Abstract
Adenoviruses (Ad) code for immunoregulatory and cytokine regulatory proteins, one of which is the early region 3, 14.7-kDa protein (Ad E3-14.7K), which has been shown to inhibit tumor necrosis factor alpha-induced apoptosis. In an effort to understand the mechanism of action of Ad E3-14.7K, we previously searched for cell proteins with which it interacted. Three Ad E3-14.7K-interacting proteins (FIP-1, -2, and -3) were isolated. FIP-1 is a small GTPase which was used in this report as bait in the yeast two-hybrid system to find other interacting cell targets. The search resulted in the isolation of a protein, which we called GIP-1 (GTPase-interacting protein) that subsequently was shown to be identical to one of the light-chain components of human dynein (TCTEL1). FIP-1 was able to bind both TCTEL1 and Ad E3-14.7K simultaneously and was necessary to form a complex in which the viral protein was associated with a microtubule-binding motor protein. The functional significance of these interactions is discussed with respect to the steps of the Ad life cycle which are microtubule associated.
Adenoviruses express several proteins which downregulate the host's immune response (32, 34). It has been postulated that these immunoregulatory and cytokine-regulatory proteins either prevent early cell lysis before the production of viral progeny during acute infection or enable the virus to establish a state of persistence or latency within the host (reviewed by Lukashok and Horwitz [17]). The adenovirus genes which code for many of these proteins are clustered in early transcription region 3 (E3). The type 2 adenovirus E3 14.7-kDa protein, named Ad E3-14.7K, is one of seven gene products expressed in this region and is a small hydrophilic protein which inhibits tumor necrosis factor alpha (TNF-α)-mediated cell killing. Its mechanism of action remains incompletely understood (8), but it has been shown that Ad E3-14.7K does not affect the number of TNF-α receptors or the affinity of TNF-α for its receptor (6, 33). Ad E3-14.7K inhibits the TNF-α-induced activation of cytoplasmic phospholipase A2, an enzyme that results in the release of arachidonic acid from the cell membrane (35). It has also been reported that Ad E3-14.7K binds to caspase 8 and prevents cell killing by inhibiting this caspase (1), but additional observations have failed to confirm the importance of this interaction (Horwitz et al., personal observations). Tufariello et al. (27, 28) demonstrated an in vivo effect of Ad E3-14.7K by showing that mice infected with a vaccinia virus expressing both Ad E3-14.7K and TNF-α had a more severe pulmonary pathology, a higher mortality rate, and higher viral titers than mice infected with a vaccinia virus expressing TNF-α alone (27, 28). These data support the conclusion that Ad E3-14.7K counteracts the inflammatory and antiviral protective effects of TNF-α in an in vivo model of viral infection.
Because the mechanism by which Ad E3-14.7K exerts its anti-TNF-α effect remains incompletely understood, we used the yeast two-hybrid system to look for proteins which interact with Ad E3-14.7K (15). Proteins from a HeLa cell (human) library were identified and named Ad E3-14.7K-interacting proteins (FIPs). BLAST sequence analysis revealed that one of these, FIP-1, was a member of a new family of low-molecular-weight (LMW) GTPases (15, 21). FIP-1 associated with various phosphorylated proteins in the presence of TNF-α, suggesting that it is involved in TNF-α signaling pathways (15). It has two regions of homology with bacterial proteases, although no proteolytic function has yet been demonstrated. FIP-1 colocalized with Ad E3-14.7K in a perinuclear structure and at the cell membrane.
A protein identical to FIP-1 was independently isolated using degenerate oligonucleotide primers from conserved sequences in other GTPases and was named RagA (21). RagA appears to be involved in the Ran/Gsp 1 GTPase pathway responsible for nucleus-cytoplasm trafficking of macromolecules (7). Ran-GTP has also recently been associated with cell cycle control by affecting aster formation induced by chromosome-binding protein RCC1 at centrosomes and controlling microtubule assembly originating from this structure during mitosis (18, 31). The involvement of FIP-1/RagA in this process has been inferred because of mutational analysis in the yeast homologues of human RCC1 (PRP20) and FIP-1 (GTR1). In order to determine cell proteins that might be involved, together with FIP-1, in these cell signaling pathways, FIP-1 was used as bait in the yeast two-hybrid system. This search identified a human protein, which we initially named GTPase-interacting protein 1 (GIP-1). However, in view of the subsequent publication of the sequences of light chains (LC) of mouse and human dyneins, GIP-1 appears to be identical to TCTEL1, the human homologue of one of the three LC (Tctex-1) of mouse dynein (11, 29). Dynein is part of the molecular motor system, which is located at the negative ends of microtubules near the nuclear membrane. Microtubules have been shown to be involved in many transport processes including movement of adenovirus, herpes simplex virus, and cytoplasmic organelles such as endosomes from the plasma membrane to the nuclear membrane (16, 23, 24, 30). Our studies show that Ad E3-14.7K can be found in a complex with GIP-1/TCTEL1 but only when FIP-1 (RagA) is present as a bridging protein. These observations appear to link two pathways: (i) the Ran-GTP effects either on nucleus-cytoplasm transport or on centrosome formation and (ii) the dynein-microtubule effects on transcytoplasmic transport of organelles, viruses, and signal transduction molecules.
