Abstract
To clarify the role of core protein phosphorylation in pregenomic-RNA encapsidation of human and duck hepatitis B viruses (HBV and DHBV, respectively), we have examined the phosphorylation states of different forms of intracellular HBV core protein and the phenotypic effects of mutations in the phosphorylation sites of HBV and DHBV core proteins. We show that HBV core protein is phosphorylated to similar extents in the form of protein dimers and after further assembly in pregenomic RNA-containing capsids. Individual and multiple substitutions of alanine and aspartic acid for serine in the phosphorylation sites of HBV core protein resulted in site-specific and synergistic effects on RNA encapsidation, ranging from 2-fold enhancement to more than 10-fold inhibition. Core protein variants with mutations in all phosphorylation sites exhibited dominant-negative effects on RNA encapsidation by wild-type protein. The results suggest that the presence of phosphoserine at position 162 of HBV core protein is required for pregenomic-RNA encapsidation, whereas phosphoserine at position 170 optimizes the process and serine might be preferable in position 155. Examination of the pregenomic-RNA-encapsidating capacities of DHBV core protein variants, in which four phosphorylation sites were jointly mutated to alanine or aspartic acid, suggests that phosphorylation of DHBV core protein at these sites may optimize pregenomic-RNA encapsidation but that its impact is much less profound than in the case of HBV. The possible mechanisms by which RNA encapsidation may be modulated by core protein phosphorylation are discussed in the context of the observed differences between the two viruses.
The DNA genome of hepadnaviruses is replicated through reverse transcription of an RNA intermediate, the pregenomic RNA (reviewed in reference 4). Replication begins with encapsidation of pregenomic RNA, a process which requires two viral proteins, core (capsid) protein and polymerase; however, the enzymatic activities of the polymerase are not essential. Sequential synthesis of a minus-strand DNA by reverse transcription and synthesis of the second DNA strand are carried out by the polymerase inside the assembled nucleocapsid.
Core protein is a phosphoprotein (9, 11, 14, 15, 17). Three phosphorylation sites have been identified in the core protein of human hepatitis B virus (HBV), located in the arginine-rich C-terminal domain of the protein (S155, S162, and S170 in subtype ayw, which are equivalent to S157, S164, and S172 in subtype adw2) (10). The core protein of duck hepatitis B virus (DHBV) has four phosphorylation sites within the 28-amino-acid C-terminal sequence (T239, S245, S257, and S259) and is also phosphorylated elsewhere (20).
The function(s) of core protein phosphorylation is unclear. In particular, conflicting evidence has been presented regarding the possible role of core protein phosphorylation in encapsidation of pregenomic RNA. There are no data directly showing when core protein becomes phosphorylated: (i) prior to nucleocapsid assembly, (ii) during assembly, (iii) after assembly, in viral capsids containing pregenomic RNA, or (iv) later, during DNA synthesis. Studies with HBV core protein phosphorylated in vitro have suggested that the protein must be unphosphorylated to encapsidate pregenomic RNA and remains unphosphorylated until a later step of DNA synthesis (7). However, polymerase was not included in these in vitro experiments, and therefore they fail to reflect the situation of viral nucleocapsid assembly. Conversely, in cells transfected with plasmids encoding the relevant viral products, individual amino acid substitutions of four phosphorylation sites in DHBV core protein (expected to functionally mimic their unphosphorylated or phosphorylated forms; Ala or Asp for Ser or Thr, respectively) had no effect on pregenomic-RNA encapsidation (21). Similar experiments with HBV suggested that core protein phosphorylation may be important for pregenomic-RNA encapsidation, with S157 being nonessential, and S164 and S172 being more important, for the process (8). However, in both studies the levels of RNA encapsidation were determined from particles in which viral DNA synthesis was allowed to proceed; therefore, some effects of mutations on encapsidation may have been masked by effects at later stages of replication.
In the present study, we have unlinked the steps of encapsidation and DNA synthesis by the provision of a form of polymerase deficient for reverse transcription, allowing RNA encapsidation but no subsequent DNA synthesis. We determined the phosphorylation state of intracellular HBV core protein in unassembled form versus that in pregenomic-RNA-containing capsids by metabolic labeling and density gradient ultracentrifugation, showing that core protein is phosphorylated prior to encapsidation and that no significant change in the overall level of phosphorylation occurs during encapsidation. Examination of the RNA-encapsidating capacities of HBV core protein variants containing one or more substitutions of Ala or Asp for Ser in the phosphorylation sites revealed a variety of site-specific effects on HBV RNA encapsidation, suggesting differential roles of the individual phosphorylation sites, consistent with an important role for phosphorylation in the encapsidation process. Examination of the pregenomic-RNA-encapsidating capacities of DHBV core protein variants, in which four phosphorylation sites were jointly mutated, suggested that phosphorylation of DHBV core protein at these sites may optimize pregenomic-RNA encapsidation but that its impact is much less profound than in the case of HBV.
MATERIALS AND METHODS
Plasmids.
Plasmid CMayw1 (a kind gift of C. Seeger) contained a 1.1-length HBVayw genome under the control of the cytomegalovirus (CMV) promoter. Plasmid HBV RT− (containing a mutation in the reverse transcriptase active center) was derived from CMayw1 by replacing the PflMI-BsrGI fragment (nucleotides [nt] 725 to 770 of HBV; the numbering of the HBVayw sequence follows the designation of Galibert et al. [3]) with the oligonucleotide pair 5′- TTGGCTTTCAGTTATATGCATGCTGTGGTATTGGGGGCCAAGTCT and 3′-ACAAACCGAAAGTCAATATACGTACGACACCATAACCCCCGGTTCAGACATG (mutated codons are in bold). Plasmid HBV RT− C− was created by digestion of HBV RT− with BsgI, followed by treatment with the Klenow fragment of DNA polymerase I and religation, which resulted in deletion of 7 nt (nt 2037 to 2043 of HBV) and creation of a frameshift, generating a termination codon in the core open reading frame (ORF) downstream of the deletion site. Plasmid HBV C− Pstop was a derivative of HBV RT− C− in which 2 stop codons created by replacing the BspEI-PshAI fragment (nt 3619 to 3688 of HBV) in HBV RT− C− with the oligonucleotide pair 5′-CCGGAAATAACTGTGTTAGACGACGAGGCAGGTCCCCTAGAAGAAGAACTCCCTCGCCTCGCAGACGA and3′-TTTATTGACACAATCTGCTGCTCCGTCCAGGGGATCTTCTTCTTGAGGGAGCGGAGCGTCTGCT, were introduced into the ORF encoding the polymerase.
