000000000000000001 010010000R001000010, where 0 indicates PCR negative, 1 indicates PCR positive, and R indicates an indeterminate result. This bar code was submitted to the SHGC RH Server v4.0, which mapped STRL33 at a distance of 13 cR10,000 (LOD, 9.46) from marker SHGC-12886. Using the multiple integrated maps at the NCBI Entrez Genomes site, we have localized the STRL33 gene to about 500 kb away from the CCR cluster in chromosome 3p21.3 and about 7.5 Mb from JVR, thus showing that STRL33 and JVR do not localize to the same position and JVR is not likely to be any of the known retroviral receptors (Fig. 2).
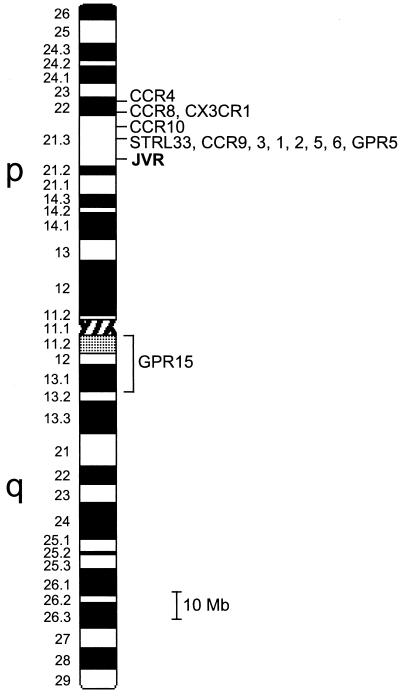
FIG. 2.
Retrovirus receptor and related G protein-coupled receptor genes on chromosome 3. Human chromosome 3 is ∼240 Mb in length, and an idiogram of this chromosome at the 550-band level is shown. Rough localizations are shown by brackets, while more precise localizations are shown by lines. Where the genes are too closely spaced to show the order on a map of this resolution, the genes are listed in order from telomere to centromere. References for the retrovirus receptor localizations are given in Table 3, and those for the related proteins are CCR9 (35), CCR10 (9, 35), and GPR5 (28). Relationships between distances (in centirays), determined by RH analysis and physical genome length, were derived primarily by using data from the multiple integrated maps of the WWW Entrez Genomes Division of the NCBI.
DISCUSSION
JSRV has recently gained prominence mainly because of the similarity of OPC to BAC in humans, suggesting that OPC can be used as an animal model to understand the process of pulmonary carcinogenesis. However, studies on JSRV have been hindered so far by the lack of a cell culture system for propagating the virus. Recently, a full-length infectious proviral molecular clone of JSRV was isolated from a natural case of OPC, and a cell culture system was developed to propagate the virus by replacing the upstream U3 with the cytomegalovirus early promoter (48). JSRV has been known to infect sheep and goats, but there is no evidence of human infection. Our studies show that the JSRV Env can be used to pseudotype MoMLV-based retroviral vectors containing MoMLV Gag-Pol proteins, thus providing a means for studying the host range of the viral Env protein. We have been able to generate packaging cell lines based on JSRV Env and MoMLV Gag-Pol proteins that can produce JSRV-pseudotype retroviral vectors at titers of up to 106 AP+ FFU/ml. Our results show that the JSRV Env promotes entry into sheep, human, monkey, bovine, dog, and rabbit cells but not mouse (laboratory or wild), rat, or hamster cells. The inability of the vector to transduce mouse or rat cells is unfortunate, as it prevents us from using rodents for studying JSRV pathogenesis or in vivo gene transfer to the lung. Poor transduction of HeLa cells suggests that the JSRV receptor (JVR) may not be constitutively expressed in all cell types.