MATERIALS AND METHODS
Cell lines.
For cotransfection and immunofluorescence labeling of cells, E293 human embryonic kidney cells and A549 human lung cells, respectively, were used. These cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. For coimmunoprecipitation experiments requiring radioactive labeling of proteins, T293 cells which express simian virus 40 T antigen were used. T293 and E293 cells were maintained in an identical manner.
Plasmid constructs.
pBMT-FIP-1, the bait, was constructed by cloning the FIP-1-8a cDNA in frame at the C terminus of the coding sequence for the LexA domain. pBMT116 was linearized by digestion with SmaI and SalI, and the FIP-1-8a cDNA was inserted between the restriction sites. Construction of pcDNA-T7 and pcDNA-FLAG from pcDNA-3 (Invitrogen) has been previously described (15). pcDNA-T7-GIP-1 was made by excising the GIP-1 cDNA from the pGAD vector at the BamHI and XhoI sites and inserting it in pcDNA-T7 linearized with BamHI and XholI. pcDNA-FLAG-FIP-1 was made by excising the FIP-1 cDNA from pBMT-FIP-1 at the EcoRI and XhoI restriction sites and cloning it into the pcDNA-FLAG plasmid with FLAG expressed at the 5′ end of the FIP-1 coding sequence. For the glutathione S-transferase (GST) assay, the pGEX-FIP-1 construct, previously described (15) was used. pCITE-GIP-1 was constructed by inserting the GIP-1 clone into the pCITE plasmid (Novagen) at the BamHI and XhoI sites. All constructs were confirmed by sequencing. The pcDNA-FLAG-RIP plasmid was obtained from David Wallach, Weizmann Institute. The pcDNA-CMV-GFP plasmid, which expresses green fluorescent protein, was previously described (12).
The yeast two-hybrid screen and specificity test.
The yeast two-hybrid screen has been previously described (15, 19, 20). The pBMT-FIP-1 was used as the bait to screen a HeLa cDNA library expressed as fusion proteins with the Gal 4 activation domain. The screening was done with Saccharomyces cerevisiae L40 yeast, which expresses reporter genes conferring histidine auxotrophy and β-galactosidase activity. The screening was carried out in histidine-deficient medium supplemented with 35 mM 3-aminotriazole, and 2 × 107 colonies were screened. Specificity of the GIP-1 clone was confirmed by cotransforming S. cerevisiae with pGAD-GIP-1 and each of four control proteins expressed in the bait vector to look for His auxotrophy and β-galactosidase activity. The proteins were the basic helix-loop-helix domain (bHLH) of mouse c-myc (mMyc), the transactivation domain (TAD) of mMyc, mouse MaxI (mMaxI), and human lamin (hLamin). GIP-1 was also tested against Ad E1B-19K and Bcl-2 to see if it interacted with known antiapoptotic proteins. hLamin, bHLH TAD, and mMaxI were generously provided by Ron DePinho, Albert Einstein College of Medicine. Bcl-2 and Ad E1B-19K were provided by R. Chinnaduri, St. Louis University, St. Louis, Mo. The HeLa cell library was provided by Greg Hannon and David Beach, Cold Spring Harbor Laboratory, and amplified according to standard protocols.
Northern blots of GIP-1.
A Northern blot of human tissues (Clontech) was probed with GIP-1 cDNA labeled with [32P]dCTP (Pharmacia) according to the manufacturer's directions.
Coimmunoprecipitation of FIP-1 and GIP-1.