A plasmid encoding wild-type HBV core protein was created by inserting a fragment of the HBV genome containing the core protein ORF (nt 1903 to 2630 of HBV) into the multicloning region of the pCI-neo vector (Promega). This plasmid was then mutated to introduce the following amino acid combinations at the three phosphorylation sites in positions 155, 162, and 170, respectively: ASS, DSS, SAS, SDS, SSA, SSD, ASD, ADS, SDD, AAA, AAD, ASD, ADS, SDD, ADA, DAA, ADD, DAD, DDA, and DDD. The mutations were created by replacing a BspEI-BglII fragment (nt 2331 to 2425 of HBV) with oligonucleotide pairs containing Ala or Asp codons in place of Ser codons in the combinations described above.
A plasmid encoding wild-type DHBV core protein was constructed by inserting the DHBV core protein ORF (nt 2547 to 414; the numbering of the DHBV sequence follows the designation of Mandart et al. [12]) into the multicloning region of the pCI-neo vector. It was then mutated by replacement of an XmaI-AvrII fragment (nt 320 to 403 of DHBV) with oligonucleotide pairs containing 4 Ala or Asp codons in positions corresponding to amino acid positions 239, 245, 257, and 259.
DHBV RT− C− was a derivative of plasmid pUC119.DHBV.CMV (18) (a generous gift from J. Summers), in which mutations resulting in YMHA-for-YMDD amino acid substitutions in the reverse transcriptase active center of polymerase were introduced using PCR, and a stop codon in the core ORF was created by substitution of an EcoRV-XbaI fragment (nt 2652 to 2662 of DHBV) with the oligonucleotide pair 5′-ATCTAAGCTT and 3′-TAGATTCGAAGATC. DHBV C− Pstop was a derivative of DHBV RT− C− in which 2 stop codons were introduced into the polymerase ORF by replacing the XmaI-XhoI fragment (nt 1177 to 1212 of DHBV) with the oligonucleotide pair 5′-CCGGTGATGATCCACTCCTGTGAAATCAGTCTCTCC and 3′-ACTACTAGGTGAGGACACTTTAGTCAGAGAGGAGCT. It also contained mutations abolishing production of viral surface proteins.
Cells and transfections.
HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), in 12.5-cm2 flasks (Falcon). LMH cells were maintained in DMEM–F-12 medium supplemented with 10% FBS, in 6-well plates (Greiner). Cells were transfected with 10 μg of DNA per flask or well using the calcium phosphate method (5). For cotransfections, 5 μg of plasmid HBV RT− C− or DHBV RT− C− was mixed with 5 μg of plasmid encoding core protein. When two plasmids encoding core protein variants were used for cotransfection, they were mixed in the ratios 1:1 or 1:4 to achieve a total of 5 μg of DNA.
Core protein phosphorylation assay.
Three days posttransfection, cells were labeled with [33P]phosphoric acid (Amersham) in phosphate-free DMEM (Gibco) supplemented with 10 nM okadaic acid (Calbiochem) for 6 h. Replicate cells were labeled with [35S]methionine-cysteine (TransLabel; ICN) in methionine- and cysteine-free minimal essential medium (ICN) for 3 h. After labeling, cells were lysed, and core protein was immunoprecipitated with a polyclonal anti-HBV core antibody (Zymed) and protein A-Sepharose (Pharmacia), resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS–12% PAGE), and transferred to polyvinyl difluoride (PVDF) membranes (DuPont) for subsequent autoradiography.
To achieve the conditions which would allow normal core protein phosphorylation and nucleocapsid assembly, radioactive labeling commenced immediately after transfection and continued for 54 h, with 1 mCi (0.3 pmol) of [33P]phosphoric acid or 0.24 mCi (0.3 pmol) of [35S]methionine-cysteine per ml, in the presence of a 1,000-fold molar excess of cold phosphate or methionine-cysteine and 5% FBS. Cells were then lysed, and precleared lysates were layered onto 10 to 60% (wt/wt) sucrose step gradients (six 600-μl steps in 50 mM Tris-HCl–50 mM NaCl–50 mM NaF–1 mM EDTA [pH 7.65]). After ultracentrifugation (in an SW60 rotor [Beckman] at 55,000 rpm for 100 min at 4°C), eight fractions were collected from the top, and core protein was immunoprecipitated from each fraction, subjected to SDS-PAGE, and transferred to a PVDF membrane for subsequent autoradiography and phosphorimager analysis (Fuji FLA 2000).
RNA encapsidation assay.
