Abstract
The functional unit of the human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins is a trimer composed of three gp120 exterior glycoproteins and three gp41 transmembrane glycoproteins. The lability of intersubunit interactions has hindered the production and characterization of soluble, homogeneous envelope glycoprotein trimers. Here we report three modifications that stabilize soluble forms of HIV-1 envelope glycoprotein trimers: disruption of the proteolytic cleavage site between gp120 and gp41, introduction of cysteines that form intersubunit disulfide bonds, and addition of GCN4 trimeric helices. Characterization of these secreted glycoproteins by immunologic and biophysical methods indicates that these stable trimers retain structural integrity. The efficacy of the GCN4 sequences in stabilizing the trimers, the formation of intersubunit disulfide bonds between appropriately placed cysteines, and the ability of the trimers to interact with a helical, C-terminal gp41 peptide (DP178) support a model in which the N-terminal gp41 coiled coil exists in the envelope glycoprotein precursor and contributes to intersubunit interactions within the trimer. The availability of stable, soluble HIV-1 envelope glycoprotein trimers should expedite progress in understanding the structure and function of the virion envelope glycoprotein spikes.
The entry of human immunodeficiency virus type 1 (HIV-1) into target cells is mediated by the viral envelope glycoproteins (53). The mature envelope glycoproteins on the virus are organized into oligomeric spikes composed of the gp120 exterior envelope glycoprotein and the gp41 transmembrane envelope glycoprotein (1, 22, 52, 54, 60). In the infected cell, the HIV-1 envelope glycoproteins are initially synthesized as an 845- to 870-amino-acid protein, depending upon the viral strain (22). N-linked, high-mannose sugars are added to this primary translation product to result in the gp160 envelope glycoprotein precursor. Oligomers of gp160 form in the endoplasmic reticulum, and several pieces of evidence suggest that these are trimers. First, X-ray crystallographic studies of fragments of the gp41 ectodomain revealed the presence of very stable, six-helix bundles (11, 53, 55). These structures were composed of a trimeric coiled coil involving N-terminal gp41 α-helices, with three C-terminal gp41 α-helices packed into the grooves formed by the three inner helices. Second, introduction of cysteine pairs at specific locations in the coiled coil resulted in the formation of intermolecular disulfide bonds between the gp160 subunits (26). The disulfide-stabilized oligomer was shown to be a trimer. Finally, the matrix proteins of HIV-1 and the related simian immunodeficiency viruses, which interact with the intravirion domains of the envelope glycoproteins, crystallize as trimers (32, 50).
Following oligomerization, the gp160 glycoprotein is transported to the Golgi apparatus, where cleavage by a cellular protease generates the gp120 and gp41 glycoproteins (1, 52, 54). The gp120 glycoprotein remains associated with the gp41 glycoprotein through noncovalent, hydrophobic interactions (30, 36). The lability of the gp120–gp41 association results in the “shedding” of some gp120 molecules from the trimer, resulting in nonfunctional envelope glycoproteins (40, 58). It has been suggested that these disassembled envelope glycoproteins result in the generation of high titers of nonneutralizing antibodies during natural HIV-1 infection (7, 45, 49). The envelope glycoprotein trimers that remain intact undergo modification of a subset of the carbohydrate moieties to complex forms before transport to the cell surface (22).
The mature envelope glycoprotein complex is incorporated from the cell surface into virions, where it mediates virus entry into the host cell. The gp120 exterior envelope glycoprotein binds the CD4 glycoprotein, which serves as a receptor for the virus (17, 33, 39). Binding to CD4 induces conformational changes in the envelope glycoproteins that allow gp120 to interact with one of the chemokine receptors, typically CCR5 or CXCR4 (2, 14, 18–20, 27; reviewed in reference 15). The chemokine receptors are 7-transmembrane, G protein-coupled receptors, and gp120 interaction with the chemokine receptors is believed to bring the viral envelope glycoprotein complex nearer to the target cell membrane and to trigger additional conformational changes in the envelope glycoproteins. Although the exact nature of these changes is unknown, mutagenic data are consistent with a role for the hydrophobic gp41 amino terminus (the “fusion peptide”) in mediating membrane fusion (8, 28, 31, 36). It has been suggested that, following interaction of the fusion peptide with the target cell membrane, formation of the 6-helical bundle by the three gp41 ectodomains would result in the spatial juxtaposition of the viral and target cell membranes (11, 53, 55). Six-helical bundles have been documented in several viral envelope glycoproteins that mediate membrane fusion and virus entry (11, 55–57). The formation of this energetically stable structure from a different and as-yet-unknown precursor structure is believed to provide the energy necessary to overcome the repulsion between the viral and cell membranes.
The HIV-1 envelope glycoproteins are inefficient in generating antibodies that neutralize the virus, especially those that can neutralize more than a limited number of HIV-1 strains (3, 16, 38; reviewed in references 6, 7, and 60). Many of the antibodies elicited by the envelope glycoproteins are not able to bind efficiently to the functional envelope glycoprotein trimer and therefore are devoid of neutralizing activity (4, 43, 45, 48, 49, 59). The lability of the envelope glycoprotein trimers, conformational flexibility in the shed gp120 glycoprotein, and the variability and glycosylation of the gp120 surface all possibly contribute to the poor neutralizing antibody responses (reviewed in references 42, 44, and 60). A better understanding of the barriers that contribute to limited neutralizing responses may suggest approaches that will present more functionally relevant epitopes to the immune system and thereby expedite vaccine development.
An understanding of the functional HIV-1 envelope glycoprotein trimer would be helpful in devising interventional approaches. Soluble forms of the HIV-1 envelope glycoproteins have been produced by deletion of the gp41 membrane-spanning region and intracytoplasmic domain, as well as modification of the proteolytic cleavage site between gp120 and gp41 (4, 21, 23–25). These soluble HIV-1 envelope glycoproteins consist mainly of dimers and tetramers (4, 21, 23–25). Here we investigate approaches designed to increase the production and stability of soluble HIV-1 envelope glycoprotein trimers.
MATERIALS AND METHODS
Envelope glycoprotein constructs.
The envelope glycoprotein expression plasmids were derived from pSVIIIenv and were constructed by PCR or by QuikChange (Stratagene) site-directed mutagenesis. The specific changes introduced into each mutant are described in Results. The sequence of the entire env open reading frame was determined for each of the mutants. Two differences between the wild-type YU2 gp120 glycoprotein and the soluble gp120 glycoprotein were noted. One of these apparently arose as a result of PCR error and converted the wild-type alanine 379 to glutamine. In addition, a single glycine residue was introduced at the C terminus of the gp120 glycoprotein, after the arginine at position 508. All of the other glycoproteins used in the study exhibited the wild-type sequence of either the YU2 R5 or the HXBc2 X4 viral envelope glycoproteins, except where modifications were deliberately introduced. Amino acid residue numbers are reported according to those of the prototypic HXBc2 sequences (35).
Envelope glycoprotein expression.