This is the first report showing the ability of JSRV Env to transduce human cells and suggesting the feasibility of developing JSRV as a prospective retroviral vector for gene transfer to the lung. Although both viral (18, 53) and nonviral (2, 31) methods have been extensively studied, efficient gene transfer to the airway epithelial cells has proved difficult (21, 22, 50). Previous studies in our laboratory have shown that amphotropic retroviral vectors can efficiently transduce the basal and secretory airway epithelial cells in vitro, but in vivo delivery resulted in no detectable transduction in the intact normal airway epithelium and a low transduction rate in the wounded epithelium (23). This low retroviral transduction in vivo is due to the low abundance of retroviral receptors and inhibition of amphotropic retroviral vector transduction by pulmonary surfactant (63) or by soluble chondroitin sulfates in pleural effusions (7). Although JSRV infects several cell types in vivo (30, 45, 47, 48), the epithelial tumor cells in the lungs of sheep have been shown to be the major sites of viral replication (45), suggesting a natural tropism of the virus for the airway epithelial cells. Furthermore, OPC can be experimentally reproduced in newborn lambs by intratracheal inoculation of concentrated lung fluid or tumor extracts collected from OPC-affected sheep (14, 37, 59), demonstrating the stability of the virus in lung fluid. We are currently studying the ability of JSRV to infect airway epithelial cells and its stability in the presence of pulmonary surfactant.
The ability of JSRV Env to promote infection of human cells in culture could be relevant to the epidemiology of human lung cancer, especially with regard to nonsmokers exposed to sheep in which OPC is endemic. Although there is no proof for JSRV involvement in human lung carcinoma, the possibility of viral etiology cannot be excluded because of the similarity of BAC to OPC and the multifocal and multiclonal nature of some BAC cases (46). Several factors could explain the absence of evidence for human infection with JSRV, such as lack of immunological reagents to detect human infections. There has been no report of any serological study to evaluate human sera for JSRV antibodies. Alternatively, the JSRV Env might be able to bind to receptors and mediate entry of the viral genome, but some of the viral replicative elements may not be functional in human cells, resulting in postentry or replication blocks. It is known that the JSRV Gag-Pol proteins are functional during viral assembly in human cells, as evidenced by the use of an infectious molecular clone of JSRV to produce the virus in 293 human epithelial cells (48). However, their functionality during reverse transcription and integration of viral DNA in human cells is unknown. Another important factor might be the presence of transcriptionally active ESRV sequences in the sheep genome, which may induce tolerance to JSRV antigens in sheep and allow the virus to propagate and establish an infection. On the contrary, humans and other animals may develop a strong immune response leading to virus clearance.
Chromosome localization provides an important alternative approach to interference analysis to determine retroviral receptor usage. The advantages of chromosome localization are that extensive cross-interference analyses between the test virus and the growing number of existing viruses need not be performed, and the technique is informative for viruses that do not exhibit strong interference to infection by viruses that use the same receptor. Utilizing the inability of JSRV Env to promote infection of hamster cells, we have used a panel of human-hamster whole-genome RH cell lines to localize the JSRV receptor (JVR) gene to the p21.3 region of human chromosome 3. Although the majority of known retroviral receptors do not localize to chromosome 3, most of the CC-chemokine receptor genes (CCRs) which have been identified as coreceptors for lentiviruses have been shown to map within the 3p21.3 region (55). Careful analysis of the mapping data revealed that JVR does not map to the same positions as most of these receptors, being ∼7 Mb away from the 285-kb cluster of CCR3, -1, -2, -5, and -6 and farther away from CCR4, CCR8, and CCR10 (Fig. 2). The lentivirus receptor CX3CR1 has recently been mapped to the 3p24 region (35), leaving one other lentivirus receptor, STRL33 (Bonzo), in question (34). Using the G3 panel of RH DNA, we have localized the STRL33 gene 500 kb telomeric to the CCR cluster in region 3p21.3 and about 7.5 Mb away from JVR. These results indicate that JVR is a new retroviral receptor in human cells.
ACKNOWLEDGMENTS
We thank Jeanette Bishop for the isolation, cloning, and sequencing of the JSRV-JS7 provirus; Christine Halbert for providing the RbTE primary rabbit tracheal epithelial cells, MF-NAN primary mouse (BALB/c) fibroblasts, and MF-H1 primary mouse (C57BL/6) fibroblasts; William Osborne for providing the sheep skin fibroblasts; and David Cox for providing the RH cell lines.