RNA encapsidation within core protein particles was examined as described previously by Yu and Summers (21) with minor modifications. Transfected cells were lysed in 200 μl of 10 mM Tris-HCl–1 mM EDTA–0.2% NP-40–1 mM phenylmethylsulfonyl fluoride (pH 8.0). Precleared lysates were adjusted to 10 mM magnesium acetate and 0.1 mg of DNase I (Type II; Sigma)/ml and incubated for 30 min at 37°C. The lysates were then precleared and electrophoresed through a 0.8% agarose gel in duplicate 10-μl samples. One replicate was transferred to a Hybond ECL membrane (Amersham) by capillary action in 10 mM Tris-HCl–150 mM NaCl–1 mM EDTA (pH 8.0). Western blotting was performed on the membrane using an anti-HBV core antibody or a rabbit antibody raised against DHBV cores produced in Escherichia coli (a kind gift of A. Jilbert), with enhanced chemiluminescence (ECL) as the method of detection (SuperSignal substrate; Pierce). Another replicate was transferred to a Hybond-N membrane (Amersham) under the same conditions, where encapsidated nucleic acid was released from core protein particles by wetting the membrane in 0.2 M NaOH–1.5 M NaCl for 30 s followed by neutralization in 0.2 M Trizma-HCl–1.5 M NaCl for 1 min. Pregenomic RNA was detected by hybridization with a 32P-labeled DNA probe prepared using the Random Primer Plus Extension Labeling System (Du Pont). Fragments of double-stranded viral DNA used for preparation of the probes (nt 2906 to 490 of HBV or nt 1294 to 2351 of DHBV) were selected so that the probes hybridized with pregenomic RNA, but not with mRNA produced by the plasmids encoding core proteins.
Total cytoplasmic RNA was extracted using an RNeasy kit (Qiagen) according to the instructions of the manufacturer. Electrophoresis of cytoplasmic RNA for Northern blotting was performed in a formaldehyde–1% agarose gel.
RESULTS
HBV core protein is phosphorylated both when unassembled and when in capsids containing pregenomic RNA.
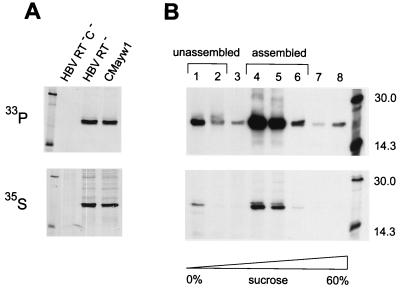
To determine at which step of HBV nucleocapsid assembly or maturation the core protein becomes phosphorylated, we first examined whether it may be phosphorylated prior to viral DNA synthesis. For this purpose we conducted [33P]phosphoric acid labeling of HepG2 cells transfected with one of three plasmids: CMayw1, HBV RT−, and HBV RT− C−. Plasmid CMayw1 produces mature HBV virions, while HBV RT− is a derivative of CMayw1 in which the encoded polymerase bears the substitution YMHA for YMDD in the reverse transcriptase active center. This substitution does not influence encapsidation of the pregenomic RNA but prevents DNA synthesis (1, 6). Plasmid HBV RT− C− is a derivative of HBV RT− containing a mutation (C−) abolishing core protein production, which served as a negative control. Replicate cells from the same transfections were labeled with [35S]methionine-cysteine to monitor core protein production. Labeled cells were then lysed, and core protein was immunoprecipitated from the lysates using an anti-HBV core antibody and analyzed by SDS-PAGE and autoradiography. As shown in Fig. 1A, core protein produced by HBV RT−, which consisted of unassembled core protein (dimers [22]) and pregenomic-RNA-containing capsids, was phosphorylated to approximately the same extent as core protein produced by CMayw1, which formed capsids containing both HBV RNA and DNA. This result clearly demonstrated that, contrary to an earlier hypothesis (7), HBV core protein is phosphorylated prior to the initiation of viral DNA synthesis.
FIG. 1.
Phosphorylation of HBV core protein. (A) HepG2 cells were transfected with the indicated plasmids and labeled with [33P]phosphoric acid for 6 h (33P) or [35S]methionine-cysteine for 3 h (35S). Labeled cells were lysed and subjected to immunoprecipitation with an anti-HBV core antibody followed by SDS-PAGE and autoradiography. (B) HepG2 cells transfected with plasmid HBV RT− were labeled with [33P]phosphoric acid or [35S]methionine-cysteine for 50 h in the presence of a 1,000-fold molar excess of cold phosphate or methionine-cysteine, respectively. Cells were then lysed, and the lysates were layered onto 10 to 60% (wt/wt) sucrose step gradients. Eight fractions collected after ultracentrifugation of the gradients were analyzed by immunoprecipitation followed by SDS-PAGE and autoradiography. Fraction numbers are shown on the top. Fractions corresponding to unassembled and assembled forms of core protein are indicated. Left lanes in panel A and right lanes in panel B, marker proteins; molecular masses in kilodaltons are indicated on the right.
Next, we determined the phosphorylation state of core protein in unassembled form versus that in pregenomic-RNA-containing capsids. HepG2 cells were transfected with HBV RT− and labeled with 33P or 35S under conditions which would allow normal core protein phosphorylation and nucleocapsid assembly (see Materials and Methods). Cells were then lysed, the lysates were sedimented over 10 to 60% (wt/wt) sucrose gradients and then fractionated, and core protein was immunoprecipitated from each fraction and analyzed by SDS-PAGE and autoradiography. The results demonstrated that both unassembled core protein and RNA-containing capsids were phosphorylated (Fig. 1B). By phosphorimager analysis of the total amounts of core protein (35S) and phosphorylated core protein (33P) in each fraction, no difference was found between the amounts of phosphate incorporated in unassembled core protein and in RNA-containing capsids in this experiment. In two other experiments a twofold difference in phosphate incorporation between the two forms of core protein was observed, with an increase in phosphorylation after assembly in one experiment and a decrease in the other (data not shown). These results demonstrate that HBV core protein is phosphorylated to similar extents in the form of protein dimers and in RNA-containing capsids.
Mutations of S155, S162, and S170 in HBV core protein affect pregenomic RNA encapsidation.
