Abstract
Experiments were performed to determine the function of a 28-nucleotide untranslated sequence lying between the envelope gene and the polypurine tract (PPT) sequence in the Moloney murine leukemia virus (Mo-MuLV) genome. A mutant virus carrying a deletion of this sequence (Mo-MuLVΔ28) replicated more slowly than wild-type (wt) virus and reverted by recombination with endogenous sequences during growth in NIH 3T3 cells. We show that this deletion did not affect the level of viral protein expression or genomic RNA packaging. Mo-MuLVΔ28 served as a helper virus as efficiently as the wt virus; in contrast, a retroviral vector harboring this mutation exhibited reduced transduction efficiency, indicating that the mutation acts not in trans but in cis. Analysis of acutely infected cells revealed that reduced levels of viral DNA were generated by reverse transcription of the Mo-MuLVΔ28 RNA as compared to the wt RNA. Analysis of DNA circle junctions revealed that plus-strand DNA of Mo-MuLVΔ28 but not wt virus often retained the PPT and additional upstream sequences. These structures suggest that aberrant 5′ ends of plus-strand DNA were generated by a failure to remove the PPT RNA primer and/or by mispriming at sites upstream of the PPT. These data demonstrate that the major role of the sequences immediately upstream of the PPT is specifying efficient and accurate plus-strand DNA synthesis.
Retroviruses convert their single-stranded genomic RNA into a double-stranded DNA molecule in a complex reaction mediated by the viral reverse transcriptase (RT) (for a general review, see reference 41). The synthesis of each DNA strand is initiated by a specific RNA primer: the minus strand is initiated by a primer tRNA taken from the host, while the plus strand is initiated by a polypurine oligonucleotide formed by the RNase H activity of RT acting on the RNA genome. The site of priming of both DNA strands must be accurate because these positions determine the 5′ ends of the completed double-stranded viral DNA; these termini, in turn, are recognized by the viral integrase and serve as its substrate in the formation of the integrated provirus. Thus, mispriming would lead to incorrect sequences at the termini of the viral DNA, resulting in a block to DNA integration and further virus replication.
The site of initiation of the plus-strand DNA, the polypurine tract (PPT), lies near the 3′ end of the genomic RNA. The PPT elements of different retroviruses and retroelements exhibit considerable sequence variation but always include a stretch of 10 to 20 purines, often flanked on the 5′ side by a T-rich block (20, 32, 33). As minus-strand DNA synthesis proceeds, the RNA genome enters into an RNA:DNA hybrid, and so the RNA becomes susceptible to the RNase H activity of RT. The PPT is relatively resistant to digestion, persisting as an oligonucleotide that remains bound to the minus-strand DNA, and is extended by RT to form plus-strand DNA. Elongation first copies the U3, R, and U5 sequences of the minus strand to form an intermediate called plus-strand strong-stop DNA, which is subsequently translocated to the 3′ end of the minus strand and extended to form the complete plus strand. The 3′-proximal nucleotide of the PPT (nt −1) thus determines the 5′ end of the 5′ long terminal repeat (LTR) (nt +1) in the completed double-stranded viral DNA. Mutations affecting the sequences of the PPT can change the cleavage site that determines the site of priming; for the Moloney murine leukemia virus (Mo-MuLV) PPT, residues −1, −2, −4, and −7 are important in determining the cleavage position (32).
A number of other functions are often contained in nearby sequences in the 3′ untranslated region (3′ UTR) of retroviral RNAs. For example, the genomes of the Mason-Pfizer monkey virus (4) and simian retroviruses type 1 and 2 (43) each contain a constitutive transport element located near the 3′ end of the RNA that is important for the export of the unspliced genomic RNA from the nucleus to the cytoplasm. This element, probably forming a highly stable RNA stem-loop, is recognized by a complex of cellular proteins that shuttle in and out of the nucleus and mediate export of the RNA. Similar elements may exist in other simple retroviruses, but none have yet been identified in the avian or mammalian viruses. In another example, the avian leukosis virus genomic RNA contains an element near the 3′ end, termed dr1, that plays a small role in nuclear RNA export but is most important for the packaging of the RNA into the virion particle. Mutations in the dr1 sequence impair virus replication and reduce RNA packaging about 10-fold (2). Similar elements have not yet been identified in the 3′ UTR of other retroviruses.
To probe the functions of the 3′ UTR of the Mo-MuLV RNA, we introduced a deletion into the complete proviral DNA that removes all the nucleotides between the translational stop codon of the envelope gene and the PPT. Analysis of the mutant virus revealed no defects in RNA transport or processing and no defects in packaging of the RNA into the genome. The mutant showed a significant reduction in viral DNA synthesis and formed abnormal circular DNAs. The structure of these molecules suggests that the mutant generates plus-strand DNAs with aberrant 5′ termini. These results extend recent findings that mutations in the region immediately upstream of the PPT in simian immunodeficiency virus (20), Mo-MuLV (33), and the pararetrovirus cauliflower mosaic virus (26) cause a reduction in viral replication and defects late in reverse transcription.
MATERIALS AND METHODS
Plasmid construction.
The pNCA plasmid contains an infectious molecular clone of Mo-MuLV (7), and pNCS is the same but contains a simian virus 40 origin of replication in the plasmid backbone (11). Overlapping PCR (16) was used to delete the 28-bp sequence immediately upstream from the PPT in a ClaI-NheI fragment containing the 3′ end of the envelope gene, the PPT, and the 5′ edge of the LTR. The mutated ClaI-NheI fragment was sequenced to verify the presence of the deletion and used to replace the corresponding sequence of pNCS. The resulting plasmid, pNCSΔ28, is identical to pNCS except for the 28-bp deletion. The pNCS and pNCSΔ28 plasmids carry an additional copy of the PPT and its upstream sequences outside the provirus. To prevent the possibility of these sequences recombining with the viral genome, the provirus was excised from the vector by digestion with NheI, purified by agarose gel electrophoresis, and ligated prior to the transfection. This procedure resulted in the formation of a circular genomic viral DNA containing a single LTR.
Mutant retroviral vectors were created by replacing the ClaI-NheI fragment of pBabePuro (24) with the corresponding wild-type (wt) or mutant fragment, generating pBabePuro(wt) and pBabePuro(Δ28), respectively. The retroviral vector encoding luciferase (pSRαLLuc) has been described (1).
Cell cultures.
Amphotropic-Phoenix (A-Phoenix) (15), AmpliGPE (39) helper cell lines, and COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing penicillin-streptomycin with 10% fetal calf serum (DMEM-FCS). NIH 3T3 and Rat2-2 cells (12) were grown in DMEM with 10% calf serum (DMEM-CS). All cells were grown at 37°C and 5% CO2.
Transformation of COS-7 cells.
About 70% confluent COS-7 cells in 90-mm plates were washed three times with phosphate-buffered salt (PBS) solution and incubated at 37°C with 2 ml of PBS containing 10 μg of plasmid DNA and 200 μg of DEAE-dextran (Pharmacia). After 30 min, 8 ml of DMEM-FCS-chloroquine cocktail was added to make a final concentration of 100 μM chloroquine. After 2.5 h at 37°C, the medium was aspirated, and the cells were incubated with DMEM containing 10% dimethyl sulfoxide (DMSO) for 2.5 min. The DMSO solution was aspirated, and the cells were incubated in DMEM-FCS for 2 days, after which the expression of the transfected plasmid was assayed.