To express the soluble HIV-1 envelope glycoproteins, a 100-mm dish of 293T cells was transfected with 4.5 μg of the pSVIIIenv plasmid expressing the mutant glycoproteins and 0.5 μg of an HIV-1 Tat-expressing plasmid. The transfection was performed using LipofectAMINE-PLUS reagent (Gibco-Life Technology) according to the manufacturer's recommendations. Sixteen hours after transfection, the cells were metabolically labeled with [35S]methionine-cysteine (NEN) in methionine- and cysteine-free Dulbecco modified Eagle medium (DMEM) for 24 h.
Sucrose density gradient centrifugation.
The oligomeric state of the envelope glycoprotein variants was investigated using sucrose density gradient centrifugation. The radiolabeled proteins in the transfected 293T cell supernatants were concentrated about 3-fold [15-fold for the less efficiently secreted gp130(−) glycoprotein] using Centriprep 30 filters (Amicon). Approximately 750 μl of the concentrated supernatants was loaded onto 10-ml 10 to 25% continuous sucrose gradients, which were centrifuged in a Beckman SW41 rotor for 20 h at 40,000 rpm at 4°C. Fractions of 1.1 ml were collected manually and precipitated using a mixture of sera from HIV-1-positive individuals and protein A-Sepharose. Precipitates were analyzed on nonreducing and reducing sodium dodecyl sulfate (SDS)-polyacrylamide gels and autoradiographed. Molecular weights of the precipitated proteins were analyzed by interpolation, using previously characterized envelope glycoprotein dimers and trimers (26) and known molecular weight standards for reference.
Molecular exclusion chromatography.
For gel filtration analysis, the soluble envelope glycoproteins were produced by transient transfection of 293T cells using Effectene reagents (Qiagen). For each protein, 30 100-mm dishes of cells were transfected. The soluble envelope glycoproteins were harvested in 5 ml of culture medium every day for 3 days after transfection. The pooled supernatants were incubated twice with 8 ml of 10% (wt/vol) protein A-agarose beads for 3 to 5 h at 4°C to deplete the culture medium of antibodies. The solution was then passed three times by gravity through an affinity column in which the F105 anti-gp120 monoclonal antibody was coupled to protein A-agarose beads. After being washed with 50 column volumes of 0.5 M NaCl in phosphate-buffered saline (PBS), the envelope glycoproteins were eluted with 10 column volumes of 3 M MgCl2 in 20 mM Tris-HCl, pH 7.2. After dialysis against 100 volumes of PBS and concentration using Centriprep filters (Amicon), the proteins were assessed by SDS-polyacrylamide gel electrophoresis for purity and amount, using a serially diluted bovine serum albumin standard.
Approximately 10 μg of the purified envelope glycoproteins in 50 μl of PBS was loaded onto a Superdex 200 gel filtration column (Pharmacia). The column was then eluted at a rate of 0.5 ml/min for 40 min, and the eluted proteins were detected by measuring optical density at 280 nm (OD280) using a Varian ProStar System (Varian Analytical Instruments). A panel of protein markers (Pharmacia) was analyzed under identical conditions to serve as molecular weight standards.
Immunoprecipitation.
Envelope glycoproteins were precipitated either by a mixture of sera from HIV-1-infected individuals or by a specific monoclonal antibody, as previously described (59). Many of the antibodies studied were used in a previous study on antibody competition mapping of HIV-1 gp120, in which they have been described (45).
Precipitation by the PK-C299 peptide.
The sequence of the PK-C299 peptide is as follows: YTHIIYSLIEQSQNQQEKNEQELLALDKWASLWNWFGGGTETSQVAPA. The underlined N terminus of this sequence corresponds to that of the DP178 peptide derived from the YU2 HIV-1 gp41 C-terminal (C34) helix (12). The underlined C terminus is derived from the C terminus of bovine rhodopsin and can be recognized by the 1D4 antibody (46). The sequence GGG was included in the peptide to introduce potential structural flexibility between the DP178 helix and the bovine rhodopsin peptide tag. PK-C299 was custom synthesized by the Protein Chemistry Core Facility at the Howard Hughes Medical Institute, Columbia University. Metabolically labeled ([35S]Met-Cys) gp130(−) and gp130(−/GCN4) glycoproteins were incubated with a 20-fold molar excess of PK-C299 for 1 h at 37°C. The glycoprotein-peptide complex was then precipitated by 5 μg of the 1D4 antibody.
Chemokine receptor binding assay.
The soluble envelope glycoproteins were tested for their ability to bind CD4 and CCR5 using a previously published protocol (34). Briefly, [35S]cysteine-methionine-labeled envelope glycoproteins were produced in transfected 293T cells. An aliquot of the cell supernatants was precipitated using pooled sera from HIV-1-infected individuals. After analysis on SDS-polyacrylamide gels, the amounts of envelope glycoproteins were measured using a PhosphorImager storage screen (Molecular Dynamics). Equivalent amounts of the envelope glycoproteins were incubated with 10 μg of soluble CD4 (sCD4)/ml at room temperature for 1 h. The mixtures were then applied to 106 Cf2Th-synCCR5 (41) cells in 6-well plates for 2 h at 37°C in the presence of 0.2% sodium azide. The unbound proteins were washed away with cold DMEM, and the bound proteins were precipitated from cell lysates by a mixture of 3 μl of pooled sera from HIV-1-infected individuals and 1 μg of the anti-gp120 monoclonal antibody C11. The precipitated proteins were analyzed on SDS-polyacrylamide gels.
RESULTS
Production and characterization of soluble HIV-1 envelope glycoproteins.
We investigated three approaches designed to stabilize soluble forms of the HIV-1 envelope glycoprotein trimers (Fig. 1). The first approach is the disruption of the proteolytic cleavage site between the gp120 and gp41 subunits by replacing arginine residues 508 and 511 with serines. The second approach is the introduction of a cysteine pair (and an adjacent glycine) into residues 576 to 578, which are located in the N-terminal (N36) gp41 helix. The introduced cysteines occupy the d and e positions of the heptad repeat and thus are located within the hydrophobic interior of the trimeric gp41 α-helical coiled coil. Identical cysteine substitutions result in the covalent cross-linking of the full-length HIV-1 gp160 envelope glycoprotein subunits (26). The third approach is the extension of the N-terminal gp41 coiled coil by the C-terminal addition of GCN4 sequences. GCN4 is a transcription factor that normally forms stable homodimers. However, introduction of hydrophobic residues at the a and d positions of the heptad repeats of the GCN4 dimerization motif increases the propensity of the protein to form trimers (29). We fused the GCN4 trimeric motif (MKQIEDKIEEILSKIYHIENEIARIKKLIGEV) C-terminal to the end (...YLRDQQLL) of the gp41 coiled coil, thereby extending the heptad repeat region and the potential stability of trimer association.