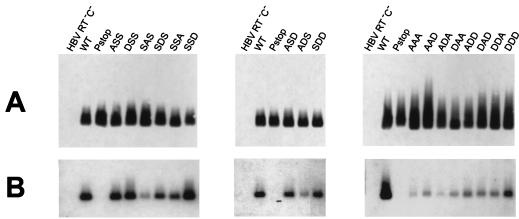
Because HBV core protein was found to be phosphorylated both before and after pregenomic-RNA encapsidation, we further examined the role of this modification in RNA encapsidation by mutagenesis of core-expressing plasmids. We created a plasmid encoding wild-type HBV core protein and a series of variants containing substitutions of Ala or Asp for Ser in the phosphorylation sites to mimic their unphosphorylated and phosphorylated forms, respectively. The following amino acid combinations were used at the three phosphorylation sites (residues 155, 162, and 170, respectively): SSS (wild type); ASS, DSS, SAS, SDS, SSA, and SSD (individual mutations); ASD, ADS, and SDD (double mutations); and AAA, AAD, ADA, DAA, ADD, DAD, DDA, and DDD (triple mutations).
HepG2 cells were cotransfected with HBV RT− C−, providing pregenomic RNA and polymerase, together with plasmids encoding wild-type or mutant core proteins, such that nucleocapsids would be formed if the core protein was competent for HBV RNA encapsidation. Note in particular that the polymerase provided in trans contained the RT− mutation ablating viral DNA synthesis, which allowed us to isolate the effects of core protein variants on RNA encapsidation from possible effects at later stages of replication. Negative controls for the encapsidation reaction were provided by transfection with HBV RT− C− alone (yielding pregenomic RNA and polymerase but no core protein) and cotransfection of a plasmid encoding wild-type core protein together with plasmid HBV C− Pstop (yielding core protein particles and pregenomic RNA, respectively, but no encapsidation due to the lack of polymerase [1]).
Three days posttransfection, the cells were lysed and HBV RNA encapsidation within core protein particles was examined as described previously for DHBV (21). Briefly, duplicate samples of cell lysates were electrophoresed through a nondenaturing agarose gel and transferred to nylon and nitrocellulose membranes by capillary action. The amount of core protein particles in each sample was estimated by ECL-Western blotting performed on the nitrocellulose membrane, followed by densitometry. The pregenomic-RNA content of the core protein particles was estimated by hybridization with a 32P-labeled HBV DNA probe performed on the nylon membrane after destruction of the particles in situ with alkali and UV fixation of RNA to the membrane. The total amount of pregenomic RNA produced in transfected cells was estimated by Northern blotting performed on cytoplasmic RNA extracted from a duplicate flask of cells from the same transfection. Radioactivity was quantitated by phosphorimager analysis.
The results demonstrated that amino acid substitutions in phosphorylation sites did not markedly affect the production of core protein particles in transfected cells (Fig. 2A), although the differences observed were consistent in repeated experiments. Therefore, in quantitation of the results we normalized the amounts of encapsidated pregenomic RNA by reference to the amounts of the corresponding core proteins. In contrast, levels of total pregenomic RNA varied within ±30% of the mean between samples in individual experiments, but these differences were not related to particular core protein variants in repeated experiments (data not shown). As such, the mean levels of total pregenomic RNA for each core protein variant were equal.
FIG. 2.
Pregenomic-RNA encapsidation by HBV core protein variants. HepG2 cells were cotransfected with plasmids expressing the indicated core protein variants and a plasmid providing pregenomic RNA and polymerase (HBV RT− C−). WT, wild-type core protein. Negative controls were cells transfected with HBV RT− C− alone (lane HBV RT− C−) and cells cotransfected with the plasmid encoding wild-type core protein and plasmid HBV C− Pstop, providing pregenomic RNA but no polymerase (lane Pstop). Pregenomic RNA encapsidation within core protein particles produced in transfected cells was analyzed by electrophoresis of the particles through a nondenaturing agarose gel followed by transfer to nylon and nitrocellulose membranes by capillary action. The amount of core protein particles in the sample was estimated by ECL-Western blotting performed on the nitrocellulose membrane (A). The HBV RNA content of the particles was estimated by hybridization with a 32P-labeled HBV DNA probe performed on a nylon membrane after destruction of the particles in situ with alkali (B).
Examination of pregenomic-RNA encapsidation by the different core protein variants revealed an interesting pattern of site-specific and synergistic effects of substitutions in phosphorylation sites on encapsidation (Fig. 2B; Table 1). Of note, individual and double substitutions in phosphorylation sites do not affect the extent of phosphorylation of the remaining sites (10).
TABLE 1.
Pregenomic RNA encapsidation by HBV core protein mutants
| HBV core protein mutant | Encapsidated HBV RNA (% of WT)a | SD | No. of quantitated exptsb |
|---|---|---|---|
| ASS | 78 | 5 | 3 |
| DSS | 57 | 13 | 3 |
| SAS | 20 | 5 | 4 |
| SDS | 56 | 13 | 4 |
| SSA | 66 | 16 | 3 |
| SSD | 193 | 30 | 3 |
| ASD | 78 | 7 | 3 |
| ADS | 39 | 4 | 3 |
| SDD | 83 | 16 | 3 |
| AAA | 10 | 7 | 3 |
| AAD | 6 | 2 | 2 |
| ADA | 2 | 2 | 2 |
| DAA | 4 | 1 | 2 |
| ADD | 11 | 3 | 2 |
| DAD | 9 | 1 | 2 |
| DDA | 8 | 1 | 2 |
| DDD | 21 | 5 | 3 |
WT, wild type.
Quantitation of some other experiments with triple mutants (data not shown) was not possible due to the low level of HBV RNA encapsidation.