Transformation of helper cell lines.
AmpliGPE helper cells (39) were transfected with plasmid DNAs by using Lipofectamine (8). Cells stably expressing the pBabePuro(wt) or pBabePuro(Δ28) vector were isolated after selection in medium containing 5 μg of puromycin per ml. A few hundred puromycin-resistant colonies were pooled after transformation with each vector. Amphotropic-Phoenix helper cells (15) were transiently transfected by the calcium phosphate method (29).
Infectivity assays.
To test helper function, COS-7 cells were transfected with 5 μg of plasmid expressing a luciferase retroviral vector and 5 μg of plasmid expressing either Mo-MuLV or Mo-MuLVΔ28. Two days later the culture supernatants were diluted as indicated, filtered through 0.45-μm filters after addition of HEPES (pH 7.4) (50 mM) and Polybrene (8 μg/ml), and used to infect Rat2-2 cells. Luciferase specific activity was determined in extracts of transfected cells (see below). To test effects on transduction in cis, A-Phoenix or AmpliGPE helper cells were transiently transfected with 5 μg of plasmid DNA expressing retroviral vector pBabePuro(wt) or pBabePuro(Δ28). A control transfection was performed without plasmid DNA (mock). Culture supernatants were diluted 1:10, filtered through 0.45-μm filters after addition of HEPES (pH 7.4) (50 mM) and Polybrene (8 μg/ml), and used to infect Rat2-2 cells. To assay the infectivity of these preparations, Rat2-2 cells were plated (2 × 105 cells per 60-mm dish) and 1 day later were infected with 2 ml of diluted viral preparations for 2 h at 37°C. Lysates of infected cells were prepared 2 days after infection and analyzed for specific luciferase activity with a luminometer (Lumat LB 9501; Berthold) and a luciferase assay system (Promega) according to the manufacturer's protocol. Total protein concentration in cell lysates was determined with a protein assay kit (Bio-Rad) according to the manufacturer's protocol. Cells infected with pBabePuro-(wt) and -(Δ28) vectors were split into medium containing 5 μg of puromycin per ml 2 days after infection and grown for a further 8 days. Drug-resistant colonies were counted after fixation, followed by Giemsa staining, and titers of the virus were calculated.
Kinetics of virus spread in NIH 3T3 and Rat2-2 cells.
Cells were plated the day before transfection at a density of 2 × 105 cells per 60-mm dish. The cells were washed three times with PBS containing 0.5 mM MgCl2 and 0.9 mM CaCl2 (PBS+) and incubated with 0.4 ml of PBS+ containing 0.5 μg of DEAE-dextran (Pharmacia) per ml and 200 ng of circularized viral DNA (see above) for 30 min at 37°C. The cells were washed once with PBS+ before addition of culture medium. The cells were split 1:10 every 3 to 4 days, and aliquots of the supernatants were collected daily and frozen at −20°C until they were assayed for RT activity.
RT assay.
Both exogenous and endogenous reactions were performed as described before (40).
Hirt extraction of infected cells.
Virus-containing supernatants (5 ml) containing Polybrene (8 mg/ml) were filtered through a 0.45-μm filter and used to infect Rat2-2 or NIH 3T3 cells at about 70% confluence in 100-mm dishes. After 3 h, another cycle of infection was performed with 5 ml of fresh virus. Hirt extracts were prepared (17) 18 h after infection, and the DNA from each dish was resuspended in 40 μl of 10 mM Tris-HCl–1 mM EDTA (pH 8) (TE).
Southern blot analysis.
Viral DNA from acutely infected cells was analyzed by Southern blot with a vacuum blotter (model 785; Bio-Rad) following the manufacturer's protocol. A radiolabeled viral DNA probe was prepared from pNCA plasmid randomly labeled with an Oligolabeling kit (Pharmacia Biotech) according to the manufacturer's protocol. The intensity of the signal was quantified with a phosphoimager (model 445 S1; Molecular Dynamics).
RNase protection assay.
RNA was extracted from cells and purified virions with RNAzolB (Tel-Test) according to the manufacturer's protocol, and RNA levels were measured with an RNase protection kit (Ambion). A 572-base XbaI-EagI fragment from Mo-MuLV, spanning the splice donor site (3), was cloned in pBluescript plasmid SK and used as a template for transcription of the riboprobe. The plasmid DNA was linearized with HindIII, and the ribopobe was transcribed with T3 RNA polymerase using the Ambion MAXIscript in vitro transcription kit according to the manufacturer's protocol.
LM-PCR.
For ligation-mediated PCR (LM-PCR), subconfluent Rat2-2 cells (10-cm dish) were infected with undiluted virus, and low-molecular-weight DNA was extracted after 18 h. The DNA was recovered and resuspended in TE (40 μl). One tenth of the sample (4 μl) was mixed with 6 μl of 3 μM dephosphorylated oligonucleotide (anchor oligonucleotide), with the arbitrary DNA sequence 5′GGAACTCAATGCACGCGT3′, and 7 μl of water, and the mixture was boiled for 3 min and cooled on ice for another 3 min. Then 2 μl of 10× T4 RNA ligase buffer and 1 μl of T4 RNA ligase (New England BioLabs) were added to make final concentrations of 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, and 20 U of enzyme. The ligation was performed at 16°C overnight, after which 6 μl from the ligation reaction mixture was amplified by PCR. The PCR conditions were as described for the circle junction analysis (see below) except that 2.5 U of native Taq DNA polymerase per reaction was used and the primer pairs were either the anchor oligonucleotide (forward primer) and oligonucleotide 4091 (reverse primer) to amplify linear ends or oligonucleotides 4091 and 5784 to amplify circle junctions.
Circle junction analysis.
Circle junction analysis was done by a modification of a procedure described previously (36). DNA from a Hirt extract was amplified by PCR with oligonucleotides 4091 (5′CTCTTTTATTGAGCTCGGG3′; nucleotides 8244 to 8226 in the Mo-MuLV map [35]) and 5784 (5′AGTCCTCCGATTGACTGAG3′; nucleotides 6 to 24 in the Mo-MuLV map). PCR was performed under the following conditions: 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 15 mM MgCl2, 0.01% (wt/vol) gelatin, 200 mM each of the four deoxynucleoside triphosphates, 0.3 mM each of the above primers, 1.25 U of native Taq DNA polymerase (Perkin-Elmer), and viral DNA (1/10 of the Hirt extract from a single dish of infected cells). The cycling conditions were 2 min at 94°C, followed by 30 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s, and one cycle of 72°C for 7 min in a GeneAmp PCR system 9700 (Perkin-Elmer). The PCR products were cloned directly from the PCR solution with a TOPO TA cloning kit (Invitrogen; K4500-01/40) according to the manufacturer's protocol, and clones were subjected to DNA sequence analysis.
Analysis of Mo-MuLVΔ28 and revertant viruses for the presence of the Δ28 deletion.