Human T293 cells were grown as a monolayer in 10-cm-diameter dishes and transfected with 4 μg of either pcDNA-FLAG-FIP-1 or pcDNA-T7-GIP-1 or with both plasmids (2 μg of each) using Lipofectamine (Gibco). Twenty-four hours after transfection, the cells were labeled with [35S]methionine and -cysteine (1 mCi/ml) in methionine- and cysteine-free medium for 3 h at 37°C. Cells were scraped from the dish, washed with ice-cold phosphate-buffered saline (PBS), and lysed with ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 1% Nonidet P-40 [NP-40], 1 mM phenylmethylsulfonyl fluoride (PMSF), aprotinin [1 μg/ml], leupeptin [1 μg/ml]) for 30 min at 4°C on a rotating platform. ATP (5 mM) was added to the solution to prevent GIP-1 precipitation (10). Samples were preincubated with protein A- and protein G-agarose (Santa Cruz Biotechnology) and briefly centrifuged at 13,000 × g; 500 μl of each sample was immunoprecipitated with 1 μg of monoclonal anti-T7 antibody (Novagen), and 500 μl of each sample was precipitated with 1 μg of monoclonal anti-FLAG antibody (Sigma) for 1 h on a rocking platform at 4°C. Forty microliters of protein A and protein G beads (50% [wt/vol] was added to the lysate, and the mixture was incubated an additional hour at 4°C. The beads were washed three times with ice-cold lysis buffer and resuspended in 40 μl of Laemmli buffer (2% sodium dodecyl sulfate [SDS], 10% glycerol, 100 mM dithiothreitol, 60 mM Tris [pH 6.8], 0.001% bromphenol blue) and boiled. After centrifugation, samples were analyzed by SDS–10% polyacrylamide gel electrophoresis (PAGE). The gel was dried and exposed to X-ray film (Kodak).
GST assay to test for specific in vitro binding between FIP-1 and GIP-1.
The GST binding assay has been previously described (2, 15). The GST–FIP-1 fusion protein was expressed in Escherichia coli and bound to glutathione beads as previously described. The single tube protein 2 T7 assay (Novagen) was used to translate and transcribe pCITE-GIP-1 into the coding sequence for [35S]methionine-labeled GIP-1 protein. An aliquot of 35S-labeled GIP-1 was incubated with an aliquot of glutathione beads with either bound GST–FIP-1 or GST alone for 1 h at 4°C. The beads were washed three times with 1 M NaCl-NETN buffer (1 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.5% NP-40, 1 mM PMSF) and then once with 0.1 M NaCl-NETN buffer. the beads were resuspended in Laemmli buffer, heat denatured, and subjected to SDS–12% PAGE followed by autoradiography.
In vitro cotranslation of GIP-1, FIP-1, and Ad E3-14.7K.
GIP-1, FIP-1 and Ad E3-14.7K were translated as T7- or FLAG-tagged pcDNA 3.1 plasmids either alone or in various combinations by the single tube protein 3 T7 assay (Novagen) according to the manufacturer's protocol. 35S-labeled proteins were incubated with anti-T7 monoclonal antibodies, anti-Ad E3-14.7K rabbit polyclonal antibodies, or anti-FIP-1 antibodies for 1 to 2 h at 4°C following precipitation with protein A- and protein G-agarose (Santa Cruz Biotechnology) for 1 h at 4°C. The beads were washed three or four times with ice-cold PBS containing 1% NP-40 and a cocktail of protease inhibitors (Boehringer). The labeled proteins were eluted by boiling for 5 min in Laemmli buffer. After SDS-PAGE, the gel was dried and exposed to X-ray film (Kodak). Polyclonal anti-FIP-1 antibodies were produced in rabbits using an N-terminal peptide linked to keyhold limpet hemocyanin.
Flow cytometric analyses of cell cycle status and apoptosis.
Transfected or untransfected cells (E293) were harvested, washed in ice-cold PBS, and fixed in 70% ethanol for 1 h. After an additional wash in PBS, 3 × 104 to 5 × 104 cells were resuspended in propidium iodide solution (10 μg/ml) with DNase-free RNase (20 u/ml) in PBS. After 30 min at 37°C in the dark followed by filtration through a 35-mm-diameter cell strainer cap (Falcon), the cell cycle status was tested by flow cytometry on a FACSort (Beckton Dickinson). The number of sub-G1 phase cells is a measure of the number of cells undergoing apoptosis (5). Apoptosis also was estimated by direct microscopy in living cells by cotransfection of various plasmids together with pcDNA-CMV-GFP as described previously (12).
RESULTS
FIP-1 interacts specifically with the cell protein GIP-1/ TCTEL1 in the yeast two-hybrid assay.
By using the yeast two-hybrid assay, we have identified a specific interaction between FIP-1 and the cell protein which we initially named GIP-1. GIP-1 was isolated in 19 of 44 colonies tested. In the yeast two-hybrid assay, GIP-1 did not interact with a battery of unrelated baits, i.e., hLamin-C, mMyc TAD, mMyc bHLH, mMaxI, Ad E1B-19K, or Bcl-2 proteins, thus confirming that the interaction between FIP-1 and GIP-1 was specific.