This work was supported by grants DK47754 (A.D.M.), HL54881 (A.D.M.), and CA59116 (J.C.D.) from the National Institutes of Health. S.K.R. was supported by institutional funding.
REFERENCES
- 1.Albritton L M, Bowcock A M, Eddy R L, Morton C C, Tseng L, Farrer L A, Cavalli-Sforza L L, Shows T B, Cunningham J M. The human cationic amino acid transporter (ATRC1): physical and genetic mapping to 13q12-q14. Genomics. 1992;12:430–434. doi: 10.1016/0888-7543(92)90431-q. [DOI] [PubMed] [Google Scholar]
- 2.Alton E, Middleton P, Caplen N, Smith S, Steele D, Munkonge F, Jeffrey P, Cieddes D, Hart S, Williamson R, Fasold K, Miller A, Dickenson P, Stevenson B, McLachlan G, Dorin J, Porteous D. Non-invasive liposome-mediated gene delivery can correct the ion transport defect in cystic fibrosis mutant mice. Nat Genet. 1993;5:135–142. doi: 10.1038/ng1093-135. [DOI] [PubMed] [Google Scholar]
- 3.Ansari-Lari M A, Muzny D M, Lu J, Lu F, Lilley C E, Spanos S, Malley T, Gibbs R A. A gene-rich cluster between the CD4 and triosephosphate isomerase genes at human chromosome 12p13. Genome Res. 1996;6:314–326. doi: 10.1101/gr.6.4.314. [DOI] [PubMed] [Google Scholar]
- 4.Bai J, Bishop J V, Carlson J O, DeMartini J C. Sequence comparison of JSRV with endogenous proviruses: envelope genotypes and a novel ORF with similarity to a G-protein-coupled receptor. Virology. 1999;258:333–343. doi: 10.1006/viro.1999.9728. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Zhu R-Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retrovirus associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barsky S H, Cameron R, Osann K E, Tomita D, Holmes E C. Rising incidence of bronchioalveolar carcinoma and its unique clinicopathologic features. Cancer. 1994;73:1163–1170. doi: 10.1002/1097-0142(19940215)73:4<1163::aid-cncr2820730407>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 7.Batra R K, Olsen J C, Hoganson D K, Caterson B, Boucher R C. Retroviral gene transfer is inhibited by chondroitin sulfate proteoglycans/glycosaminoglycans in malignant pleural effusions. J Biol Chem. 1997;272:11736–11743. doi: 10.1074/jbc.272.18.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battini J-L, Rasko J E J, Miller A D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonini J A, Martin S K, Dralyuk F, Roe M W, Philipson L H, Steiner D F. Cloning, expression, and chromosomal mapping of a novel human CC-chemokine receptor (CCR10) that displays high-affinity binding for MCP-1 and MCP-3. DNA Cell Biol. 1997;16:1249–1256. doi: 10.1089/dna.1997.16.1249. [DOI] [PubMed] [Google Scholar]
- 10.Carney D N, De Leij L. Lung cancer biology. Semin Oncol. 1988;15:199–214. [PubMed] [Google Scholar]
- 11.Chattopadhyay S K, Lander M R, Gupta S, Rands E, Lowy D R. Origin of mink cytopathic focus-forming (MCF) viruses: comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981;113:465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- 12.Clayton F. The spectrum and significance of bronchioalveolar carcinomas. Pathol Annu. 1988;23:361–394. [PubMed] [Google Scholar]
- 13.Combadiere C, Ahuja S K, Murphy P M. Cloning, chromosomal localization, and RNA expression of a human beta chemokine receptor-like gene. DNA Cell Biol. 1995;14:673–680. doi: 10.1089/dna.1995.14.673. [DOI] [PubMed] [Google Scholar]
- 14.DeMartini J C, Rosadio R H, Sharp J M, Russell H I, Lairmore M D. Experimental coinduction of type D retrovirus-associated pulmonary carcinoma and lentivirus-associated lymphoid interstitial pneumonia in lambs. J Natl Cancer Inst. 1987;79:167–177. [PubMed] [Google Scholar]