Viral DNAs extracted from acutely infected cells were amplified by PCR with the primers 5′CTCCTAATGATTTTGCTCTTCGGACCC3′ and 5′TTCCATCTGTTCCTGACC3′, located upstream and downstream of the ClaI and the NheI sites, respectively. The PCR products, with expected lengths of 298 bases for the wt virus or 270 bases for the Mo-MuLVΔ28, were cloned and sequenced. The PCR conditions and cloning of the PCR products were as described for the circle junction analysis.
RESULTS
A 28-bp deletion in the 3′ UTR of Mo-MuLV impairs replication.
The genomic RNA of Mo-MuLV contains a 28-nucleotide untranslated sequence between the 3′ end of the envelope gene and the start of the PPT sequence at the edge of the 3′ LTR. This sequence contains a near-perfect inverted repeat and thus could form a stem-loop hairpin structure in the RNA. To evaluate the significance of this sequence, we deleted the 28 bases from a wt clone of Mo-MuLV, generating the Mo-MuLVΔ28 mutant virus (Fig. 1A). Rat2-2 cells were transfected with plasmid DNAs containing either the wt or the Mo-MuLVΔ28 virus to transiently initiate virus expression, and the spread of virus in the cultures was monitored by measuring the RT activity present in the cell culture supernatants on subsequent days. The Mo-MuLVΔ28 virus consistently showed a 6-day delay in the appearance of detectable RT activity compared with the wt virus (Fig. 1B). To test the rate of viral spread in a different setting, virus stocks were first generated by transformation of COS-7 cells with the same plasmids. The virus preparations were normalized to equal RT activities and used to infect NIH 3T3 cells, and the rate of virus replication in these cultures was determined by RT assays. A similar delay in appearance of virus was observed (data not shown).
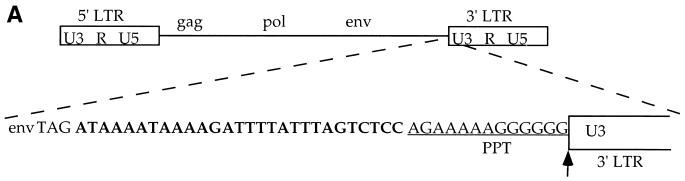
FIG. 1.
Structure and replication of Mo-MuLV and Mo-MuLVΔ28. (A) Schematic representation of the organization of the Mo-MuLV genomic DNA (upper drawing), and an expanded view of the PPT with the upstream untranslated sequence (lower drawing). The U3, R, and U5 regions in the LTRs are shown (open boxes). The PPT sequence is underlined, and the sequence of the 28 bases deleted in Mo-MuLVΔ28 is shown in bold letters. This sequence is located 3′ to the envelope stop codon (envTAG) and 5′ to the PPT sequence. The vertical arrow indicates the RNase H cleavage site, necessary for the formation of the correct 5′ LTR edge during reverse transcription. (B) Viral spread in Rat2-2 cells. Rat 2-2 cells were transiently transfected with DNA expressing the indicated viruses, and the cultures were passaged at 1:20 dilution every 3 to 4 days. The decrease seen at day 15 is due to a transfer the previous day. An exogenous RT assay was performed on supernatants collected on the indicated days after transfection. A control transfection was performed without adding plasmid DNA (mock).
The late appearance of replicating virus after the delay could reflect the true phenotype of the mutant or could result from the generation of revertant viruses, which often arise by recombination with endogenous virus-like sequences (5, 23, 34). To distinguish between these two possibilities, the supernatant from the Mo-MuLVΔ28-transfected Rat2-2 cultures (day 20 after transfection) was used to infect naive Rat2-2 cells, and 18 h later the unintegrated viral DNA was isolated from the infected cells (17). The region of the viral DNA that harbors the deletion was amplified by PCR, cloned, sequenced, and found to retain the Δ28 deletion (data not shown). In two such clones sequenced, no nucleotide changes from the parental mutant virus were detected in the vicinity of the deletion. These data suggest that Mo-MuLVΔ28 can replicate but at a slower rate than the wt virus.
A different result occurred when this experiment was repeated using NIH 3T3 cells. Virus arising in these cells after a delay were used in an acute infection, and the unintegrated DNAs were cloned and sequenced as before. These viral DNAs had reverted to the wt sequence. Of two clones sequenced, both had restored the entire 28-bp wt sequence; whereas one contained no additional changes, the other one contained two A-to-G substitutions in the adjacent sequences of the 3′ LTR. The reappearance of the wt sequence in these revertants is most likely due to recombination with endogenous retrovirus-like elements very similar to Mo-MuLV present in the mouse genome. To rule out the possibility that these reverted viruses originated from a contamination of the transfected DNA, the experiments were repeated with a single transfection mixture used to initiate viral spread in both Rat2-2 cells and NIH 3T3 cells. As before, wt virus was generated only in NIH 3T3 cells. These experiments indicate that the Δ28 virus can replicate slowly in Rat cells without reversion but that it tends to revert in NIH 3T3 cells. The reversion is probably facilitated by the high copy number of provirus-like sequences in the mouse genome.
The 28-bp deletion acts in cis and not in trans.
The 28-bp deletion could act by impairing viral gene expression, through effects on either mRNA stability or mRNA translation. To address this possibility, we tested the ability of Mo-MuLVΔ28 and the wt virus to provide helper function in trans for the transfer of a retroviral vector carrying the luciferase reporter gene. COS-7 cells were transfected with vector plus helper DNAs, virus was collected, and the efficiency of transfer was assessed by measuring luciferase levels after infection of Rat2-2 cells. Both viruses pseudotyped the retroviral vector with similar efficiencies (Table 1). Similar results were obtained with a green fluorescence protein-containing vector (data not shown). These results demonstrate that the mutation had no effect on expression of viral proteins required in trans. In addition, pools of 293 cells stably expressing either the wt or the mutated virus released similar levels of RT activity into the culture medium (data not shown). These data suggest that the 28-base deletion does not affect the level of production of functional viral proteins.
TABLE 1.
Mo-MuLVΔ28 is as efficient a helper virus as Mo-MuLV
| Expt and virus | Luciferase activity (106 relative light units/μg of total protein)
|
Normalized infectivitya | |
|---|---|---|---|
| Transfected cells | Infected cells | ||
| 1 | |||
| Mo-MuLV | 214 | 31 (1:2)b | 0.14 |
| 11 (1:10) | 0.05 | ||
| Mo-MuLVΔ28 | 231 | 42 (1:2) | 0.18 |
| 12 (1:10) | 0.05 | ||
| 2 | |||
| Mo-MuLV | 129 | 27 (1:2) | 0.21 |
| 13 (1:10) | 0.10 | ||
| Mo-MuLVΔ28 | 159 | 32 (1:2) | 0.20 |
| 8 (1:10) | 0.05 | ||
Specific luciferase activity in infected cells divided by specific luciferase activity in transfected cells.
Dilution factor of the supernatants used for infection.