In SDS-PAGE, GIP-1 migrated approximately as a 14-kDa protein. Analysis of the sequence by the BLAST program revealed that GIP-1 was 113 amino acids in length and had 93% identity and 97% homology with the mouse Tctex-1 protein (11) (Fig. 1). When the sequence of the human homologue named TCTEL1 was published (29), we found that GIP-1 and TCTEL1 were identical.
FIG. 1.
Comparison of the sequence of a human library-derived clone of GIP-1 and mouse Tctex-1. The cDNA sequence was derived from clones from the yeast two-hybrid screening. Polypeptide primary structures were deduced by using conventional genetic codons through the computerized program GCG (Genetics Computer Group, Madison, Wis.). GIP-1 was found to be identical to TCTEL1 (29).
GIP-1/TCTEL1 mRNA is expressed in many human tissues.
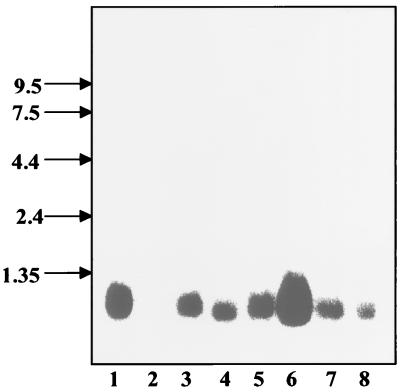
A Northern blot of eight human tissues was probed with a GIP-1/TCTEL1 cDNA probe, and the mRNA of the protein was detected in all tissues except human brain, with the highest levels in skeletal muscle. The probed band measured approximately 1 kb, which is consistent with the size of the full-length GIP-1/TCTEL1 clone (Fig. 2).
FIG. 2.
GIP-1/TCTEL1 is expressed ubiquitously in human tissues. Northern blots of human tissues purchased from Clontech were hybridized with a 32P-labeled cDNA probe of GIP-1/TCTEL1. GIP-1/TCTEL1 was expressed in seven of the eight human tissues but at different levels in the various organs tested. Lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas.
FIP-1 binds specifically to GIP-1/TCTEL1 in vitro and in vivo and does not require the addition of TNF-α.
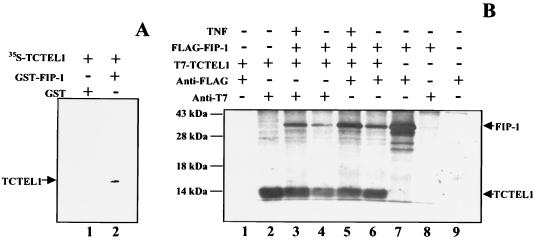
The interaction between FIP-1 and GIP-1/TCTEL1 revealed by the yeast complementation assay was further confirmed by in vitro and in vivo binding assays. The GST–FIP-1 fusion protein bound to glutathione beads retained GIP-1/TCTEL1 specifically, while there was no binding between GST alone and GIP-1/TCTEL1 (Fig. 3A).
FIG. 3.
Interactions between FIP-1 and GIP-1/TCTEL1. (A) FIP-1 was expressed as a GST fusion protein and was bound to glutathione-Sepharose beads. FIP-1-containing beads were incubated with [35S]methionine-labeled transcription and translation product GIP-1/TCTEL1. After being washed, the beads were resuspended in SDS-containing sample buffer, boiled, and subjected to SDS-PAGE followed by autoradiography. The radioactive GIP-1/TCTEL1 protein was retained by the GST–FIP-1 protein (lane 2) and not by GST alone (lane 1). (B) Coimmunoprecipitations of FLAG–FIP-1 and T7–GIP-1/TCTEL1 from T293 cells cotransfected with both of the corresponding plasmids and labeled with [35S]methionine. Cell lysates were prepared in the presence of 5 mM ATP and coimmunoprecipitated with monoclonal antibodies recognizing either T7- or FLAG-tagged proteins. Cells in lane 9 were mock transfected; those in lanes 3 and 5 were also treated with TNF-α (20 ng/ml for 20 min). The molecular mass markers are on the left, and relevant proteins are on the right.
Coimmunoprecipitation was used to confirm in vivo binding between FIP-1 and GIP-1/TCTEL1. The complex formed by the tagged proteins FLAG–FIP-1 bound to T7–GIP-1/TCTEL1 was immunoprecipitated by antibodies to both FLAG and T7 in the presence of ATP, which is required to maintain GIP-1/TCTEL1 in its soluble form (Fig. 3B). The interaction of GIP-1/TCTEL1 and FIP-1 was not dependent on the addition of TNF-α. Analysis of the cells overexpressing the immunofluorescent FIP-1 and GIP-1/TCTEL1 had shown that both proteins colocalized in the cytoplasm of transiently transfected cells (data not shown).