In contrast to the case of DHBV (21), all individual mutations in phosphorylation sites (ASS, DSS, SAS, SDS, SSA, and SSD) affected HBV RNA encapsidation, clearly demonstrating that serine residues in positions 155, 162, and 170 play a role in this process. In comparison with wild-type core protein, the SSD variant encapsidated 1.9-fold more HBV RNA, while the SAS variant encapsidated 5-fold less HBV RNA, and other mutations caused more-moderate decreases in RNA encapsidation. Pairwise comparison of the effects of Ala and Asp substitutions at each site showed that the ASS mutant encapsidated HBV RNA better than DSS protein, whereas the SAS and SSA mutants contained approximately threefold less HBV RNA than their SDS and SSD counterparts.
Selected double mutations in phosphorylation sites, in which the individual mutations which resulted in relatively efficient HBV RNA encapsidation were combined, were also examined. These pairwise combinations resulted in small-to-moderate decreases in RNA encapsidation by the ASD, ADS, and SDD mutant proteins in comparison with wild-type protein.
The observed RNA-encapsidating capacity of each double mutant was compared with that predicted from the combination of individual mutations. Predictions of RNA-encapsidating capacities of core proteins bearing multiple substitutions in the phosphorylation sites were made by multiplying the observed effects of individual substitutions. For example, individual substitutions of Ala for S155 and Asp for S170 resulted in RNA-encapsidating capacities of the corresponding proteins equal to 0.78 and 1.93 of the wild-type capacity, respectively. Therefore, their combined effects were predicted to result in the ASD protein having an RNA-encapsidating capacity equal to 1.51 (0.78 × 1.93) of the wild-type capacity. The levels of RNA encapsidation by the ADS and SDD proteins were similar to those predicted on the basis of the individual substitutions, but the combination ASD had a synergistic negative effect resulting in a nearly twofold difference from that expected.
All triple substitutions in the phosphorylation sites (AAA, AAD, ADA, DAA, ADD, DAD, DDA, and DDD, representing all possible combinations of alanine and aspartic acid residues) resulted in strong inhibition of pregenomic-RNA encapsidation. The amounts of RNA encapsidated by the AAA, AAD, ADA, DAA, ADD, DAD, and DDA proteins were very similar (between 2 and 11% of that for wild-type protein, which is within the range of variability in the measurements of such small amounts of RNA). DDD protein contained slightly more RNA (21% of the wild-type amount). The AAA, AAD, ADA, DAA, DAD, and DDA proteins were expected to encapsidate viral RNA inefficiently: on the basis of the effects of individual mutations, we estimated that they would contain 10, 30, 29, 8, 22, and 21% of the wild-type level of RNA, respectively. However, the RNA-encapsidating capacities of four of these mutants (AAD, ADA, DAD, and DDA) were even lower than predicted (at least twofold lower). This suggested synergistic inhibition of RNA encapsidation by these amino acid combinations, as in the case of the double mutation ASD. Such synergistic inhibition of HBV RNA encapsidation was even more evident with the ADD and DDD mutations, because the predicted levels of RNA encapsidation for these mutants were 84 and 61% of the wild-type level, respectively (seven- and threefold higher than those observed), and moreover, the corresponding double amino acid substitutions resulted in relatively efficient RNA encapsidation (ASD, ADS, and SDD versus ADD, and SDD versus DDD).
There were three possible reasons for the synergistic negative effects of triple mutations on RNA encapsidation in comparison with double mutations: (i) at least one of three phosphorylation sites must be charged only in a portion of core protein molecules, (ii) at least one must change charge in the process of encapsidation, or, alternatively, (iii) amino acid substitutions in every phosphorylation site must have disrupted protein folding. To test the first two options, we initially examined the possible site specificity of these two requirements. Our data showing that a fixed charge in amino acid position 170 stimulates HBV RNA encapsidation when two other phosphorylation sites are not substituted (the SSD protein) and provides a level of RNA encapsidation close to that of the wild type when either one of the other two sites is also substituted (the ASD and SDD proteins) lead to a conclusion that constitutive phosphorylation of S170 is fully compatible with the encapsidation process. In contrast, all individual amino acid substitutions in positions 155 and 162 resulted in reductions in the RNA-encapsidating capacity of core protein (the ASS, DSS, SAS, and SDS proteins). Double mutation of these sites (ADS) also had a negative effect on RNA encapsidation. These data, together with the fact that double mutants ASD and SDD, in which either S155 or S162 was unsubstituted, encapsidated viral RNA with nearly wild-type efficiency, suggested that if diversity or change of charge at one of three phosphorylation sites is crucial for RNA encapsidation, then it is sufficient that either of these two sites fulfills these conditions. Therefore, we examined the role of diversity in charge at position 155 of core protein in RNA encapsidation. We coexpressed the ADD and DDD proteins in three different ratios to see if any of the combinations would restore RNA encapsidation to the level of the SDD protein (83% of the wild-type level). To examine the possibility that change of charge is required for encapsidation, we coexpressed the AAA and DDD proteins with wild-type protein to test whether a proportion of wild-type core protein would rescue RNA encapsidation.
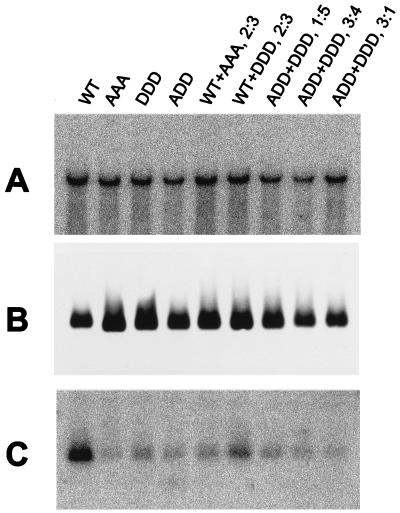
Three-plasmid cotransfections did not influence the level of production of pregenomic RNA in comparison with two-plasmid cotransfections (Fig. 3A). Total amounts of core proteins in each cotransfection corresponded to the ratios in which the core-producing plasmids were mixed and the different levels of protein production by each plasmid (Fig. 3B). Therefore, the ratios of core protein variants produced in transfected cells were calculated as follows: wild type to AAA, 2:3; wild type to DDD, 2:3; ADD to DDD, 1:5; 3:4, and 3:1. All the combinations of proteins encapsidated only small amounts of pregenomic RNA, similar to those encapsidated by individual triple mutants, and even coassembly with wild-type protein resulted in only a marginal increase in RNA encapsidation (Fig. 3C).