The alternative possibility is that the 28-bp deletion might affect the replication of the viral genome in cis. To directly test this possibility, the wt and mutant sequences were introduced into a retroviral vector encoding the puromycin resistance gene to generate the pBabePuro(wt) and pBabePuro(Δ28) vector, respectively. These vector DNAs were expressed in helper cell lines that provide viral proteins in trans, and the titer of the transducing virus was measured after infection of Rat2-2 cells by counting the resulting puromycin-resistant colonies. Transient expression of these vectors in two different cell lines (A-Phoenix and AmpliGPE) revealed that pBabePuro(Δ28) consistently produced approximately 10-fold-lower titers than pBabePuro(wt) (Table 2). To rule out the possibility that the reduction in the titer was due to a difference in the efficiency of transient transfection, the experiment was repeated by generating pools of several hundred AmpliGPE colonies that stably express these vectors. The virus produced by the pBabePuro(Δ28) pools showed a similar reduction in titer compared with the wt pools (Table 2). These experiments demonstrate that the Δ28 mutation acts in cis to impair virus replication.
TABLE 2.
Δ28 mutation reduces retroviral vector titer
| Vector | Titer (CFU)
|
||
|---|---|---|---|
| A-Phoenix (transient) | AmpliGPE (transient) | AmpliGPE (stable) | |
| pBabePuro(wt) | 2.9 × 102 | 1.5 × 104 | 1 × 105 |
| pBabePuro(Δ28) | 32 | 1.4 × 103 | 7 × 103 |
| None | 0 | 0 | 0 |
The 28-base deletion does not affect RNA packaging.
One possible explanation for the slower replication of Mo-MuLVΔ28 is a reduced level of packaged viral RNA. Indeed, mutations in the 3′ UTR between the 3′ end of env and the LTR of the avian sarcoma/leukosis virus have been shown to cause a reduction in genomic RNA packaging (2). To examine the levels of viral RNA packaged into virions, 293T cells were transfected with plasmids expressing either Mo-MuLV or Mo-MuLVΔ28, and virus particles were harvested from the culture medium. RNAs were isolated from the producer cells and from virus particles and normalized by RT assays, and the levels of viral RNA were determined by RNase protection assays with a riboprobe that detects both unspliced and spliced viral RNA forms (Fig. 2A). The wt and mutant viruses exhibited similar levels of spliced and unspliced viral RNAs both in the expressing cells and in the purified virions (Fig. 2B). Similar results were obtained in stably transfected virus-producing Rat2-2 cells. Thus, MuLVΔ28 packages its genomic RNA as efficiently as wt virus.
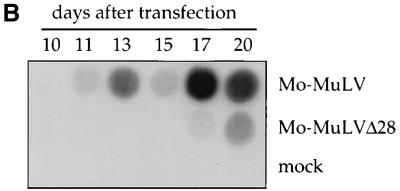
FIG. 2.
RNase protection assay to monitor viral RNA levels. (A) Schematic representation of the riboprobe and the regions protected from RNase digestion. The upper line shows the organization of the 5′ portion of the Mo-MuLV genome. The 5′ LTR (open box), the transcription start site (arrow), the splice donor (SD) site, and the flanking restriction sites that were used to clone the riboprobe template (XbaI and EagI) are indicated. Lower lines show the full-length riboprobe with flanking plasmid sequences (wavy lines) and the two fragments protected by the unspliced and spliced viral RNA. The lengths of the different products are indicated. (B) Cellular RNA was extracted from 293T cells transiently producing either Mo-MuLV or Mo-MuLVΔ28. Virions from culture supernatants were purified through 25 to 45% sucrose step gradients, followed by an additional purification step through a 25% sucrose cushion. RNA was extracted from virions, and the amounts were normalized by RT activity. Total cellular RNA and virion RNAs were used for the RNase protection assay. The positions of the full-length riboprobe and its protected fragments (unspliced and spliced) are indicated at the right (the prominent band above the spliced RNA product is probably derived by cleavage of the unspliced RNA). The viruses are indicated above the panels, and the source of the RNA is shown below the panels. Controls included riboprobe mixed only with yeast RNA with and without RNase (digested and undigested, respectively) and 293T cells transfected without plasmid DNA (mock). As a quantitation control, reactions with 1:5 and 1:10 dilutions of the wt viral RNA sample (Mo-MuLV 1:5 and 1:10) were also performed.
Mo-MuLVΔ28 virions carry out the early stages of reverse transcription as efficiently as wt virions.
To examine the ability of Mo-MuLVΔ28 to carry out reverse transcription, we used the endogenous RT reaction assay in which the encapsidated genomic RNA serves as the template for the virion-associated RT. Virions of Mo-MuLV and Mo-MuLVΔ28 were purified from culture supernatants of transiently transfected 293T cells, and the levels were normalized by quantitative exogenous RT assay. The virions were then incubated with NP-40 and radiolabeled nucleotides, and the formation of minus-strand strong-stop DNA (−sssDNA) was measured by electrophoresis and autoradiography. Similar levels of −sssDNA were formed by both the wt and mutant viruses (Fig. 3A). To test for the formation of longer DNA products, reaction mixtures were incubated for extended times, and the products were analyzed by electrophoresis on alkaline agarose gels followed by autoradiography. The mutant and control virions produced a similar heterogeneous distribution of DNAs, appearing as a smear in the gels (Fig. 3B). These data indicate that the viral RNA is packaged in a functional form by the mutant and that the early stages of reverse transcription are carried out with normal efficiency. Similar results were obtained with purified virions produced from COS-7 cells and with levels of virions normalized by Western blot analysis with antibodies against RT (data not shown).
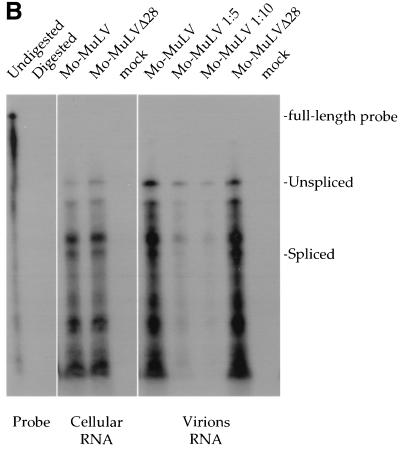
FIG. 3.
Endogenous RT assay. Virions were purified as described in the legend to Fig. 2B, and the endogenous RT assay was performed in the presence of 0.05% NP-40. (A) To detect the formation of −sssDNA, assays were performed for 15 min, and the products were separated on an 8% acrylamide–urea gel. (B) To monitor synthesis of longer DNA products, reactions were performed for 9 h, and the products were separated on a 1% alkaline agarose gel. The positions of migration of the −sssDNA, extended products, and DNA markers are shown on the left. As a control, 293T cells were transfected without plasmid DNA (mock).
Δ28 deletion reduces the yield of viral full-length genomic DNA.