FIP-1 promoted complex formation between GIP-1/TCTELI and Ad E3-14.7K.
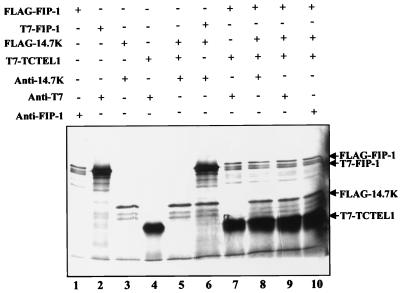
The in vitro system was used for studying the interactions between GIP-1/TCTEL1, FIP-1, and Ad E3-14.7K. When GIP-1/TCTEL1 and Ad E3-14.7K were cotranslated in the reticulocyte lysate system and immunoprecipitated by an antibody to Ad E3-14.7K, no GIP-1/TCTEL1 was detected in the immunoprecipitate (Fig. 4, lane 5). In contrast, transcription and translation in vitro of Ad E3-14.7K and FIP-1 together followed by immunoprecipitation with an antibody to Ad E3-14.7K resulted in the detection of FIP-1 and Ad E3-14.7K in the immunoprecipitate (Fig. 4, lane 6) as had been shown previously in vivo (15). Because FIP-1 binds to both GIP-1/TCTEL1 (Fig. 4, lane 7) and Ad E3-14.7K but the latter two proteins do not bind directly, it was possible to show that FIP-1 promoted the formation of a complex containing both GIP-1/TCTEL1 and Ad E3-14.7K (Fig. 4, lanes 8 to 10).
FIG. 4.
In vitro binding of GIP-1/TCTEL1, FIP-1, and Ad E3-14.7K. After in vitro transcription and translation of the GIP-1/TCTEL1, FIP-1, and Ad E3-14.7K, the 35S-labeled proteins were precipitated with anti-Ad E3-14.7K, anti-T7, or anti-FIP-1 antibodies and tested by SDS–12% PAGE. The relevant proteins are indicated on the right.
Overexpression of GIP-1/TCTEL-1 does not affect cell cycle progression or lead to cell death.
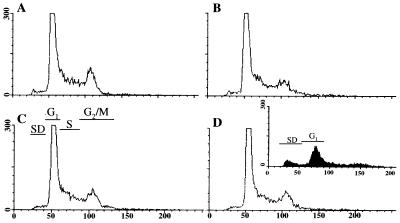
Because GIP-1/TCTEL1 formed a complex with the Ad E3-14.7K inhibitor of TNF-α cytolysis, we studied whether GIP-1/TCTEL1 would either cause or prevent apoptosis. When GIP-1/TCTEL1 was overexpressed in 293 cells, the cells maintained their normal morphology both in medium with 10% serum and in the absence of serum, as detected by coexpression with a green fluorescent protein expressing plasmid (data not shown). This was confirmed by fluorescence-activated cell sorter analysis of propidium iodide-stained transfected cells, which showed few cells with subdiploid amounts of DNA and no cell cycle differences between FIP-1-transfected, GIP-1/TCTEL1-transfected, and cotransfected cells (Fig. 5A to D). As a positive control, FIP-3-transfected cells, some of which do undergo apoptosis (13), are shown in the inset of Fig. 5D. An increase in the number of cells with subdiploid amounts of DNA and a decrease in the number of cells in G1 are shown. When GIP-1/TCTEL1 was overexpressed in combination with the receptor-interacting protein (RIP), the apoptotic effects of RIP were not reversed (data not shown). These results suggest that the GIP-1/TCTEL1 is not directly involved in TNF-α apoptotic pathways.
FIG. 5.
Analysis of cell cycle of GIP-1/TCTEL1- and FIP-1-transfected cells. E293 cells were transfected with pcDNA-FIP-1 (B), pcDNA-GIP-1/TCTEL1 (C), and pcDNA-FIP-1 plus pcDNA-GIP-1/TCTEL1 (D), and after 4 to 5 h cells were maintained with medium with 5% fetal calf serum. Control cells (A) did not contain any of the three plasmids described above. At 24 h posttransfection, cells were stained with propidium iodide and analyzed by fluorescence-activated cell sorter. The percentage of transfected cells in each of the panels was determined to be between 70 and 80% by cotransfection with 0.15 μg of the pcDNA-GFP plasmid as described previously (12). The inset (D) shows that transfections with pcDNA-FIP-3, which cause apoptosis (13), result in an increase in the number of cells containing subdiploid (SD) amounts of DNA and a decrease in the number in G1 or other phases of the cell cycle. Axes: x, intensity of fluorescence (amount of DNA); y, number of cells (in hundreds). The SD, G1, S, and G2/M phases of the cell cycle are indicated.