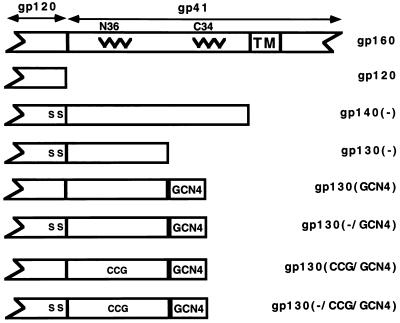
FIG. 1.
Structure of the soluble HIV-1 envelope glycoprotein variants. The partial structures of the soluble gp120, gp140, and gp130 envelope glycoproteins used in this paper are shown beneath the corresponding segment of the wild-type gp160 glycoprotein. The junction of gp120 and gp41, the positions of the N-terminal (N36) and C-terminal (C34) gp41 α-helices, and the transmembrane (TM) region are shown. The DP178 peptide used in this study contains sequences corresponding to those of the C34 helix. The positions of the serine substitutions for arginine 508 and arginine 511 are shown (S S). The position of the Cys-Cys-Gly substitution in the N-terminal helical segment (residues 576 to 578) is shown (CCG). The position of the GCN4 trimeric peptide is also shown.
Most of the envelope glycoproteins used in this study (Fig. 1) were derived from the primary R5 HIV-1 isolate YU2. Stop codons were introduced into the env gene (GenBank accession number M93258), resulting in termination of the proteins N-terminal to the natural gp160 transmembrane (TM) region. Thus, the majority of the envelope glycoproteins were secreted into the medium of expressing cells. To express soluble gp120, the YU2 envelope glycoprotein was terminated after arginine 508. Soluble gp130 glycoproteins contained all of the YU2 envelope glycoprotein residues up to and including leucine 593, which is located near the C terminus of the gp41 coiled coil. The soluble gp140 glycoproteins, which were derived from the YU2 and HXBc2 HIV-1 isolates, were terminated after leucine 669, which is located N-terminal to the membrane-spanning region. The presence of modifications designed to increase trimer stability in the mutants is indicated in parentheses in the mutant name. Thus, a minus sign in parentheses indicates the R-to-S changes at positions 508 and 511 affecting gp120–gp41 proteolytic processing; CCG indicates the substitution of the cysteine pair and a glycine residue at positions 576 to 578; and GCN4 indicates the presence of the GCN4 trimeric peptide at the carboxyl terminus of the protein. Our initial studies indicated that, in contrast to the results seen for the membrane-anchored gp160 envelope glycoprotein, the CCG substitutions were not sufficient to cross-link the subunits of either soluble gp130 or gp140 glycoproteins (data not shown). Therefore, in this report we focus on the effects of the cleavage site modification and CCG substitution in the context of soluble glycoproteins containing the GCN4 trimeric motif, which was found to have a major stabilizing influence (see below).
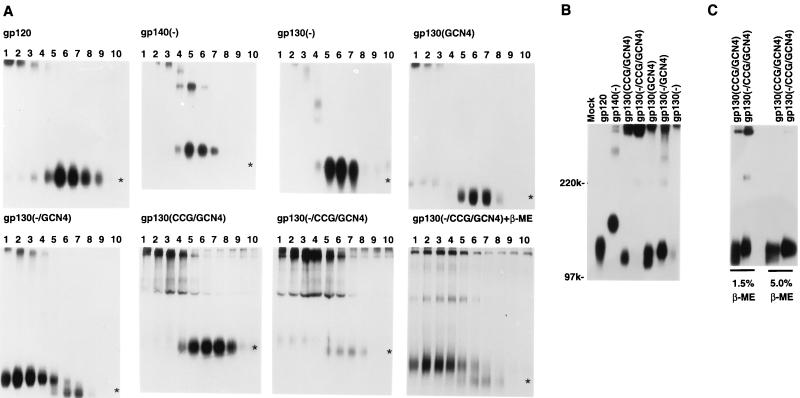
Radiolabeled envelope glycoproteins were produced transiently in transfected 293T cell supernatants, which were concentrated and analyzed by sucrose density gradient centrifugation. Gradient fractions were collected and precipitated by a mixture of sera from HIV-1-infected individuals. The immunoprecipitates were analyzed on SDS-polyacrylamide gels under reducing and nonreducing conditions. Autoradiographs of some of these gels are shown in Fig. 2A. The gp120 glycoprotein, which is known to be monomeric, sedimented primarily in fractions 6 and 7 of the gradient. A small amount of the gp120 glycoprotein was found in fractions 1 to 3 and migrated on the nonreducing gels as high-molecular-weight forms. These slowly migrating forms of gp120 were not observed on reducing gels (data not shown) and probably represent previously described, aberrantly cross-linked envelope glycoproteins (47). The gp140(−) glycoproteins from the YU2 and HXBc2 HIV-1 isolates both exhibited mostly monomeric forms that sedimented in fractions 5 to 7, although some higher-order forms were observed in fractions 2 to 4 (Fig. 2A and data not shown). Likewise, the gp130(−) glycoprotein sedimented mostly as a monomer in fractions 5 to 7, although higher-order forms could be seen in fractions 2 to 4. The gp130(GCN4) glycoprotein sedimented similarly to the gp130(−) glycoprotein. The majority of the gp130(GCN4) glycoprotein sedimented as a monomer in the sucrose gradient (fractions 5 to 7) and migrated with an apparent molecular size of approximately 120 kDa. The small portion of the gp130(GCN4) glycoprotein that sedimented more rapidly (fractions 1 to 3) exhibited two molecular sizes (approximately 130 and greater than 350 kDa) on nonreducing SDS-polyacrylamide gels. This pattern suggests that uncleaved gp130(GCN4) is oligomeric, whereas the proteolytically cleaved gp130(GCN4) glycoprotein is monomeric. That the lack of cleavage between the gp120 and gp41 moieties stabilizes the oligomer is supported by the sedimentation pattern of the gp130(−/GCN4) glycoprotein. The majority of this protein sedimented in fractions 1 to 4, with the proteins in these fractions exhibiting molecular sizes of 130 and greater than 350 kDa. A small portion of the gp130(−/GCN4) glycoprotein was cleaved to a form of approximately 120 kDa, which sedimented as a monomer in fractions 5 to 8. Thus, the faster-sedimenting fractions of the gp130(GCN4) and gp130(−/GCN4) glycoproteins, which presumably represent higher-order oligomers, contain only uncleaved proteins.
FIG. 2.
Characterization of the soluble HIV-1 envelope glycoprotein variants. (A) Sucrose density gradient fractions containing the soluble HIV-1 envelope glycoproteins are shown. Fraction 1 was collected from the bottom of the gradient; fraction 10 represents the top of the gradient. Fractions were precipitated by a mixture of sera from HIV-1-infected individuals. Precipitates were analyzed on SDS-polyacrylamide gels under non-reducing conditions, except for the samples [gp130(−/CCG/GCN4) + β-ME] shown in the bottom right panel, which were treated with 1.5% β-ME before gel analysis. Asterisks indicate the expected position of a gp120 monomer in each gel. (B) Radiolabeled envelope glycoproteins were precipitated from transfected cell supernatants by a mixture of sera from HIV-1-infected individuals and were analyzed under nonreducing conditions (in the absence of β-ME). (C) Precipitates of radiolabeled envelope glycoproteins were formed as for panel B and were analyzed on SDS-polyacrylamide gels after treatment with either 1.5% β-ME for 3 min at 100°C or 5% β-ME for 10 min at 100°C.