FIG. 3.
Pregenomic-RNA encapsidation by coassembling HBV core protein variants. HepG2 cells were cotransfected with plasmids expressing the indicated core protein variants in the indicated ratios and plasmid HBV RT− C−. (A) The amount of total pregenomic RNA produced in transfected cells was estimated by Northern blotting performed on cytoplasmic RNA. (B and C) Pregenomic RNA encapsidation within core protein particles was analyzed as described in the legend to Fig. 2, using duplicate flasks of cells from the same transfections as in panel A. (B) Immunostaining of core protein particles. (C) Detection of encapsidated pregenomic RNA.
The inability of the ADD and DDD mutants to complement each other for RNA encapsidation demonstrated that if diversity in fixed charge or phosphorylation state between core protein molecules played a role in the process, it was not the major reason for the failure of triple mutants to support RNA encapsidation. The dominant-negative effect of DDD protein on RNA encapsidation by wild-type core protein also led to this conclusion. The results were less conclusive with respect to the other two possibilities. If encapsidation requires a change in the extent of core phosphorylation during the process, then the fact that coexpression of triple mutants with wild-type protein in a 3:2 ratio did not rescue RNA encapsidation suggests that more than 40% of core protein molecules are required to undergo such a change in phosphorylation state. Our initial result showing that unassembled core protein and RNA-containing capsids were phosphorylated to similar extents does not exclude such a possibility, because a relatively small change in the extent of phosphorylation (such as that between 2 and 3 phosphoserines) could have been obscured by the variability in the labeling experiments (around twofold in three experiments). Therefore, the dominant-negative effects of the AAA and DDD proteins on RNA encapsidation by wild-type core protein suggest either (i) a requirement for the majority of core protein molecules to change phosphorylation state during the encapsidation process or (ii) the disruption of protein folding by the triple substitutions such that it obstructs RNA encapsidation in coassembly with wild-type protein.
Multiple mutations in phosphorylation sites of DHBV core protein affect pregenomic-RNA encapsidation.
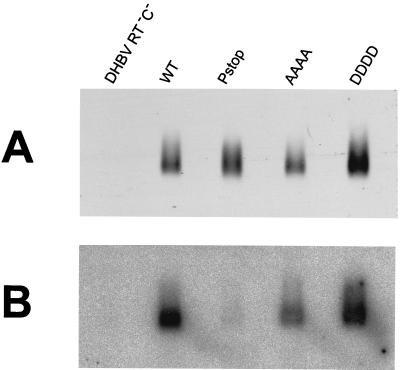
The remarkable difference between HBV and DHBV in the effects of individual mutations in phosphorylation sites of the core protein on pregenomic-RNA encapsidation (our data and reference 21) prompted us to compare the roles of core protein phosphorylation in RNA encapsidation in the two viruses in more detail. We created plasmids which encoded DHBV core proteins in which all the C-terminal phosphorylation sites (T239, S245, S257, and S259) were replaced with alanine (AAAA) or aspartic acid (DDDD) residues. The pregenomic RNA-encapsidating capacities of the variant proteins were tested in cotransfection experiments similar to those described above for HBV. Briefly, LMH cells were cotransfected with mixtures of plasmids encoding core protein variants (wild type, AAAA, or DDDD) and plasmid DHBV RT− C−, which contained, under the control of the CMV promoter, a 1.2-length DHBV genome with mutations abolishing reverse transcription and core protein production. Transfection with DHBV RT− C− alone and cotransfection of a plasmid encoding wild-type core protein with plasmid DHBV C− Pstop were used as negative controls, as above.
Pregenomic-RNA encapsidation within core protein particles produced in transfected cells was examined using the method described above for HBV. The results demonstrated that multiple mutations in phosphorylation sites diminished the pregenomic-RNA-encapsidating capacity of DHBV core protein, but not to the same extent as for HBV (Fig. 4). The DHBV AAAA and DDDD mutant proteins encapsidated 35 and 60% of wild-type levels of pregenomic RNA, respectively, whereas their HBV counterparts, the AAA and DDD proteins, encapsidated 10 and 21% of wild-type levels, respectively.
FIG. 4.
Pregenomic-RNA encapsidation by DHBV core protein variants. LMH cells were cotransfected with plasmids expressing the indicated core protein variants and a plasmid providing pregenomic RNA and polymerase (DHBV RT− C−). WT, wild-type core protein. Negative controls were cells transfected with DHBV RT− C− alone (lane DHBV RT− C−) and cells cotransfected with the plasmid encoding wild-type core protein and plasmid DHBV C− Pstop, providing pregenomic RNA but no polymerase (lane Pstop). Pregenomic-RNA encapsidation within core protein particles produced in transfected cells was analyzed as described in the legend to Fig. 2, except that a DHBV DNA probe was used for hybridization (B) and an anti-DHBV core antibody was used for immunostaining (A).
DISCUSSION
The results obtained in this study demonstrated that HBV core protein is phosphorylated to similar extents in the form of protein dimers and after further assembly in pregenomic-RNA-containing capsids. While this does not formally exclude the possibility that core protein dimers may assemble into capsids while in a transiently dephosphorylated state, we believe that it is more likely that core protein is phosphorylated throughout the encapsidation process.