DISCUSSION
Using a yeast two-hybrid assay, we have determined that FIP-1, a LMW GTPase, binds specifically to GIP-1. Although there is no direct interaction between GIP-1 and Ad E3-14.7K, the complex containing FIP-1 and GIP-1 is also able to bind the Ad E3-14.7K protein. BLAST analysis revealed that GIP-1 has a very high degree of homology with the mouse Tctex-1 protein, which has been identified as one of the LC of cytoplasmic dynein (10, 11, 29).
The mouse Tctex-1 protein is encoded in the t complex, a large region on chromosome 17 which is implicated in transmission ratio distortion. This is a process whereby a chromosomal haplotype is not inherited in a Mendelian fashion, but rather is preferentially passed on to the offspring. In mice, heterozygous males pass the t haplotype to 99% of their offspring and males homozygous for the t haplotype are sterile. The exact mechanism of transmission ratio distortion remains unknown. In 1996, King et al. determined that Tctex-1 is the 14-kDa LC of cytoplasmic dynein (10). It therefore seems that Tctex-1, either alone or in association with cytoplasmic dynein, plays a role in the transmission of ratio disorder and male sterility in mice.
Dyneins are energy-dependent motor proteins. Axonal dynein mediates flagellar movement, while cytoplasmic dynein is involved in the translocation of cellular organelles. Cytoplasmic dynein is a multimer made up of two heavy chains, a 74-kDa intermediate chain, and three LC molecular masses of 8, 14, and 22 kDa. King et al. isolated, purified, and characterized the 14-kDa LC, thus determining that it is a 113-amino-acid protein. GIP-1/TCTEL1 is also 113 amino acids long and has 93% homology with the mouse Tctex-1 protein; therefore, we conclude that GIP-1 is the 14-kDa LC of dynein.
The concept that Ad E3-14.7K associates with FIP-1, which associates with the LC of dynein, is indeed provocative. There have been a number of reports of adenovirus type 5 associating with microtubules and cytoskeletal elements. It has been shown by electron microscopy that the adenovirion binds to microtubules, and it was hypothesized that this represents a mechanism to vectorially transfer virions from the cell membrane to the nucleus (4). These early studies were confirmed and extended to show that binding was specifically between the hexon protein and microtubules (16). Further data suggested that cytoplasmic dynein plays a role in translocating adenovirus to the cell nucleus (P. L. Leopold, G. Kreitzer, S. Rempel, K. K. Pfister, and R. Crystal, Abstr. 1st Ann. Meet. Am. Soc. Gene Ther., p. 178a, 1998). The rate of movement of adenoviruses toward the negative ends of the microtubules, which are nearest the nuclear membrane and which contain dynein with the TCTEL1 subunit, was determined recently (23). This study showed that even early after infection there was some bidirectional movement toward the positive microtubule end, which is placed near the plasma membrane and which is the site of the exit of adenovirus from the endosome. However, in the first hour postinfection, the net vectorial movement during a productive infection is toward the nucleus. In addition to the binding of Ad E3-14.7K to a component of microtubules mediated by the FIP-1 cell protein, another adenovirus E3 protein, gp19K, which interacts with class I major histocompatibility complex (MHC-I) molecules, has been reported to bind directly to microtubules (3). This was proposed as a mechanism for anchoring or retaining MHC-I molecules in the endoplasmic reticulum and downregulating cell surface expression of this immunoregulatory protein (3).
The conceptual problem with proposing that Ad E3-14.7K is involved in the transport of virus across the cytoplasm during viral entry is that this virally encoded protein is not a structural component of the virion. Ad E3-14.7K is synthesized only after entry of the viral DNA into the nucleus, viral mRNA transcription, and subsequent translation. Unless viral entry during natural infections can be sequential over 10 to 12 h (25), it is unlikely that Ad E3-14.7K plays a functional role in viral entry or transport across the cytoplasm. However, during exit of the virus from its assembly site in the nucleus, Ad E3-14.7K may function, perhaps together with the ADP (adenovirus death protein), another E3 protein that has been implicated in the exit of the virus from the nucleus and viral spread (26). Alternatively, Ad E3-14.7K may function in the cytoplasm-nucleus-shuttling of the myriad of viral macromolecules that undergo intracellular migration during viral infection. Some of these steps are virus specific, such as the exit of viral mRNA from the nucleus; however, the retention of host mRNA in the nucleus during the late phase of the adenovirus cycle has been reported to be a function of adenovirus E1 and E4 (23).