- 15.Dunn K J, Yuan C C, Blair D G. A phenotypic host range alteration determines RD114 virus restriction in feline embryonic cells. J Virol. 1993;67:4704–4711. doi: 10.1128/jvi.67.8.4704-4711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Federsppiel B, Melhado I G, Duncan A M V, Delaney A, Schappert K, Clark-Lewis I, Jirik F R. Molecular cloning of the cDNA and chromosomal localization of the gene for a putative seven-transmembrane segment (7-TMS) receptor isolated from human spleen. Genomics. 1993;16:707–712. doi: 10.1006/geno.1993.1251. [DOI] [PubMed] [Google Scholar]
- 17.Fields-Berry S C, Halliday A L, Cepko C L. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flotte T R, Affione S A, Conrad C, McGrath S A, Solow R, Oka H, Zietlin P L, Guggino W B, Carter B J. Stable in vivo expression of the cystic fibrosis transmembrane regulator with adeno-associated virus vector. Proc Natl Acad Sci USA. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia J V, Jones C, Miller A D. Localization of the amphotropic murine leukemia virus receptor gene to the pericentromeric region of human chromosome 8. J Virol. 1991;65:6316–6319. doi: 10.1128/jvi.65.11.6316-6319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavalchin J, Fan N, Waterbury P G, Corbett E, Faldaz B D, Peshick S M, Poiesz B J, Papsidero L, Lane M J. Regional localization of the putative cell surface receptor for HTLV-I to human chromosome 17q23.2-17q25.3. Virology. 1995;212:196–203. doi: 10.1006/viro.1995.1468. [DOI] [PubMed] [Google Scholar]
- 21.Goldman M J, Lee P S, Yang J S, Wilson J M. Lentiviral vectors for gene therapy of cystic fibrosis. Hum Gene Ther. 1997;8:2261–2268. doi: 10.1089/hum.1997.8.18-2261. [DOI] [PubMed] [Google Scholar]
- 22.Grubb B R, Pickles R J, Ye H, Yankaskas J R, Vick R N, Engelhardt J F, Wilson J M, Johnson L G, Boucher R C. Inefficient gene transfer by adenovirus vector to cystic fibrosis airway epithelia of mice and humans. Nature. 1994;371:802–806. doi: 10.1038/371802a0. [DOI] [PubMed] [Google Scholar]
- 23.Halbert C L, Aitken M L, Miller A D. Retroviral vectors efficiently transduce basal and secretory airway epithelial cells in vitro resulting in persistent gene expression in organotypic culture. Hum Gene Ther. 1996;7:1871–1881. doi: 10.1089/hum.1996.7.15-1871. [DOI] [PubMed] [Google Scholar]
- 24.Halbert C, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hecht S J, Carlson J O, DeMartini J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and unaffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 26.Hecht S J, Sharp J M, DeMartini J C. Retroviral aetiopathogenesis of ovine pulmonary carcinoma: a critical appraisal. Br Vet J. 1996;152:395–409. doi: 10.1016/s0007-1935(96)80034-0. [DOI] [PubMed] [Google Scholar]
- 27.Hecht S J, Stedman K, Carlson J, DeMartini J C. Distribution of endogenous type B and D retroviral sequences in ungulates and other mammals. Proc Natl Acad Sci USA. 1996;93:3297–3302. doi: 10.1073/pnas.93.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heiber M, Docherty J M, Shah G, Nguyen T, Cheng R, Heng H H Q, Marchese A, Tsui L-C, Shi X, George S R, O'Dowd B F. Isolation of three novel human genes encoding G protein-coupled receptors. DNA Cell Biol. 1995;14:25–35. doi: 10.1089/dna.1995.14.25. [DOI] [PubMed] [Google Scholar]
- 29.Heiber M, Marchese A, Nguyen T, Heng H H Q, George S R, O'Dowd B F. A novel human gene encoding a G-protein-coupled receptor (GPR15) is locate on chromosome 3. Genomics. 1996;32:462–465. doi: 10.1006/geno.1996.0143. [DOI] [PubMed] [Google Scholar]