To determine if potential intersubunit disulfide bonds could be formed and if these might contribute to trimer stability, the gp130(CCG/GCN4) and gp130(−/CCG/GCN4) proteins were studied. The gp130(CCG/GCN4) glycoprotein sedimented similarly to the gp130(GCN4) glycoprotein, with a substantial portion of the protein present in fractions 5 to 7 and a portion of the protein in fractions 1 to 4. As was seen for the gp130(GCN4) glycoprotein, the more slowly sedimenting fractions consisted mainly of an apparently cleaved gp120 glycoprotein. The faster-sedimenting portion of the gp130(CCG/GCN4) protein exhibited a molecular size greater than 350 kDa under nonreducing conditions. Unlike the case for the gp130(GCN4) protein, no 130-kDa band was evident in fractions 1 to 3 of the gp130(CCG/GCN4) protein. This indicates that the substitution of the cysteine pair in the gp41 ectodomain resulted in covalent cross-linking of some of the gp130(CCG/GCN4) envelope glycoprotein subunits. The majority of the gp130(−/CCG/GCN4) glycoprotein sedimented in fractions 2 to 4 and migrated with an apparent molecular size greater than 350 kDa. A portion of these high-molecular-weight glycoproteins could be reduced by treatment with 1.5% β-mercaptoethanol (β-ME), and interestingly, these reduced products of the gp130(−/CCG/GCN4) oligomer consisted almost exclusively of the uncleaved 130-kDa glycoprotein (Fig. 2A, bottom right panel). A small amount of the gp130(−/CCG/GCN4) was apparently cleaved and sedimented in fractions 5 to 8, consistent with a monomer. Almost all of the greater-than-350-kDa forms of the gp130(CCG/GCN4) and gp130(−/CCG/GCN4) glycoproteins could be reduced by boiling in 5% β-ME to proteins with an apparent molecular size of 130 kDa (see Fig. 2C).
These results indicate that the inclusion of GCN4 sequences and the disruption of proteolytic cleavage contribute to the formation and stabilization of soluble, higher-order oligomers. The presence of the cysteine pair in the gp41 coiled coil allows intersubunit disulfide bonds to form, covalently stabilizing these oligomers.
Molecular exclusion chromatography of soluble HIV-1 envelope glycoproteins.
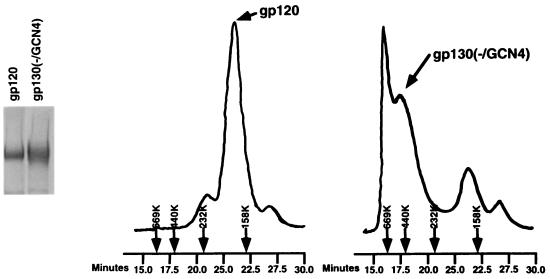
The YU2 gp120 and gp130(−/GCN4) glycoproteins were analyzed by molecular exclusion chromatography on a Superdex 200 column (Fig. 3). The gp120 glycoprotein exhibited an apparent molecular size of 157 kDa. A major form of the gp130(−/GCN4) protein exhibited a molecular size of 410 kDa. Smaller amounts of the gp130(−/GCN4) protein were eluted at the position corresponding to the gp120 glycoprotein. Also, some higher-molecular-weight aggregates were apparent for this glycoprotein. A substantial fraction of the gp130(−/GCN4) glycoprotein exhibits a molecular weight consistent with that of a trimer.
FIG. 3.
Molecular exclusion chromatography of soluble envelope glycoproteins. The YU2 gp120 and gp130(−/GCN4) glycoproteins were analyzed on a Superdex 200 column and compared with molecular size standards, the positions of which are indicated by arrows. Elution times are also shown. The aggregate peak for the gp130(−/GCN4) glycoprotein elutes before the highest molecular size standard used (660 kDa).
Immunoprecipitation of soluble HIV-1 envelope glycoproteins.
The radiolabeled supernatants containing the soluble HIV-1 envelope glycoprotein variants were also studied by direct immunoprecipitation by a mixture of sera from HIV-1-infected individuals. The precipitates were analyzed on SDS-polyacrylamide gels run under nonreducing conditions (Fig. 2B). The results indicate that, under these conditions, the glycoproteins with the cysteine pair [gp130(CCG/GCN4) and gp130(−/CCG/GCN4)] exhibited the greatest proportion of high-molecular-weight forms. Consistent with the results from the sucrose gradients, the low-molecular-weight species of the gp130(CCG/GCN4) protein appears to be a cleaved gp120. The vast majority of the gp130(−/CCG/GCN4) protein migrated as a high-molecular-weight species, underscoring the efficacy of the cysteine cross-linking in this context. This higher-order form of the gp130(−/CCG/GCN4) glycoprotein migrated on SDS-polyacrylamide gels at a molecular size of approximately 400 kDa, which was determined by comparing its migration with those of protein molecular weight markers and envelope glycoprotein oligomers of known sizes (26) (Fig. 2B). This estimated molecular size was consistent with the notion that the gp130(−/CCG/GCN4) glycoprotein forms a trimer. The gp130(−/GCN4) protein migrated primarily as a 130-kDa product, although some higher-molecular-size forms were evident. Consistent with proteolytic cleavage, the gp130(GCN4) protein migrated as 130- and 120-kDa species. The gp140(−) protein migrated primarily as a 140-kDa product, although high-molecular-weight forms consistent with dimers, trimers, and other higher-order forms could be discerned. No high-molecular-weight species of the gp120 glycoprotein were evident.
To demonstrate that disulfide bond formation contributed to the stabilization of oligomers, the precipitated gp130(CCG/GCN4) and gp130(−/CCG/GCN4) glycoproteins were boiled in the presence of β-ME prior to analysis on SDS-polyacrylamide gels (Fig. 2C). Boiling for 3 min in 1.5% β-ME resulted in partial disruption of the higher-molecular-weight forms of these glycoproteins. Boiling for 10 min in 5% β-ME almost completely reduced the gp130(CCG/GCN4) and gp130(−/CCG/GCN4) oligomers to lower-molecular-weight forms. The majority of the monomeric forms produced upon reduction of both these proteins migrated as a 130-kDa species, consistent with the idea that lack of proteolytic cleavage promotes trimer stability. These results indicate that the gp130(CCG/GCN4) and gp130(−/CCG/GCN4) glycoproteins maintain the higher-order forms on SDS-polyacrylamide gels through the formation of disulfide bonds.
Recognition of soluble envelope glycoproteins by the DP178 gp41 peptide.