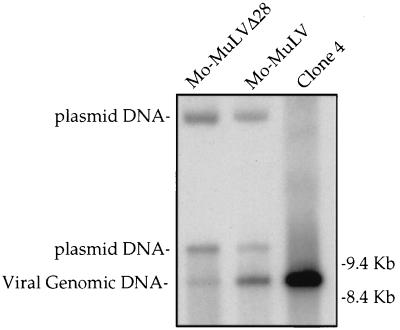
We next examined the amount of the full viral DNA genome generated by reverse transcription in vivo. Mo-MuLV and Mo-MuLVΔ28 were transiently expressed in COS-7 cells, and supernatants with equal amounts of RT activity were used to infect naive Rat2-2 cells. Low-molecular-weight DNA was extracted from the Rat2-2 cells 18 h after infection by the Hirt extraction method, and the viral DNAs were analyzed by Southern blotting with the complete viral genome as a probe. This analysis revealed a reduction in the linear genome levels of Mo-MuLVΔ28 compared with Mo-MuLV (Fig. 4). This reduction was about sixfold, as quantitated with a phosphoimager. Thus, the Δ28 deletion appears to reduce the yield of full-length linear viral genomic DNA, indicating that some step in reverse transcription is inefficient.
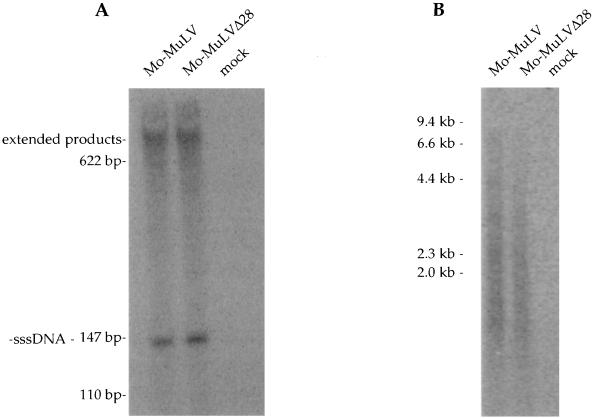
FIG. 4.
Southern blot analysis of unintegrated viral DNA synthesized after acute infection. COS-7 cells were transfected with plasmids expressing either Mo-MuLV or Mo-MuLVΔ28, and 2 days later culture supernatants were collected, normalized to contain equal amounts of RT activity, and used to infect Rat2-2 cells. Parallel cultures were also infected with wt virus from chronically infected NIH 3T3 cells (clone 4) to distinguish the reverse-transcribed linear DNA product from the transfected DNA plasmid. Low-molecular-weight DNA was isolated 18 h after infection with the indicated viruses and analyzed by Southern blot. The positions of migration of the linear viral genomic DNA and two forms of the contaminating plasmid DNA are indicated on the left, and those of the DNA markers are shown on the right.
Mo-MuLVΔ28 generates aberrant 5′ plus-strand DNA ends.
The Δ28 deletion is located immediately adjacent to the PPT and might affect the efficiency or the position of plus-strand DNA priming. The site of initiation of plus-strand DNA synthesis near the PPT ultimately determines the 5′ end of the linear double-stranded viral DNA. We first attempted to amplify and clone the 5′ ends of the plus-strand DNAs by LM-PCR. Rat2-2 cells were infected with equal amounts of either wt or mutated virus, and after 18 h, low-molecular-weight DNA was extracted and denatured. The 5′ ends of the DNA were ligated to an anchor oligonucleotide using T4 RNA ligase, and the ligation products were amplified by PCR using the anchor oligonucleotide as a forward primer and an oligonucleotide derived from the U3 region as a reverse primer. The expected LM-PCR product could only be obtained from Hirt extracts of wt-infected cells and not from Mo-MuLVΔ28-infected cells (Fig. 5). The failure to obtain LM-PCR products from Mo-MuLVΔ28-infected cells may be due either to a lower yield of linear plus-strand DNA by Mo-MuLVΔ28 or to aberrant structures at the 5′ ends of plus-strand DNA that cannot be amplified (see Discussion).
FIG. 5.
Gel electrophoresis of amplified linear 5′ viral DNA ends and LTR-LTR circle junctions. Mo-MuLV and Mo-MuLVΔ28 produced by chronically infected Rat2-2 cells were used to infect naive Rat2-2 cells. Low-molecular-weight DNA was extracted by the Hirt method 18 h postinfection, and aliquots were used to amplify the 5′ viral DNA ends by LM-PCR. Another aliquot of the same extracts was used to amplify LTR-LTR circle junctions. The PCR products were separated on a 2% agarose gel containing ethidium bromide. The expected sizes for the amplified 5′ linear viral DNA end and for the LTR-LTR circle junctions are 447 and 567 bp, respectively. Mock, no virus added; M, MspI-digested pBR322 plasmid DNA size markers.
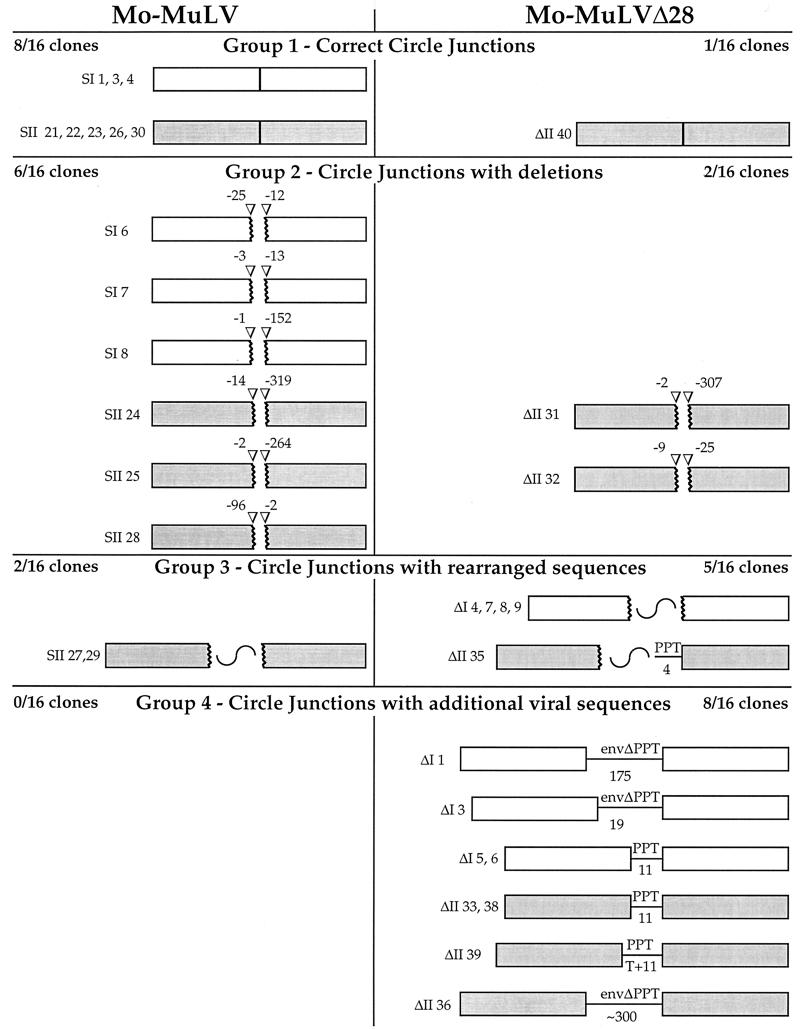
Since no amplified DNA was detected from the mutant-infected cells, a second approach was used to identify the 5′ termini. This method is based on the fact that the termini of a portion of the viral linear DNAs are joined in the nucleus to form a two-LTR circular DNA. Thus, the sequences originally present at the termini can be examined by amplifying and sequencing the unique LTR-LTR junction region of these circular DNAs. In contrast to the LM-PCR approach, circle junction amplification resulted in readily detectable products for both viruses (Fig. 5). Importantly, the amplified LTR-LTR junctions from Mo-MuLVΔ28-infected cells migrated slightly more slowly than those of the wt, suggesting the presence of additional sequences (Fig. 5). To characterize these sequences, the LTR-LTR junctions were amplified by PCR and cloned directly into plasmids, and clones chosen at random were sequenced. This procedure was repeated in two independent experiments. In the first experiment, viruses were collected from transiently transfected COS-7 cells, and in the second experiment the viruses were obtained from chronically infected Rat2-2 cells. Figure 6 shows a schematic representation of 32 clones of circle junctions (16 of the wt and 16 of the mutated virus).