- 30.Holland M J, Palmarini M, Garcia-Goti M, Gonzalez L, McKendrick I, de las Heras M, Sharp J M. Jaagsiekte retrovirus is widely distributed in both T and B lymphocytes and in mononuclear phagocytes of the sheep with naturally and experimentally acquired pulmonary adenomatosis. J Virol. 1999;73:4004–4008. doi: 10.1128/jvi.73.5.4004-4008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyde S, Gill D, Higgins C, Trezise A, Macvinish L, Cuthbert A, Ratcliff R, Evans M, Colledge W. Correction of the ion transport defect in cystic fibrosis transgenic mice by gene therapy. Nature. 1993;362:250–255. doi: 10.1038/362250a0. [DOI] [PubMed] [Google Scholar]
- 32.Ives J C, Buffler P A, Greenberg S D. Environmental associations and histopathological patterns of carcinoma of the lung: the challenge and dilemma in epidemiological studies. Am Rev Respir Dis. 1983;128:195–209. doi: 10.1164/arrd.1983.128.1.195. [DOI] [PubMed] [Google Scholar]
- 33.Kaelbling M, Eddy R, Shows T B, Copeland N G, Gilbert D J, Jenkins N A, Klinger H P, O'Hara B. Localization of the human gene allowing infection by gibbon ape leukemia virus to human chromosome region 2q11-q14 and to the homologous region on mouse chromosome 2. J Virol. 1991;65:1743–1747. doi: 10.1128/jvi.65.4.1743-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loetscher M, Amara A, Oberlin E, Brass N, Legler D F, Loetscher P, D'Apuzzo M, Meese E, Rousset D, Virelizer J-L, Baggioloini M, Arenzana-Seisdedos F, Moser B. TYMSTR, a putative chemokine receptor selectively expressed in activated T cells, exhibits HIV-1 coreceptor function. Cell Biol. 1997;7:652–660. doi: 10.1016/s0960-9822(06)00292-2. [DOI] [PubMed] [Google Scholar]
- 35.Maho A, Bensimon A, Vassart G, Parmentier M. Mapping of the CCXCR1, CX3CR1, CCBP2 and CCR9 genes to the CCR cluster within the 3p21.3 region of human genome. Cytogenet Cell Genet. 1999;87:265–268. doi: 10.1159/000015443. [DOI] [PubMed] [Google Scholar]
- 36.Marchese A, Docherty J M, Nguyen T, Heiber M, Cheng R, Heng H H Q, Tsui L-C, Shi X, George S R, O'Dowd B F. Cloning of human genes encoding novel G protein-coupled receptors. Genomics. 1994;23:609–618. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- 37.Martin W B, Scott F M, Sharp J M, Angus K W, Norval M. Experimental production of sheep pulmonary adenomatosis (jaagsiekte) Nature. 1976;264:183–185. doi: 10.1038/264183a0. [DOI] [PubMed] [Google Scholar]
- 38.Miller A D, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for entry. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 41.Miller A D, Wolgamot G. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Dowd B F, Heiber M, Chan A, Heng H H Q, Tsui L-C, Kennedy J L, Shi X, Petronis A, George S R, Nguyen T. A human gene that shows identity with the gene encoding the angiotensin receptor is located on chromosome 11. Gene. 1993;136:355–360. doi: 10.1016/0378-1119(93)90495-o. [DOI] [PubMed] [Google Scholar]
- 44.Palmarini M, Cousens C, Dalziel R G, Bai J, Stedman K, DeMartini J C, Sharp J M. The exogenous form of jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmarini M, Dewar P, de las Heras M, Inglis N F, Dalziel R G, Sharp J M. Epithelial tumor cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 46.Palmarini M, Fan H, Sharp J M. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 47.Palmarini M, Holland M J, Cousens C, Dalziel R G, Sharp J M. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J Gen Virol. 1996;77:2991–2998. doi: 10.1099/0022-1317-77-12-2991. [DOI] [PubMed] [Google Scholar]