The association of Ad E3-14.7K, an LMW GTPase and the LC of dynein brings together a number of molecules that could control the formation of the microtubule spindle and centrosomes, which are important in mitosis. Indeed, the net effect of adenovirus infection in human cells is the inhibition of mitosis, although there are a number of adenovirus early gene products, primarily from E-1 (E1A and E1B), which promote entry of the cells into the S phase (9, 22).
The functional significance of our finding that Ad E3-14.7K can be complexed with dynein is still unknown, although we hypothesize that it is one step in the mechanism of action of Ad E3-14.7K. It is also possible that the FIP-1–GIP-1/TCTEL1 complex may be involved in transporting the Ad E3-14.7K protein itself to its destined site of action, or in mediating signals initiated by Ad E3-14.7K in its role as an inhibitor of TNF-α-induced cell death. We have shown that Ad E3-14.7K is capable of inhibiting cell death induced by a number of molecules on the TNF-α-induced cell signaling pathway (13, 14). These molecules include the TNF-α receptor, RIP, and a recently isolated protein (FIP-3/IKKγ), all of which can induce cell death after transfection (13). However, overexpression of GIP-1/TCTEL1 did not cause cell death, nor did it counteract the effect of RIP, a mediator of cell death, suggesting that the Ad E3-14.7K protein may have an additional role other than as a modulator of TNF-α cytolysis.
ACKNOWLEDGMENTS
S. Lukashok and L. Tarassishin contributed equally to the results.
This research was supported by NIH grant RO1 CA72963 (M.S.H., L.T.), the Oncology Research Faculty Development Program (L.T.), and Cancer Center of the Albert Einstein College of Medicine grant P30-CA13330.
We gratefully acknowledge the assistance of Michael Cammer of the Analytical Imaging Facility, David Gebhard of the FACS Facility, and W.S.M. Wold (St. Louis University School of Medicine, St. Louis, Mo.) for antibodies against Ad E3-14.7K.
REFERENCES
- 1.Chen P, Tian J, Kovesdi I, Bruder J T. Interaction of the adenovirus 14.7-kDa protein with FLICE inhibits Fas ligand-induced apoptosis. J Biol Chem. 1998;273:5815–5820. doi: 10.1074/jbc.273.10.5815. [DOI] [PubMed] [Google Scholar]
- 2.Chittenden T, Livingston D M, Kaelin W G., Jr The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell. 1991;65:1073–1082. doi: 10.1016/0092-8674(91)90559-h. [DOI] [PubMed] [Google Scholar]
- 3.Dahllof B, Wallin M, Kvist S. The endoplasmic reticulum retention signal of the E3/19K protein of adenovirus-2 is microtubule binding. J Biol Chem. 1991;266:1804–1808. [PubMed] [Google Scholar]
- 4.Dales S, Chardonnet Y. Early events in the interaction of adenoviruses with HeLa cells. IV. Association with microtubules and the nuclear pore complex during vectorial movement of the inoculum. Virology. 1973;56:465–483. doi: 10.1016/0042-6822(73)90050-0. [DOI] [PubMed] [Google Scholar]
- 5.Darzynkiewicz Z. Cell cycle analyses by flow cytometry. In: Celis J E, editor. Cell biology: a laboratory handbook. Vol. 1. San Diego, Calif: Academic Press; 1994. pp. 261–271. [Google Scholar]
- 6.Gooding L R, Sofola I O, Tollefson A E, Duerksen-Hughes P J, Wold W S M. The adenovirus E3-14.7K protein is a general inhibitor of tumor necrosis factor-mediated cytolysis. J Immunol. 1990;145:3080–3086. [PubMed] [Google Scholar]
- 7.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Horton T M, Ranheim T S, Aquino L, Kusher D I, Saha S K, Ware C F, Wold W S M, Gooding L R. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J Virol. 1991;65:2629–2639. doi: 10.1128/jvi.65.5.2629-2639.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horwitz M S. Intermediates in the synthesis of type 2 adenovirus deoxyribonucleic acid. J Virol. 1971;8:675–683. doi: 10.1128/jvi.8.5.675-683.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King S M, Dillman III J F, Benashski S E, Lye R J, Patel-King R S, Pfister K K. The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J Biol Chem. 1996;271:32281–32287. doi: 10.1074/jbc.271.50.32281. [DOI] [PubMed] [Google Scholar]