FIG. 6.
Schematic representation of LTR-LTR circle junction analysis. Boxes represent LTRs. White and gray boxes represent clones obtained from two separate experiments. Broken boxes represent LTRs with truncations. The number of bases missing from the edge of the LTRs is shown above arrowheads. Broken boxes with a wavy line represent truncated LTRs with inserts of rearranged plasmid or unidentified sequences. Straight lines represent viral sequences retained at the LTR-LTR junction. The number of base pairs in these retained sequences is indicated below the line. T+11, an untemplated T plus 11 bp.
The 16 clones of the wt Mo-MuLV could be divided into three different groups based on the sequences found at the LTR-LTR junctions. The first group contained eight clones harboring intact LTR-LTR junctions, with unaltered 5′ and 3′ ends of the U3 and U5 sequences, respectively (Fig. 6, group 1). The second group consisted of six clones with LTR-LTR junctions in which the LTRs were truncated to different extents, lacking sequences at the 5′ ends of U3 and the 3′ ends of U5 (Fig. 6, group 2). The loss of sequences at the ends in the LTR-LTR junction is commonly observed even in wt virus (18, 36). The third group contained two clones with truncated LTRs and with unidentified adjacent sequences, which probably resulted from amplification of rare recombination events (Fig. 6, group 3).
For the Δ28 mutant virus, the circle junction analysis gave a strikingly different result. Only 1 of the 16 clones (clone ΔII 40) had an intact LTR-LTR junction (Fig. 6, group 1). More importantly, another major and unique group of clones was identified in which the intact LTRs were retained and flanked additional viral sequences. This group included 8 of the 16 clones (Fig. 6, group 4). In five of these clones (ΔI 5 and 6 and ΔII 33, 38, and 39), the additional sequence consisted of 11 nucleotides derived from the PPT sequence (an additional thymidine was found at the 5′ end of this extra sequence in clone ΔII 39, and its source is not clear). The other three clones had even longer sequences that included the PPT and part of the envelope gene. These sequences varied in their length, consisting of an additional 19 bases (clone ΔI 3), 175 bases (clone ΔI 1), and about 300 bases (clone ΔI 36) (the sequencing reaction did not reach the LTR edge, and the length of the insert was estimated by restriction analysis). All three of these clones contained the expected Δ28 deletion, confirming that they indeed originated from Mo-MuLVΔ28. The other clones, like the wt, could be divided into two groups, in which the LTR-LTR junctions contained either truncated LTR ends (Fig. 6 group 2, clones ΔII 31 and 32) or truncated LTRs flanking additional sequences which originated from either the plasmid backbone, inverted viral sequences, or unknown sources (Fig. 6, group 3, clones ΔI 4, 7, 8, and 9 and ΔII 35). These rearranged clones probably represent amplification of rare recombination events.
The observations that Mo-MuLVΔ28 produces few DNAs with perfect LTR-LTR junctions (1 of 16 clones, compared with 8 of 16 clones for the wt) and that half of the mutant clones had additional sequences attached to the 5′ end of the 5′ LTR (whereas none did for the wt) strongly suggest that formation of the correct 5′ end of plus-strand DNA is impaired in this virus.
DISCUSSION
The experiments described here show that a mutation near the PPT causes slower viral replication, impairs transfer of viral genomes to new cells in cis, and causes a reduction in the level of viral genomic DNA synthesized in infected cells. The most remarkable feature of the mutant virus was a high frequency of extra sequences at the LTR-LTR junctions of circular viral DNAs. These extra sequences were of various lengths and derived from the 3′ portion of the genome, contiguous with the 5′ end of the 3′ LTR. In contrast, no structures similar to these were observed for the wt virus in this or other studies (36). While deletions and insertions at both LTR edges have been reported for nucleocapsid mutants (14), the asymmetric retention of sequences adjacent only to the 3′ LTR is unique to the Mo-MuLVΔ28 virus.
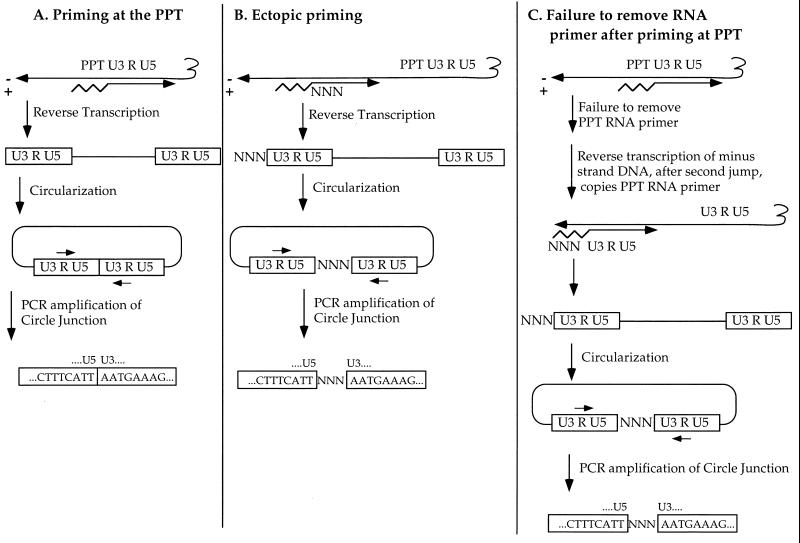
How did these additional sequences arise? Previous reports have described additional nucleotides at the LTR edges. For example, analysis of a circular DNA of an Mo-MuLV variant with a mutation at the U5-primer binding site border revealed a tRNA sequence at the LTR-LTR junction; these sequences probably resulted from a failure to remove the tRNA primer from the 5′ end of the minus-strand DNA (6). Other studies observed the presence of additional sequences that resulted from a nontemplated nucleotide addition (18, 22, 25, 27, 28, 37). However, the extra sequences found in our analysis are unique in that they are asymmetric, derived from viral sequences just upstream of the PPT. The simplest explanation is that the circles arose from linear DNAs with aberrant 5′ ends of the plus-strand DNA. We propose that these additional sequences are the result of either nonlegitimate plus-strand priming at a site upstream from the PPT, the inefficient removal of a short RNA primer, or both (Fig. 7).
FIG. 7.