The cross-linking of the soluble envelope glycoprotein subunits by cysteines placed at the internal d and e positions of the gp41 heptad repeat implies that at least part of the N-terminal helical coiled coil can be formed in these proteins. To examine this further, we asked whether the DP178 peptide, which corresponds in sequence to the C-terminal α-helix of the gp41 ectodomain (C34 in Fig. 1), could interact with the soluble trimers. The DP178 peptide forms a helix that packs into a hydrophobic groove formed on the outer surface of the trimeric N-terminal gp41 coiled coil (12). The [35S]Met-Cys-labeled supernatants, which were adjusted to contain the same amounts of the gp130(−) and gp130(−/GCN4) glycoproteins, were precipitated either by a mixture of sera from HIV-1-infected individuals or by the PK-C299 peptide–1D4 antibody mixture. The PK-C299 peptide is composed of the DP178 HIV-1 gp41 sequence fused to a C9 peptide tag corresponding to the C terminus of bovine rhodopsin. The C9 peptide epitope is recognized by the 1D4 antibody. Equivalent amounts of the gp130(−) and gp130(−/GCN4) glycoproteins were precipitated by the mixture of sera from HIV-1-infected individuals (Fig. 4, left panel). The gp130(−/GCN4) glycoprotein was precipitated much more efficiently than the gp130(−) glycoprotein by the PK-C399–1D4 antibody mixture (Fig. 4, right panel). Thus, the soluble gp130(−/GCN4) glycoprotein, which forms stable trimers, can be recognized more efficiently by the DP178 peptide than a similar glycoprotein that is primarily monomeric.
FIG. 4.
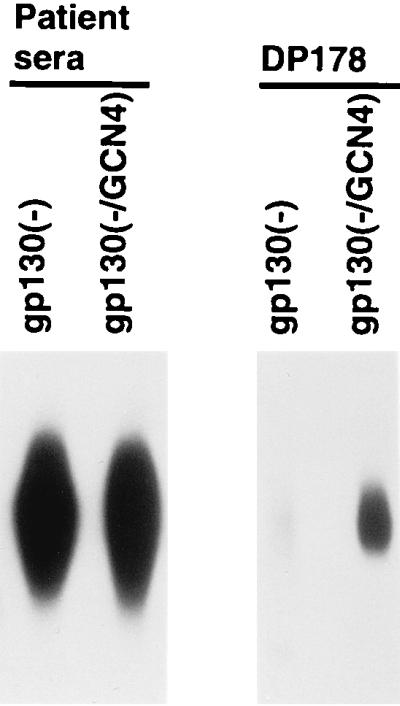
Recognition of the soluble HIV-1 envelope glycoprotein variants by the DP178 gp41 peptide. Radiolabeled envelope glycoproteins in transfected cell supernatants were normalized for the amount of envelope protein and then precipitated either with a mixture of sera from HIV-1-infected patients (patient sera) or with a mixture of the 1D4 antibody and the PK-C299 peptide (DP178). The PK-C299 peptide contains the residues of the DP178 peptide, which corresponds to the C-terminal gp41 helical region (C34 in Fig. 1), and the residues of the C9 peptide, which are recognized by the 1D4 antibody.
Recognition of the soluble HIV-1 envelope glycoproteins by antibodies.
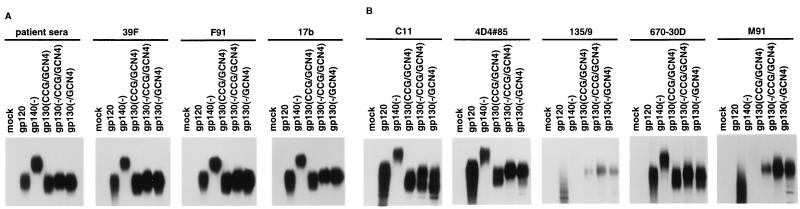
The structural integrity of three of the soluble glycoproteins [gp130(CCG/GCN4), gp130(−/CCG/GCN4), and gp130(−/GCN4)] that exhibited some trimeric forms was examined by precipitation by a panel of antibodies directed against different epitopes on the HIV-1 envelope glycoproteins. The soluble YU2 gp120 and gp140(−) glycoproteins, which are mainly monomeric, were included in this study for comparison. Radiolabeled supernatants containing similar levels of all five glycoproteins were precipitated either by a mixture of sera from HIV-1-infected individuals or by a monoclonal antibody. Precipitates were analyzed on SDS-polyacrylamide gels after complete reduction in 5% β-ME. Figure 5A shows that the mixture of HIV-1-infected patient sera precipitated similar levels of all five glycoproteins. The gp130(CCG/GCN4) glycoprotein migrated as proteolytically cleaved gp120 and uncleaved gp130 forms. The recognition patterns of the 39F, 17b, and F91 antibodies were similar to that of the mixture of sera. The 39F antibody recognizes a conformation-dependent structure in the third variable (V3) loop of the HIV-1 gp120 glycoprotein (J. Robinson, personal communication). The 17b antibody binds to a discontinuous gp120 epitope that is induced by CD4 binding and that overlaps the conserved chemokine receptor binding region. The 48d antibody, which recognizes a related, overlapping epitope, was also tested and exhibited a similar pattern of recognition (data not shown). The F91 antibody recognizes a discontinuous gp120 epitope overlapping the CD4 binding site. Two other antibodies directed against CD4 binding site (CD4BS) epitopes on gp120, F105 and IgG1b12, were also tested and exhibited similar recognition profiles (data not shown). These results suggest that the discontinuous epitopes recognized by several anti-gp120 monoclonal antibodies are present and accessible on the soluble trimers. These results were confirmed when the rapidly sedimenting fractions of the gp130(CCG/GCN4), gp130(−/CCG/GCN4), and gp130(−/GCN4) proteins were first prepared on sucrose gradients and then immunoprecipitated (data not shown).
FIG. 5.
Recognition of the soluble HIV-1 envelope glycoprotein variants by monoclonal antibodies. Immunoprecipitation of the radiolabeled envelope glycoproteins was performed with a mixture of sera from HIV-1-infected individuals (patient sera) or with monoclonal antibodies. The precipitates were analyzed on SDS-polyacrylamide gels under reducing conditions. (A) The envelope glycoproteins were precipitated by a panel of anti-gp120 monoclonal antibodies that recognize discontinuous gp120 epitopes. The 39F antibody recognizes the gp120 V3 variable loop, the F91 antibody recognizes the gp120 CD4 binding site, and the 17b antibody recognizes a CD4-induced gp120 epitope. (B) The envelope glycoproteins were precipitated by monoclonal antibodies against epitopes in the C1 and C5 regions of gp120: a discontinuous epitope involving the C1 and C5 regions (C11), and linear gp120 epitopes within the first (C1) conserved region (4D4#85 and 135/9) or within the fifth (C5) conserved region (670-30D and M91).