- 11.Lader E, Hae-Sook H, O'Neill M, Artzt K, Bennett F M. tctex-1: a candidate gene family for a mouse t complex sterility locus. Cell. 1989;58:969–979. doi: 10.1016/0092-8674(89)90948-3. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Horwitz M S. Use of green fluorescent protein in studies of apoptosis of transfected cells. Biotechniques. 1997;23:1026–1029. doi: 10.2144/97236bm12. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Kang J, Friedman J, Tarassishin L, Ye J, Kovalenko A, Wallach D, Horwitz M S. Identification of a cell protein (FIP-3) as a modulator of NF-kappaB activity and as a target of an adenovirus inhibitor of tumor necrosis factor alpha-induced apoptosis. Proc Natl Acad Sci USA. 1999;96:1042–1047. doi: 10.1073/pnas.96.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Kang J, Horwitz M S. Interaction of an adenovirus E3-14.7kDa protein with a novel tumor necrosis factor alpha inducible cellular protein containing leucine zipper domains. Mol Cell Biol. 1998;18:1601–1610. doi: 10.1128/mcb.18.3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Kang J, Horwitz M S. Interaction of an adenovirus 14.7-kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J Virol. 1997;71:1576–1582. doi: 10.1128/jvi.71.2.1576-1582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luftig R B, Weihing R R. Adenovirus binds to rat brain microtubules in vitro. J Virol. 1975;16:696–706. doi: 10.1128/jvi.16.3.696-706.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashok S, Horwitz M S. Adenovirus persistence. In: Ahmed R, Chen I, editors. Persistent viral infections. Chichester, England: John Wiley & Sons, Ltd.; 1997. pp. 147–164. [Google Scholar]
- 18.Pennisi E. Nuclear transport protein does double duty in mitosis. Science. 1999;284:1260–1261. doi: 10.1126/science.284.5418.1260. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 20.Schreiber-Agus N, Chin L, Chen K, Torres R, Thomson C T, Sacchettini J C, DePinho R A. Evolutionary relationships and functional conservation among vertebrate Max-associated proteins: the zebra fish homolog of Mxi1. Oncogene. 1994;9:3167–3177. [PubMed] [Google Scholar]
- 21.Schurmann A, Brauers A, Mabmann S, Becker W, Joost H G. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270:28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 22.Shenk T. Adenoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2111–2148. [Google Scholar]
- 23.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. Microtubule-dependent plus- and minus-end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai A W, Chuang J Z, Bode C, Wolfrum U, Sung C H. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 25.Thomas G P, Mathews M B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980;22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 26.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tufariello J, Cho S, Horwitz M S. The adenovirus E3 14.7-kilodalton protein which inhibits cytolysis by tumor necrosis factor increases the virulence of vaccinia virus in a murine pneumonia model. J Virol. 1994;68:453–462. doi: 10.1128/jvi.68.1.453-462.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tufariello J, Cho S, Horwitz M S. The adenovirus E3 14.7K protein, an antagonist of tumor necrosis factor cytolysis, increases the virulence of vaccinia virus in SCID mice. Proc Natl Acad Sci USA. 1994;91:10987–10991. doi: 10.1073/pnas.91.23.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T K, Fujiwara T, Shimizu F, Okuno S, Suzuki M, Takahashi E, Nakamura Y, Hirai Y. Cloning, expression, and mapping of TCTEL1, a putative human homologue of murine Tcte1, to 6q. Cytogenet Cell Genet. 1996;73:153–156. doi: 10.1159/000134329. [DOI] [PubMed] [Google Scholar]
- 30.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 31.Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- 32.Wold W S, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 33.Wold W S M, Gooding L R. Adenovirus region E3 proteins that prevent cytolysis by cytotoxic T cells and tumor necrosis factor. Mol Biol Med. 1989;6:433–452. [PubMed] [Google Scholar]
- 34.Wold W S M, Tollefson A E, Hermiston T W. The E3 transcription unit of adenovirus. Curr Top Microbiol Immunol. 1995;199:237–274. doi: 10.1007/978-3-642-79496-4_13. [DOI] [PubMed] [Google Scholar]
- 35.Zilli D, Voelkel-Johnson C, Skinner T, Laster S M. The adenovirus E3 region 14.7 kDa protein, heat and sodium arsinate inhibit the TNF-induced release of arachidonic acid. Biochem Biophys Res Commun. 1992;188:177–183. doi: 10.1016/0006-291x(92)92366-6. [DOI] [PubMed] [Google Scholar]