Schematic model for the formation of aberrant 5′ ends of plus-strand DNA. In all panels, the tRNA primer is shown as a squiggly line, the RNA primer for plus-strand DNA is shown as a zigzag line, and minus- and plus-strand DNAs are shown as long horizontal arrows marked − and +, respectively. The sequence of the wt LTR-LTR junction is shown inside the LTRs (open boxes), and additional nucleotides are shown between these boxes (NNN). (A) Normal priming at the PPT site followed by removal of the PPT RNA primer; completion of DNA synthesis results in intact LTRs, which are joined during circularization to form a perfect inverted repeat. (B) Priming at a site upstream of the PPT. The extra plus-strand sequences (NNN) are retained in the completed linear DNA and, upon circularization, lie between the two LTRs. (C) Priming at the PPT site but failure to remove the RNA primer. The primer is copied in the final stages of minus-strand DNA synthesis, the extra sequences are retained, and upon circularization the sequences lie between the two LTRs.
In one mechanism, priming which starts 5′ to the PPT will generate plus-strand DNA with additional sequences attached to the U3 region (Fig. 7A and B). This mechanism allows for the addition of relatively short or long sequences, depending on how far from the 5′ end of U3 the priming occurs. Mutations in the PPT itself have been shown to affect the efficiency and accuracy of priming of several viruses in vitro. Point mutations in human immunodeficiency virus type 1 at positions −2 and −4 relative to the site of priming showed the importance of two guanine residues for primer production (31). The 3′-proximal nucleotides are most important, since an artificial sequence containing six guanines could be recognized and cleaved correctly. A short RNA-DNA hybrid with a sequence different from the wt was found to serve as a primer, but the site of cleavage and initiation was imprecise (30). Mutations in the PPT have also been shown to affect priming in vivo; between 9 and 29 nucleotides near the PPT of Rous sarcoma virus were required for replication (38), and point mutations in the Mo-MuLV PPT caused a delay in virus replication and reduced efficiency of plus-strand priming (33). Thus, the mutation studied here might well impair proper priming and reveal inefficient priming at upstream sites.
The other explanation for the extra sequences is a failure to remove the RNA primer by RNase H, resulting in extension of the minus-strand DNA to copy the RNA primer as a template at the final stages of minus-strand DNA synthesis (Fig. 7C). This could explain the relatively short PPT sequences found attached to U3 in some clones but is not a likely mechanism in the case of long inserts of hundreds of bases, because RNase H normally cleaves the RNA every 10 to 13 nucleotides (13). Defects in primer removal have been seen in other settings; imprecise removal of tRNA and PPT primers by RNase H has been observed for the yeast Ty1 retrotransposon (25). The idea that the Δ28 mutation interferes with the removal of the RNA primer is also supported by the experiment in which we attempted to use LM-PCR to amplify the 5′ ends of plus-strand DNA. While we were able to amplify these ends from cells infected with wt virus, we were unable to do so for the Mo-MuLVΔ28-infected cells (Fig. 5). Because T4 RNA ligase can ligate oligonucleotides to both DNA and RNA molecules, the inability to PCR-amplify the 5′ ends of Mo-MuLVΔ28 might be explained by the formation of a sequence composed of the anchor oligonucleotide plus RNA plus the LTR, which Taq polymerase would be unable to amplify.
What are the signals for accurate priming and for timely removal of the RNA primer? The RNase H domain of RT is thought to produce the primer by digesting the viral genomic RNA as it enters into an RNA-DNA duplex, leaving a purine-rich RNA fragment with a defined 3′ end. RNase H should then remove the RNA primer soon after the initiation of plus-strand DNA synthesis, so that it would not normally be used as a template in the final stages of minus-strand DNA synthesis. It is not known how the RNase H domain specifically recognizes the PPT sequence to achieve the above tasks. Previous in vitro studies demonstrated that the PPT per se is essential for the priming of the plus-strand DNA and for correct cleavage of this RNA primer from the newly synthesized DNA strand. It was suggested that RNA-DNA hybrids in general, and the purine stretch in particular, form a unique structure that is somehow recognized by RNase H (9, 10, 30). However, the region upstream of the PPT may be recognized and bound by the DNA polymerase domain of RT in order to correctly position the RNase H domain to cleave the 3′ end of PPT (19, 42).
Although experiments carried out in vitro show that the PPT sequence is the key element required for plus-strand priming, this work and two other recently published papers (20, 33) demonstrate the importance of additional upstream sequences for reverse transcription and plus-strand synthesis. Examination of simian immunodeficiency virus revealed that the sequence upstream of the PPT is rich in thymidines; this region, conserved in most retroviruses, was accordingly named the U box (20). Mutations in this sequence impaired replication, and the mutant viruses gave rise to revertants with additional alterations in the region. The most common compensatory mutation recovered in these revertants was the acquisition of thymidines immediately upstream of the PPT, indicating their importance at this position. However, another set of revertants included the addition of a stretch of adenines that extended the PPT. Similar reversion events were also observed for Mo-MuLV mutants (33). Presumably, either of these compensatory mutations can partially restore normal replication.
The 28-bp sequence that was deleted from Mo-MuLV in this work contains a stretch of adenines upstream of a stretch of thymidines. We observed that these two stretches, together with the two last bases of the envelope stop codon, have the potential to form a stem structure (except for one adenine in each of the 13-base-long stretches). However, there is no direct evidence for the existence of such an RNA stem structure, especially if the structure to be recognized by the RNase H is in the form of an RNA-DNA hybrid. Moreover, a deletion in the envelope gene that included all of the adenine stretch and part of the thymidine stretch was made in an attempt to minimize the length of Mo-MuLV-based retroviral vectors (21). This deletion did not reduce the titer of a retroviral vector, indicating that at least in this assay, the adenine stretch, and thus the presence of complementary stretches, is not important for PPT function.
Recently, Robson and Telesnitsky described substitution mutations in the region upstream from the Mo-MuLV PPT that caused a delay in replication and in some cases reduction in the use of PPT (33). These substitution mutations may have impaired replication in ways similar to the deletion mutation described here. However, the substitution mutations caused a more severe delay in replication than the deletion mutation, suggesting that subtly altered sequences in the region upstream of the PPT may be more deleterious to replication than the absence of the wt sequence. These investigators were unable to detect longer plus-strand DNAs by primer extension on linear viral DNA, but the levels of the products may have been low and below the limit of detection. Since any extended DNAs would be defective for integration, they may tend to be diverted to the pathway leading to the formation of circular species; the accumulation of these DNAs may have facilitated their detection in our assays.
The fact that some of the substitution mutations in the sequence upstream from the PPT had a relatively modest effect on PPT usage but severe effects on viral replication suggests that this region might have other functions; one possibility is that it might act as an RNA export element (33). In Mason-Pfizer monkey virus, a constitutive RNA export element located in the 3′ UTR was found to be important for the efficient export of unspliced viral RNA from the nucleus to the cytoplasm (4). Mutations in such an element should in principle reduce expression of the Gag and Pol proteins, which are encoded by the unspliced viral message. In our experiments, transient or stable expression of Mo-MuLVΔ28 did not show reduced amounts of Gag or RT compared with amounts in the wt controls, suggesting that the deleted sequence does not serve as an RNA export signal. Also, the sixfold reduction in the level of Mo-MuLVΔ28 genomic DNA is in a good agreement with the roughly 10-fold reduction in titer observed for the vector pBabePuro(Δ28). Thus, the lower efficiency and accuracy of the reverse transcription of Mo-MuLVΔ28 are probably the main cause of its slower replication.