Further investigation of the role of HBV core protein phosphorylation in pregenomic-RNA encapsidation, by examination of the RNA-encapsidating capacities of core protein variants containing individual and multiple substitutions of Ala or Asp for Ser in the phosphorylation sites, revealed a variety of site-specific and synergistic effects of the mutations on HBV RNA encapsidation, ranging from 2-fold enhancement to more than 10-fold inhibition. In the following interpretation of these results, we took into account the fact that limitations of Ala and Asp as substitutes for serine and phosphoserine, respectively, could contribute in some cases to the negative effect on HBV RNA encapsidation. For example, alanine cannot form the hydrogen bonds which could be formed by the hydroxyl group of serine, while aspartic acid residues carry one negative charge instead of the two of a phosphate group, have a different sterical structure, and cannot form the same hydrogen bonds as phosphoserine. However, in the instances when Asp substitution resulted in a much higher level of HBV RNA encapsidation than Ala substitution, we conclude that phosphoserine in that position is preferable to serine, because Asp is a much less conservative change for Ser than is Ala.
Therefore, our results suggest that phosphorylation of S162 in at least a proportion of core protein molecules is required for HBV RNA encapsidation, because the SAS protein containing an individual alanine substitution in this position encapsidated fivefold less HBV RNA than wild-type protein and nearly threefold less than its aspartic acid counterpart (the SDS variant). The roles of S155 and S170 in encapsidation appear to be less crucial than that of S162, because individual mutations in these positions caused much more moderate effects on RNA encapsidation (within twofold of the level in wild-type protein). Phosphoserine in position 170 appears to increase the RNA-encapsidating capacity of core protein, because an individual aspartic acid substitution in this position resulted in nearly twofold-increased levels of RNA encapsidation in comparison with the level for wild-type protein and a threefold increase in comparison with that for the alanine counterpart, and its combination with an Asp substitution in position 162, which individually caused a decrease in RNA encapsidation, resulted in a nearly wild-type RNA-encapsidating capacity of the mutant protein (SDD). Serine might be preferable to phosphoserine in position 155; however, the difference between individual alanine and aspartic acid substitutions in this case is much less convincing than those for the other two phosphorylation sites.
The effects of selected double amino acid substitutions in the phosphorylation sites on RNA encapsidation generally confirmed the results obtained using individual mutations, with nearly wild-type levels of RNA detected within capsids formed by two of three mutant proteins. However, one combination (ASD) had a synergistic negative effect on RNA encapsidation. Such synergistic inhibition of RNA encapsidation by multiple substitutions in the phosphorylation sites was much more pronounced when all three phosphorylation sites were mutated, resulting in a low RNA-encapsidating capacity of the mutant proteins, which could not be improved by coassembly by two mutant proteins or by a mutant protein with wild-type protein.
The dominant-negative effects of the proteins with triple mutations on pregenomic-RNA encapsidation by wild-type protein were similar to the effects previously demonstrated for HBV core protein with the N-terminal pre-C extension (16) and for DHBV core proteins with C-terminal additions (19). The inhibitory role of HBV pre-C–core protein was suggestively explained by the alteration of the protein structure, which renders capsids unable to incorporate pregenomic RNA (16). Our results also suggest that the disruption of protein folding created by three amino acid substitutions may be the cause of the dominant-negative effect of the mutant proteins on HBV RNA encapsidation. However, our data do not exclude an alternative explanation, that HBV RNA encapsidation may require a change in the extent of phosphorylation in the majority, or in all, of the core protein molecules during the process.
The above conclusions regarding the differential roles of the phosphorylation sites in HBV core protein in pregenomic-RNA encapsidation largely agree with the conclusion of Lan and coauthors that S157 is nonessential and S164 and S172 are more important for the process (8); however, our results suggest a much more important role for S162 than for S170 and call into question the insignificance of S155 due to the observed synergistic effects of the three sites (the different numbering of amino acids reflects the differences between the virus strains; adw2 was used in reference 8, and ayw was used in this study). There is also a striking discrepancy between the results obtained in the two studies using dicarbonic amino acids as substitutes for phosphoserine. The triple glutamic acid (EEE) substitutions in the phosphorylation sites used by Lan et al. resulted in close to wild-type levels of HBV RNA encapsidation by the mutant protein, in contrast to the dominant-negative effect on RNA encapsidation of the DDD variant in this study. Since glutamic acid seems unlikely to have any advantages over aspartic acid as a substitute for phosphoserine, we see two possible explanations for the discrepancy in the results. One is that RNA encapsidation has different requirements for the arginine-rich domains of core proteins in different viral strains (adw2 and ayw). The other is that this discrepancy relates to the linkage between RNA encapsidation and reverse transcription, which results in degradation of the encapsidated RNA by RNase H activity. We have used RT− forms of polymerase in these studies to dissociate encapsidation from later events, so that the amount of encapsidated RNA is related only to the efficiency of encapsidation. The experiments of Lan and colleagues were conducted using wild-type polymerase (8). In the presence of wild-type polymerase, those core proteins which are competent for encapsidation and also for the support of reverse transcription will contain amounts of RNA reflecting a balance between the efficiencies of encapsidation and reverse transcription. Because EEE mutations were found to abolish reverse transcription (8), we believe that the amount of encapsidated RNA could be overestimated compared to that in wild-type capsids, in which a proportion of the RNA would be degraded.
In the case of DHBV, AAAA and DDDD substitutions in core protein diminished pregenomic-RNA-encapsidating capacity, and the DDDD mutant encapsidated nearly twofold more DHBV RNA than the AAAA mutant; however, the effects of both mutations were much less profound than those of analogous substitutions in HBV. These results, together with the previously published data showing that individual mutations of the phosphorylation sites in DHBV core protein have no effect on pregenomic-RNA encapsidation (21), suggest that phosphorylation of DHBV core protein at these sites may optimize pregenomic-RNA encapsidation but that its impact is much less profound than in the case of HBV.