The first (C1) and fifth (C5) conserved regions which represent the N and C termini, respectively, of the gp120 envelope glycoprotein, have been implicated in the interaction with the gp41 ectodomain (17, 30, 59). To investigate the integrity and accessibility of gp120 epitopes from these regions on the soluble envelope glycoproteins, the radiolabeled glycoproteins were precipitated by antibodies that recognize epitopes with C1 and/or C5 components (Fig. 5B). The C11 antibody, which recognizes a discontinuous gp120 epitope composed of C1 and C5 elements, was able to precipitate all five of the soluble envelope glycoproteins. However, there were some qualitative and quantitative differences between the recognition pattern of the C11 antibody and those of the antibodies described above. As reported previously (59), the gp140(−) glycoprotein was recognized by the C11 antibody less efficiently than the soluble gp120 glycoprotein, in contrast to the recognition pattern observed for the mixture of sera from HIV-1-infected individuals. The proteolytically cleaved, monomeric forms of the gp130(CCG/GCN4), gp130(−/CCG/GCN4), and gp130(−/GCN4) glycoproteins were more efficiently precipitated by the C11 antibody than by the serum mixture. The efficient precipitation of the gp130(−/CCG/GCN4) and gp130(−/GCN4) monomers by the C11 antibody was particularly noteworthy because proteolytically cleaved monomers represented only a small fraction of these two glycoprotein preparations. Although the trimeric, uncleaved forms of these glycoproteins could be precipitated by the C11 antibody, the precipitation of these forms was relatively less efficient than that observed for the previously discussed antibodies. These results suggest that the C11 antibody can access the gp130 trimers but binds less efficiently to the trimers than to the gp120 monomer. This impression was confirmed by precipitation of monomers and trimers prepared on sucrose density gradients (data not shown).
The 522-149, #45, and M90 antibodies, which are directed against discontinuous C1 gp120 epitopes, precipitated the uncleaved gp130 glycoproteins efficiently (data not shown). Similar results were obtained with the A32 antibody, which recognizes a discontinuous C1–C4 gp120 epitope (data not shown). As previously reported (59), the efficiency with which the gp140(−) glycoprotein was precipitated by these antibodies was somewhat decreased relative to that observed for the mixture of sera from HIV-1-infected individuals (data not shown).
The recognition of the soluble glycoproteins by antibodies that bind linear HIV-1 gp120 epitopes in the C1 region was also examined. The 4D4#85 antibody, which recognizes an epitope within residues 35 to 50 of gp120, efficiently precipitated the gp120, gp130(CCG/GCN4), gp130(−/CCG/GCN4), and gp120(−/GCN4) glycoproteins and precipitated the gp140(−) glycoprotein less efficiently (Fig. 5B). The 133/290 and 135/9 antibodies recognize epitopes that span gp120 residues 61 to 70 and 111 to 120, respectively (45). These antibodies only inefficiently precipitated faster-migrating, presumably underglycosylated forms of the HIV-1 gp120 glycoprotein (Fig. 5B and data not shown), consistent with the findings of earlier studies indicating the poor exposure of these linear epitopes on native gp120 (31). The gp140(−) glycoprotein was not precipitated by these antibodies. The 133/290 and 135/9 antibodies precipitated only the uncleaved, trimeric forms of the gp130 glycoprotein variants (Fig. 5B and data not shown). The monomeric, cleaved forms of the gp130 glycoproteins were not recognized by these antibodies. The tendency of the 133/290 and 135/9 antibodies to precipitate the trimeric forms of the gp130(−/CCG/GCN4) and gp130(CCG/GCN4) glycoproteins preferentially over monomeric forms was even more pronounced when sucrose density-purified fractions of these proteins were tested (data not shown).
The recognition of the soluble glycoproteins by antibodies that bind linear gp120 epitopes in the C5 region was also examined. The 670-30D antibody, which recognizes an epitope encompassing gp120 residues 498 to 504, precipitated all five proteins efficiently (Fig. 5B) (61). A similar result was obtained with the 1331A antibody, which is directed against the same C5 region (data not shown). The M91 and CRA-1 antibodies recognize gp120 residues 461 to 470, which span the boundary of the V5 and C5 regions (45). These antibodies precipitated only faster-migrating, presumably less-glycosylated forms of soluble gp120, consistent with the findings of previous studies indicating that native gp120 is not recognized efficiently by these antibodies (44). The gp140(−) glycoprotein was not precipitated by either antibody (Fig. 5B and data not shown). The M91 and CRA-1 antibodies preferentially recognized the trimeric, uncleaved forms of the gp130 glycoproteins. This preference was confirmed by precipitation of monomeric and trimeric preparations of these glycoproteins purified on sucrose gradients (data not shown).
Chemokine receptor binding.
The ability of the gp130(−/GCN4) and gp120 glycoproteins to bind CCR5-expressing cells in the absence or presence of sCD4 was examined (Fig. 6). Both glycoproteins bound CCR5-expressing cells in the presence of sCD4 but not in its absence. The binding of the gp120 glycoprotein was more efficient than that of the gp130(−/GCN4) glycoprotein. Nonetheless, these results indicate that the gp130(−/GCN4) glycoprotein retains the ability to bind sCD4 and that CD4 binding enhances the ability to bind CCR5.
FIG. 6.
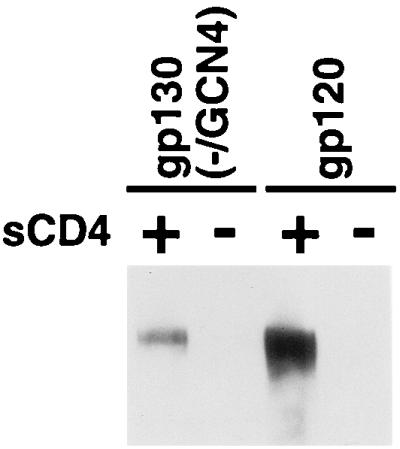
CCR5 binding of the soluble HIV-1 envelope glycoproteins. Equivalent amounts of radiolabeled soluble envelope glycoproteins were incubated with Cf2Th/synCCR5 cells in the absence or presence of sCD4. The bound proteins were precipitated and analyzed on SDS-polyacrylamide gels, the autoradiographs of which are shown.
DISCUSSION
Previous studies have produced soluble HIV-1 envelope glycoproteins lacking a transmembrane region and containing a modified proteolytic cleavage site between gp120 and gp41 (4, 21, 23–25). These soluble glycoproteins were expressed in mammalian cells using vaccinia virus vectors and were reported to form dimers and tetramers (4, 21, 23–25). Equivalent gp140(−) constructs made in this study from both the primary YU2 and laboratory-adapted HXBc2 HIV-1 isolates were mainly monomeric. The reasons for this difference are unknown. The formation of gp140(−) oligomers may depend upon the high levels of expression achieved by vaccinia virus vectors; higher concentrations of the gp140(−) glycoproteins could allow weak interactions favoring dimer and tetramer formation.