- 48.Palmarini M, Sharp J M, de las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perk K, Hod I. Sheep lung carcinoma: an endemic analogue of a sporadic human neoplasm. JNCI. 1982;69:747–749. [PubMed] [Google Scholar]
- 50.Pickles R J, McCarty D, Matsui H, Hart P J, Randell S H, Boucher R C. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol. 1998;72:6014–6023. doi: 10.1128/jvi.72.7.6014-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quade K. Transformation of mammalian cells by avian myelocytomatosis virus and avian erythroblastosis virus. Virology. 1979;98:461–465. doi: 10.1016/0042-6822(79)90569-5. [DOI] [PubMed] [Google Scholar]
- 52.Rasko J E J, Battini J-L, Gottschalk R J, Mazo I, Miller A D. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenfeld M A, Yoshimura K, Trapnell B C, Yoneyama K, Rosenthal E R, Dalemans W, Fukayama M, Bargon J, Stier L E, Stratford-Perricaudet S, Perricaudet M, Guggino M W B, Pavirani A, Lecocq J, Crystal R G. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68:143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- 54.Samson M, Edinger A L, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms R W, Parmentier M. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol. 1998;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 55.Samson M, Soularue P, Vassart G, Parmentier M. The genes encoding the human CC-chemokine receptors CC-CKR1 to CC-CKR5 (CMKBR1-CMKBR5) are clustered in the p21.3-p24 region of chromosome 3. Genomics. 1996;36:522–526. doi: 10.1006/geno.1996.0498. [DOI] [PubMed] [Google Scholar]
- 56.Samson M, Stordeur P, Labbe O, Soularue P, Vassart G, Parmentier M. Molecular cloning and chromosomal mapping of a novel human gene, ChemR1, expressed in T lymphocytes and polymorphonuclear cells and encoding a putative chemokine receptor. Eur J Immunol. 1996;26:3021–3028. doi: 10.1002/eji.1830261230. [DOI] [PubMed] [Google Scholar]
- 57.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, Lee R, Maratukulam A, O'Connor K, Perkins S, Piercy M, Qin F, Reif T, Sanders C, She X, Sun W-L, Tabar P, Voyticky S, Cowles S, Fan J-B, Mader C, Quackenbush J, Myers R M, Cox D R. An STS-based radiation hybrid map of the human genome. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 58.Tajima Y, Tashiro K, Camerini D. Assignment of the possible HTLV receptor gene to chromosome 17q21-q23. Somat Cell Mol Genet. 1997;23:225–227. doi: 10.1007/BF02721374. [DOI] [PubMed] [Google Scholar]
- 59.Verwoerd D W, Williamson A L, De Villiers E M. Aetiology of jaagsiekte: transmission by means of subcellular fractions and evidence for the involvement of a retrovirus. Onderstepoort J Vet Res. 1980;47:275–280. [PubMed] [Google Scholar]
- 60.Wei C-M, Gibson M, Spear P G, Scolnick E M. Construction and isolation of a transmissible retrovirus containing the src gene of Harvey murine sarcoma virus and the thymidine kinase gene of herpes simplex virus type 1. J Virol. 1981;39:935–944. doi: 10.1128/jvi.39.3.935-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeitlin P L, Lu L, Rhim J, Cutting G, Stetten G, Kieffer K A, Craig R, Guggino W B. A cystic fibrosis bronchial epithelial cell line: immortalization by Adeno-12-SV40 infection. Am J Respir Cell Mol Biol. 1991;4:313–319. doi: 10.1165/ajrcmb/4.4.313. [DOI] [PubMed] [Google Scholar]
- 63.Zsengeller Z K, Halbert C, Miller A D, Wert S E, Whitsett J A, Bachurski C J. Keratinocyte growth factor stimulates transduction of the respiratory epithelium by retroviral vectors. Hum Gene Ther. 1999;10:341–353. doi: 10.1089/10430349950018797. [DOI] [PubMed] [Google Scholar]