Our results, combined with published data, provide new information for elucidation of the function of phosphorylation in the encapsidation process. The previous study of Nassal (13) demonstrated that coexpression of HBV core protein lacking the arginine-rich domain together with wild-type core protein resulted in wild-type levels of pregenomic RNA encapsidation. This means that intermolecular interactions in nucleocapsid assembly do not require the presence of the arginine-rich domain in every core protein molecule. Moreover, the C-terminal part of the domain including S170 can be deleted without any effect on viral RNA encapsidation (13) (Fig. 5). However, our results demonstrate that, when the arginine-rich domain is present, its amino acid sequence in every assembling molecule of core protein (or at least in the majority of molecules) is crucial for RNA encapsidation, and S170 has a role in this process as well as the two other phosphorylation sites.
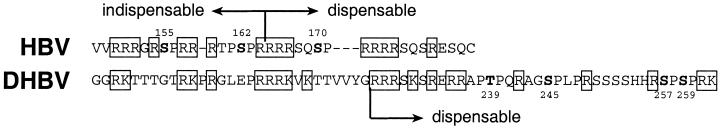
FIG. 5.
The C-terminal domains of HBV and DHBV core proteins. Phosphorylation sites are boldfaced (10, 20); arginine and lysine residues are boxed. Amino acid numbering refers to HBVayw and DHBV 16 (3, 12). Arrows indicate which portions of each core protein are dispensable and which are indispensable for RNA encapsidation (2, 13, 17).
Our hypothesis, therefore, is that phosphorylation plays a dual conformational role in HBV RNA encapsidation. When unphosphorylated, the arginine-rich domain assumes a conformation which obstructs the encapsidation of pregenomic RNA (perhaps in the same way as when all phosphorylation sites are mutated). One role of phosphorylation, therefore, is to create conformational changes which render the protein structure permissive for encapsidation. The N-terminal part of the arginine-rich domain in a proportion of core protein molecules is involved in the interaction with pregenomic RNA and/or polymerase in nucleocapsid assembly, which is indicated by its indispensability for the process (2) (Fig. 5). The second role of phosphorylation may be, therefore, to provide the proper conformation and exposure of the N-terminal part of the domain for intermolecular interactions. The role of phosphoserine 162 might go beyond providing the correct folding of the arginine-rich domain, because individual alanine substitution in this position strongly inhibits RNA encapsidation: it might also participate in ionic intermolecular interactions, since aspartic acid, which is permissive for encapsidation, can substitute for phosphoserine for this type of interaction only.
The difference between HBV and DHBV in the impact of substitutions in phosphorylation sites on pregenomic-RNA encapsidation is likely to be caused by the difference in the locations of these sites in viral core proteins. Phosphorylation sites T239, S245, S257, and S259 of DHBV core protein are located in the C-terminal part of the arginine-rich domain, which is absent in HBV core protein (Fig. 5, aligned with MacVector [Stratagene]). This part of DHBV core protein can be deleted without any effect on viral RNA encapsidation (17). We believe, therefore, that the conformational changes in the arginine-rich domain of DHBV core protein induced by phosphorylation of T239, S245, S257, and S259 (20) may optimize pregenomic-RNA encapsidation but are not crucial for the process. However, it must be noted that not all phosphorylation sites in DHBV core protein have been identified; therefore, the role of core protein phosphorylation in DHBV RNA encapsidation needs further examination.
ACKNOWLEDGMENTS
These studies were supported in part by the Research Fund of the Macfarlane Burnet Centre for Medical Research.
We thank C. Seeger and J. Summers for supplying plasmids, A. Jilbert for the antibody, and A. Jaworowski for critical reading of the manuscript.
REFERENCES
- 1.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beames B, Lanford R E. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology. 1993;194:597–607. doi: 10.1006/viro.1993.1299. [DOI] [PubMed] [Google Scholar]
- 3.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 4.Ganem D. Hepadnaviridae: the viruses and their replication. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 2703–2737. [Google Scholar]
- 5.Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 6.Hirsch R C, Lavine J E, Chang L, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 7.Kann M, Gerlich W H. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol. 1994;68:7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan Y T, Li J, Liao W-Y, Ou J-H. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology. 1998;259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- 9.Lanford R E, Notvall L. Expression of hepatitis B virus core and precore antigens in insect cells and characterization of a core-associated kinase activity. Virology. 1990;176:222–233. doi: 10.1016/0042-6822(90)90247-o. [DOI] [PubMed] [Google Scholar]
- 10.Liao W, Ou J-H. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida A, Ohnuma H, Tsuda F, Yoshikawa A, Hoshi Y, Tanaka T, Kishimoto S, Akahane Y, Miyakawa Y, Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J Virol. 1991;65:6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugh J, Zweidler A, Summers J. Characterization of the major duck hepatitis B virus core particle protein. J Virol. 1989;63:1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roossinck M J, Siddiqui A. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J Virol. 1987;61:955–961. doi: 10.1128/jvi.61.4.955-961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaglioni P P, Melegari M, Wands J R. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlicht H J, Batrenschlager R, Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summers J, Smith P M, Huang M J, Yu M S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Weizsacker F, Kock J, Wieland S, Offensperger W-B, Blum H E. Dominant negative mutants of the duck hepatitis B virus core protein interfere with RNA pregenome packaging and viral DNA synthesis. Hepatology. 1999;30:308–315. doi: 10.1002/hep.510300139. [DOI] [PubMed] [Google Scholar]
- 20.Yu M, Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol. 1994;68:2965–2969. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, Summers J. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol. 1994;68:4341–4348. doi: 10.1128/jvi.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]