In contrast to the gp140(−) glycoproteins, some of the soluble gp130 glycoproteins created in this study form relatively stable trimers. The functionally relevant form of the HIV-1 envelope glycoproteins is thought to be a trimer (10, 11, 26, 53, 55, 60). Several lines of evidence indicate that the gp130(−/GCN4) molecules are trimeric. First, the gp130(−/GCN4) glycoprotein, but not the gp130(−) glycoprotein, reacts with the DP178 peptide. The DP178 peptide intercalates into the hydrophobic grooves formed by a trimeric coiled coil (11, 12, 53, 55) but would not be expected to bind efficiently to dimeric or tetrameric coiled coils. Second, the placement of the cysteine-cysteine-glycine residues at gp41 positions 576 to 578 would favor intersubunit disulfide bond formation only in the context of a trimeric coiled coil (26). In dimeric or tetrameric coiled coils, the distances between the Cβ atoms of the cysteines would be unfavorable for cross-linking of the subunits. Finally, gel filtration analysis indicates that the molecular weight of the gp130(−/GCN4) glycoprotein is consistent with that expected for a trimer.
Our work provides insights into the factors that influence the stability of soluble HIV-1 envelope glycoprotein trimers. These insights, in addition to providing practical guidance for producing tractable, soluble trimers, might also apply to the native, membrane-anchored envelope glycoprotein complex. The extension of the N-terminal gp41 coiled coil by the trimeric GCN4 sequence was required for the efficient production of stable, soluble trimers. The success of this approach suggests that, within the trimers, elements of the N-terminal coiled coil are formed and participate in intersubunit contacts. This assertion is supported by the observation that intersubunit disulfide bonds form when cysteines are placed in the d and e positions of the N-terminal gp41 heptad repeat (26). These positions are located on the inner, hydrophobic face of the trimeric coiled coil and are separated by distances that are acceptable for disulfide bond formation (11, 53, 55). That the N-terminal gp41 coiled coil is well formed along its length in the soluble gp130 trimers is supported by the ability of DP178, which corresponds to the C-terminal α-helix in the gp41 ectodomain, to bind these oligomers. C-terminal α-helical gp41 peptides have been shown to bind within a long hydrophobic groove created by the interaction of two N-terminal gp41 helices in the trimeric coiled coil (10). The accommodation of the N-terminal gp41 coiled coil within a stable, soluble trimer supports a model in which this coiled coil exists, at least in part, within the complete HIV-1 envelope glycoprotein precursor and contributes to oligomerization. Disulfide bonds among the trimer subunits form as a result of the introduction of the cysteine pair at positions 576 and 577 of the complete HIV-1 gp160 envelope glycoprotein precursor, further supporting this model (26). Perhaps the coiled coil of the HIV-1 fusion protein, unlike that of influenza virus hemagglutinin, does not need to undergo extensive conformational changes from a precursor state in order to form (5, 9).
Besides the addition of trimeric GCN4 sequence, another factor that exhibited a major influence on the stability of soluble HIV-1 envelope glycoprotein trimers was proteolytic cleavage at the gp120–gp41 junction. Regardless of the means by which the trimer subunits were associated, including the presence of covalent disulfide bonds in the gp41 subunit, cleaved proteins were monomeric. This was unexpected because at least a portion of the cleaved, membrane-associated HIV-1 envelope glycoproteins retains trimeric structure on native virions. The basis for this difference is unknown, but it may simply reflect the greater lability of soluble envelope glycoprotein trimers or, alternatively, it might be due to differences in the accommodation of the cleaved segments in the two contexts.
In the case of the gp130(−/GCN4) and gp130(−/CCG/GCN4) glycoproteins, a low level of proteolytic cleavage was observed despite alteration of two basic residues N-terminal to the cleavage site. It is uncertain whether cleavage occurred precisely at the natural site in these mutants, although the recognition of the cleaved gp120 glycoprotein by antibodies against gp120 C-terminal regions was comparable to that of wild-type gp120. Efforts to reduce the observed residual cleavage by further alteration of basic residues near the natural cleavage site did not succeed (data not shown).
Covalent linkage of the trimeric subunits through disulfide bond formation resulted in extremely stable oligomers that remained associated on SDS-polyacrylamide gels run under nonreducing conditions or in the presence of 1.5% β-ME. Although covalent linkage was neither necessary nor sufficient for the production of stable soluble trimers, it may prove useful in circumstances where trimers are subjected to harsh conditions.
During natural infection, the humoral response to the HIV-1 envelope glycoproteins consists of both nonneutralizing and neutralizing antibodies. Many of the nonneutralizing antibodies appear to be generated against shed, monomeric gp120 glycoproteins and do not bind efficiently to the functional envelope glycoprotein trimer (48). The vast majority of the gp120 epitopes, including all of the neutralization epitopes examined, were present on the soluble gp130 trimers, where their exposure was similar to that seen on the monomeric gp120 glycoprotein. Differences between monomeric and trimeric envelope glycoproteins involved epitopes in the first (C1) and fifth (C5) conserved regions of gp120, which have been previously implicated in the interaction with gp41. The C11 antibody, which recognizes a discontinuous gp120 epitope with C1 and C5 components, precipitated the proteolytically cleaved, monomeric forms of the soluble glycoproteins more efficiently than any of the other antibodies studied. This is consistent with the idea that the C11 antibody was generated to a monomeric, soluble gp120 glycoprotein shed from virions or infected cells during natural HIV-1 infection (45).
Recognition of the gp130 proteins by some antibodies directed against linear C1 and C5 epitopes was actually increased relative to recognition of the gp120 or gp140(−) monomers. Previous studies of the native HIV-1 gp120 monomer suggested that C1 and C5 sequences at the very N and C termini of the protein, respectively, were well exposed, whereas more-interior N- and C-terminal residues were less accessible to antibodies (44, 45). This is consistent with the known involvement of the interior C1 and C5 sequences in secondary structural elements of the gp120 core domains (37). In our study, recognition of the mature, fully glycosylated gp120 monomer by antibodies against interior C1 regions (residues 61 to 70 and 111 to 120) and an interior C5 region (residues 461 to 470) was minimal (43). By contrast, these regions were accessible to antibodies on the trimeric, but not the monomeric, forms of the soluble gp130 glycoproteins. These observations suggest that, in the formation of these trimers, the N- and C-terminal regions of gp120 are extended into more-exposed conformations than those assumed in the gp120, gp140, and gp130 monomers. Interestingly, the influenza virus HA1 glycoprotein N and C termini, which make extensive contacts with the HA2 transmembrane protein, also exhibit extended structures in the trimeric hemagglutinin complex (13).
Another explanation for the differential recognition of the monomers and trimers by the C1-directed antibodies 133/290 and 135/9 and by the C5-directed antibodies M91 and CRA-1 is a potential difference in the glycosylation of monomeric and trimeric soluble glycoproteins. Although these antibodies did not recognize the fully glycosylated gp120 monomer, they did precipitate a faster-migrating form of gp120 that is presumably incompletely glycosylated. It is possible that soluble trimers are glycosylated differently than the monomeric proteins, contributing to better recognition by these antibodies.