In summary, the region upstream from the PPT of Mo-MuLV is apparently most important for reverse transcription of the viral RNA. No other replication function, including export of the viral RNA from the nucleus or packaging of the RNA genome into virion particles, was affected by the deletion of this sequence. Removal of this region results in reduced levels of viral DNA synthesis and in the formation of aberrant circular DNAs with extra viral sequences at the LTR-LTR junction. Thus, the data suggest that the major function of this short untranslated sequence adjacent to the 5′ end of the PPT is to contribute to the accurate priming of the plus-strand DNA in vivo.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R01 CA 30488 from the National Cancer Institute. E.B. is an Associate and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
We thank Alice Telesnitsky for sharing data prior to publication, Guangxia Gao and Matthew Evans for helpful discussions, and Sharon Boast and Kenia de los Santos for technical assistance.
REFERENCES
- 1.An D S, Koyanagi Y, Zhao J Q, Akkina R, Bristol G, Yamamoto N, Zack J A, Chen I S. High-efficiency transduction of human lymphoid progenitor cells and expression in differentiated T cells. J Virol. 1997;71:1397–1404. doi: 10.1128/jvi.71.2.1397-1404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aschoff J M, Foster D, Coffin J M. Point mutations in the avian sarcoma/leukosis virus 3′ untranslated region result in a packaging defect. J Virol. 1999;73:7421–7429. doi: 10.1128/jvi.73.9.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1986;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colicelli J, Goff S P. Structure of a cloned circular retroviral DNA containing a tRNA sequence between the terminal repeats. J Virol. 1986;57:674–677. doi: 10.1128/jvi.57.2.674-677.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 8.Fassati A, Takahara Y, Walsh F S, Dickson G. Production of high titre helper-free recombinant retroviral vectors by lipofection. Nucleic Acids Res. 1994;22:1117–1118. doi: 10.1093/nar/22.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedoroff O, Salazar M, Reid B R. Structure of a DNA:RNA hybrid duplex. Why RNase H does not cleave pure RNA. J Mol Biol. 1993;233:509–523. doi: 10.1006/jmbi.1993.1528. [DOI] [PubMed] [Google Scholar]
- 10.Fedoroff O Y, Ge Y, Reid B R. Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J Mol Biol. 1997;269:225–239. doi: 10.1006/jmbi.1997.1024. [DOI] [PubMed] [Google Scholar]
- 11.Gao G, Goff S P. Replication defect of Moloney murine leukemia virus with a mutant reverse transcriptase that can incorporate ribonucleotides and deoxyribonucleotides. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao G, Goff S P. Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol Biol Cell. 1999;10:1705–1717. doi: 10.1091/mbc.10.6.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerard G F. Mechanism of action of Moloney murine leukemia virus RNA-directed DNA polymerase associated RNase H (RNase H I) Biochemistry. 1981;20:256–265. doi: 10.1021/bi00505a005. [DOI] [PubMed] [Google Scholar]
- 14.Gorelick R J, Fu W, Gagliardi T D, Bosche W J, Rein A, Henderson L E, Arthur L O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 16.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 177–188. [Google Scholar]
- 17.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 18.Hong T, Drlica K, Pinter A, Murphy E. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J Virol. 1991;65:551–555. doi: 10.1128/jvi.65.1.551-555.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes S H, Hostomsky Z, Le Grice S F, Lentz K, Arnold E. What is the orientation of DNA polymerases on their templates? J Virol. 1996;70:2679–2683. doi: 10.1128/jvi.70.5.2679-2683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilyinskii P O, Desrosiers R C. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 1998;17:3766–3774. doi: 10.1093/emboj/17.13.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S H, Yu S S, Park J S, Robbins P D, An C S, Kim S. Construction of retroviral vectors with improved safety, gene expression, and versatility. J Virol. 1998;72:994–1004. doi: 10.1128/jvi.72.2.994-1004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulkosky J, Katz R A, Skalka A M. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J Acquir Immune Defic Syndr. 1990;3:852–858. [PubMed] [Google Scholar]
- 23.Martinelli S C, Goff S P. Rapid reversion of a deletion mutation in Moloney murine leukemia virus by recombination with a closely related endogenous provirus. Virology. 1990;174:135–144. doi: 10.1016/0042-6822(90)90062-v. [DOI] [PubMed] [Google Scholar]
- 24.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mules E H, Uzun O, Gabriel A. In vivo Ty1 reverse transcription can generate replication intermediates with untidy ends. J Virol. 1998;72:6490–6503. doi: 10.1128/jvi.72.8.6490-6503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noad R J, Al-Kaff N S, Turner D S, Covey S N. Analysis of polypurine tract-associated DNA plus-strand priming in vivo utilizing a plant pararetroviral vector carrying redundant ectopic priming elements. J Biol Chem. 1998;273:32568–32575. doi: 10.1074/jbc.273.49.32568. [DOI] [PubMed] [Google Scholar]
- 27.Olsen J C, Swanstrom R. A new pathway in the generation of defective retrovirus DNA. J Virol. 1985;56:779–789. doi: 10.1128/jvi.56.3.779-789.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel P H, Preston B D. Marked infidelity of human immunodeficiency virus type 1 reverse transcriptase at RNA and DNA template ends. Proc Natl Acad Sci USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powell M D, Levin J G. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pullen K A, Rattray A J, Champoux J J. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1993;268:6221–6227. [PubMed] [Google Scholar]
- 32.Rattray A J, Champoux J J. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J Mol Biol. 1989;208:445–456. doi: 10.1016/0022-2836(89)90508-1. [DOI] [PubMed] [Google Scholar]
- 33.Robson N D, Telesnitsky A. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J Virol. 1999;73:948–957. doi: 10.1128/jvi.73.2.948-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartzberg P, Colicelli J, Goff S P. Recombination between a defective retrovirus and homologous sequences in host DNA: reversion by patch repair. J Virol. 1985;53:719–726. doi: 10.1128/jvi.53.3.719-726.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 36.Smith C M, Potts W B, 3rd, Smith J S, Roth M J. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology. 1997;229:437–446. doi: 10.1006/viro.1997.8454. [DOI] [PubMed] [Google Scholar]
- 37.Smith J S, Kim S Y, Roth M J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J Virol. 1990;64:6286–6290. doi: 10.1128/jvi.64.12.6286-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorge J, Hughes S H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982;43:482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahara Y, Hamada K, Housman D E. A new retrovirus packaging cell for gene transfer constructed from amplified long terminal repeat-free chimeric proviral genes. J Virol. 1992;66:3725–3732. doi: 10.1128/jvi.66.6.3725-3732.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 41.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 42.Wohrl B M, Georgiadis M M, Telesnitsky A, Hendrickson W A, Le Grice S F. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 43.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]