Surprisingly, the formation of stable gp130 trimers was not sufficient to render all of the nonneutralizing gp120 epitopes inaccessible to antibodies. In fact, the linear C1 and C5 epitopes that are accessible only on the soluble gp130 trimers are not thought to be available for antibody binding in the context of the functional virion spike and, consequently, are not neutralization targets. Some of these differences between soluble gp130 glycoproteins and membrane-associated, native envelope glycoprotein trimers may be due to the presence of the glycosylated, C-terminal portion of the gp41 ectodomain or the viral membrane in the latter. Alternatively, the soluble gp130 glycoproteins may be trapped in a conformation different from that normally assumed by the envelope glycoproteins on the virion spike. Future studies of these and other issues will be expedited by the availability of stable, tractable forms of HIV-1 envelope glycoprotein trimers.
ACKNOWLEDGMENTS
We thank Susan Zolla-Pazner, James Robinson, and Michael Page for antibodies, Nicholas F. Pileggi for the synthesis of the PK-C299 peptide, Bernard Moss for helpful discussions, and Sheri Farnum and Yvette McLaughlin for manuscript preparation.
This work was supported by NIH grants AI24755, AI31783, and AI39420 and by a Center for AIDS Research grant (AI28691). We also acknowledge the support of the G. Harold and Leila Mathers Foundation, the Friends 10, Douglas and Judith Krupp, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Alan J S, Coligan J E, Barin F, Sodroski J, Rosen C A, Haseltine W A, Lee T-H, Essex M. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–1094. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Berman P W, Gray A M, Wrin T, Vennari J C, Eastman D J, Nakamura G R, Francis D P, Gorse G, Schwartz D H. Genetic and immunologic characterization of viruses infecting MN-rgp120-vaccinated volunteers. J Infect Dis. 1997;176:384–397. doi: 10.1086/514055. [DOI] [PubMed] [Google Scholar]
- 4.Broder C C, Earl P L, Long D, Abedon S T, Moss B, Doms R W. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc Natl Acad Sci USA. 1994;91:11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl. A):S87–S98. [PubMed] [Google Scholar]
- 7.Burton D R, Moore J P. Why do we not have an HIV vaccine and how can we make one? Nat Med. 1998;4(Suppl. 5):495–498. doi: 10.1038/nm0598supp-495. [DOI] [PubMed] [Google Scholar]
- 8.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 (HIV-1) gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 10.Chan D C, Chutkowski C T, Kim P S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 12.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Lee K H, Steinhauer D A, Stevens D J, Skehel J J, Wiley D C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 14.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 15.Choe H, Martin K A, Farzan M, Sodroski J, Gerard N P, Gerard C. Structural interactions between chemokine receptors, gp120 Env and CD4. Semin Immunol. 1998;10:249–257. doi: 10.1006/smim.1998.0127. [DOI] [PubMed] [Google Scholar]
- 16.Connor R I, Korber B T, Graham B S, Hahn B H, Ho D D, Walker R D, Neumann A U, Vermund S H, Mestecky J, Jackson S, Fenamore E, Cao Y, Gao F, Kalams S, Kunstman K J, McDonald D, McWilliams N, Trkola A, Moore J P, Wolinsky S M. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Earl P L, Doms R W, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earl P L, Moss B, Doms R W. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earl P L, Koenig S, Moss B. Biological and immunological properties of the human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earl P L, Moss B. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res Hum Retrovir. 1993;9:589–594. doi: 10.1089/aid.1993.9.589. [DOI] [PubMed] [Google Scholar]
- 25.Earl P L, Broder C, Long D, Lee S, Peterson J, Chakrabarti S, Doms R, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol. 1998;72:7620–7625. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 28.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 30.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W A, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 34.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L, Choe H, Sodroski J. Adaptation of a CCR5-using, primary HIV-1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korber B, Foley B, Kuiken C, Pillai S, Sodroski J. Numbering positions in HIV relative to HXB2. Human Retroviruses and AIDS. Los Alamos, N. Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 36.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 37.Kwong P D, Wyatt R, Robinson J, Sweet R, Sodroski J, Hendrickson W. Structure of an HIV-1 gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola J R, Snyder S W, Weislow O S, Belay S M, Belshe R B, Schwartz D H, Clements M L, Dolin R, Graham B S, Gorse G J, Keefer M C, McElrath M J, Walker M C, Wagner K F, McNeil J G, McCutchan F E, Burke D S. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 39.McDougal J S, Nicholson J K, Cross G D, Cort S P, Kennedy M S, Mawle A C. Binding of the human retrovirus HTLV-III/LAV/ARV/HIV to the CD4 (T4) molecule: conformation dependence, epitope mapping, antibody inhibition, and potential for idiotypic mimicry. J Immunol. 1986;137:2937–2944. [PubMed] [Google Scholar]
- 40.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirzabekov T, Bannert N, Farzan M, Hofmann W, Kolchinsky P, Wu L, Wyatt R, Sodroski J. Enhanced expression, native purification and characterization of CCR5, a principal HIV-1 coreceptor. J Biol Chem. 1999;274:28745–28750. doi: 10.1074/jbc.274.40.28745. [DOI] [PubMed] [Google Scholar]
- 42.Montefiori D C, Evans T G. Toward an HIV type 1 vaccine that generates potent, broadly cross-reactive neutralizing antibodies. AIDS Res Hum Retrovir. 1999;15:689–698. doi: 10.1089/088922299310773. [DOI] [PubMed] [Google Scholar]
- 43.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore J P, Sattentau Q, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oprian D D, Molday R S, Kaufman R J, Khorana G. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owens R J, Compans R W. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology. 1990;179:827–833. doi: 10.1016/0042-6822(90)90151-g. [DOI] [PubMed] [Google Scholar]
- 48.Parren P W, Burton D R, Sattentau Q J. HIV-1 antibody—debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 49.Parren P W, Mondor I, Naniche D, Ditzel H J, Klasse P J, Burton D R, Sattentau Q J. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 51.Richardson T M, Jr, Stryjewski B L, Broder C C, Hoxie J A, Mascola J R, Earl P L, Doms R W. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J Virol. 1996;70:753–762. doi: 10.1128/jvi.70.2.753-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robey W G, Safai B, Oroszlan S, Arthur L Q, Gonda M A, Gallo R C, Fischinger P J. Characterization of envelope and core gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–595. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- 53.Tan K, Lee J-H, Wang J-H, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veronese F D, DeVico A L, Copeland T D, Oroszlan S, Gallo R S, Sarngadharan M G. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 55.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 56.Weissenhorn W, Calder L J, Wharton S A, Skehel J J, Wiley D C. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc Natl Acad Sci USA. 1998;95:6032–6036. doi: 10.1073/pnas.95.11.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 58.Willey R L, Martin M A, Peden K W. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 61.Zolla-Pazner S, O'Leary J, Burda S, Gorny M K, Kim M, Mascola J, McCutchan F. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]