Abstract
Coxsackievirus B3 (CVB3) infections can cause myocarditis in humans and are implicated in the pathogenesis of dilated cardiomyopathy. The natural genetic determinants of cardiovirulence for CVB3 have not been identified, although using strains engineered in the laboratory, cardiovirulence determinants have been identified in the CVB3 5′ nontranslated region (5′NTR) and capsid. The myocarditic phenotypes of two CVB3 clinical isolates were determined using an established murine model of inflammatory heart disease. The 5′NTRs and capsid proteins of the noncardiovirulent CVB3/CO strain and cardiovirulent CVB3/AS strain were examined to determine their influence on the cardiovirulence phenotype. Six intratypic chimeric viruses were constructed in which 5′NTR and capsid sequences of the infectious cDNA copy of the cardiovirulent CVB3/20 genome were replaced by homologous sequences from CVB3/CO or CVB3/AS. Chimeric strains were tested for cardiovirulence by inoculation of C3H/HeJ mice. Sections of hearts removed at 10 days postinoculation were examined for evidence of myocarditis by light microscopy and assayed for the presence of virus. Replacement of the CVB3/20 capsid coding region by that from the homologous region of CVB3/CO resulted in no change in the cardiovirulent CVB3/20 phenotype, with virus recoverable from the heart at 10 days postinoculation. However, recombinant virus containing the CVB3/CO 5′NTR alone or the 5′NTR and capsid sequences together were not myocarditic, and infectious virus was not recovered from the myocardium. Chimeric viruses containing the CVB3/AS 5′NTR alone, capsid sequence alone, or both together preserved the myocarditic phenotype. These data support the 5′NTR as the primary site in the determination of the natural cardiovirulence phenotype of CVB3.
Coxsackievirus B3 (CVB3), one of six CVB serotypes, is a member of the genus Enterovirus within the family Picornaviridae. The genome of CVB3, like that of other enteroviruses, is a single-stranded, messenger sense, polyadenylated RNA molecule 7,400 nucleotides (nt) in length. The single open reading frame, flanked 5′ and 3′ by nontranslated regions (NTRs), is subdivided into three regions, P1 to P3 (51), that encode a polyprotein of 2,185 amino acids (aa). The P1 region encodes the viral capsid proteins VP1 (281 aa), VP2 (263 aa), VP3 (238 aa), and VP4 (69 aa). The P2 and P3 regions encode the nonstructural viral proteins important for processing of the polyprotein, replication, and translation. The near atomic structure of the CVB3 virion has been solved (31), demonstrating a structure similar to that of poliovirus type 1 (18) and rhinovirus (38).
Coxsackie B viruses have been identified as a major cause of human viral myocarditis. They are also thought to play a significant role in the development of dilated cardiomyopathy (reviewed in reference 1). In general, neonates and children suffer more severe clinical syndromes due to CVB. In one study, of 41 infants who succumbed to CVB infection, approximately 95% demonstrated clinical or histologic evidence of myocarditis that contributed to the outcome (20). The predicted mortality rate for CVB infections among infants less than 3 months of age is 3.9/100,000 (20). Over a 4-year period, 45% of infant cases reported to the CDC Enteroviral Surveillance Program were attributed to CVB infection (reviewed in reference 30).
The enteroviral capsid has been shown to contain determinants contributing to the pathogenic phenotype of CVB4 (3, 34), CVB3 (22), and the polioviruses (PVs) (2, 7, 25, 26, 50). Interestingly, the sites determining the virulence phenotypes of these viruses do not colocalize to a single capsid region or even a single capsid protein. Determinants have been found in all four capsid proteins and are not necessarily located at surface-exposed residues of the virion. A noncardiovirulent antibody escape mutant derived from the highly cardiovirulent CVB3/H3 strain (49) was found to contain a single amino acid substitution (Asn→Asp) at position 165 of VP2 (22). When Asp165 was substituted for the Asn165 in VP2 of the parental cardiovirulent CVB3 strain, the myocarditic phenotype was significantly attenuated. Conversely, a change to Asn165 in VP2 of the antibody escape mutant reverted this strain to the cardiovirulent phenotype (22).
Specific nucleotide(s) within the 5′NTR are also known to alter the virulence phenotype of the PVs (reviewed in reference 29), CVB1 (35), and CVB3 (48). A U→C mutation at nt 234 within the CVB3 5′NTR results in attenuation of the cardiovirulent phenotype in mice (48). Replacement of the cardiovirulent CVB3/M or CVB3/20 5′NTRs with that from CVB3/0 attenuates the resultant viruses for myocarditis (24, 48). Subsequent analysis of multiple clinical CVB3 isolates as well as other enteroviruses demonstrated that nt 234 is always U regardless of the cardiovirulence phenotype of the virus (6, 37), consistent with 234C being an artificial mutation. Zhang et al. (53) isolated an attenuated CVB3 strain (p14V1) following multiple passages of cardiovirulent CVB3/Nancy (19) in human dermatofibroblasts. Sequence analysis of the 5′NTR revealed a single nucleotide change at position 690 (A→U) (53). Insertion of 690U into the cardiovirulent parental virus did not alter the myocarditic phenotype, demonstrating that this mutation does not affect the cardiovirulence phenotype. Following passage of p14V1 in scid mice hearts, a revertant to cardiovirulence was isolated (4). Sequence comparison of the 5′NTR and capsid coding region of this revertant to the attenuated p14V1 and cardiovirulent CVB3/Nancy strains suggested that amino acid 155 in VP1 might play a role in attenuation. However, experiments demonstrating this have not been reported.
The studies of genomic determinants of virulence for these and the majority of enteroviruses have relied on strains engineered by physiochemical or biologic means in the laboratory. To begin to examine the natural genetics of cardiovirulence in clinical CVB3 strains, we used two phenotypically and genotypically distinct CVB3 clinical isolates. Using reverse transcription-PCR (RT-PCR) to obtain the 5′NTR and P1 coding regions from CVB3/AS and CVB3/CO RNAs, we constructed intratypic chimeric viral genomes in the CVB3/20 background to test the hypothesis that these genomic regions encoded determinants of the viral cardiovirulent phenotype.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells (American Type Culture Collection, Manassas, Va.) were maintained as monolayers in minimal essential medium (MEM) supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 25.5 mM sodium bicarbonate, and 50 μg of gentamicin/ml. The complete sequence and characterization of the full-length infectious cDNA clone of CVB3/20 have been described previously (46). The CVB3/AS and CVB3/CO strains were isolated from stool samples of patients with viral encephalitis. CVB3/AS was isolated from a 10-year-old male in 1977, and the CVB3/CO strain was isolated from a 5-year-old male in 1978. The cardiovirulence phenotypes of CVB3/AS and CVB3/CO have been defined previously in C3H/HeJ and CD-1 mice (6, 14, 47). Aliquots of low-passage viral stocks were obtained by inoculation of nearly confluent HeLa cell monolayers at a multiplicity of infection (MOI) of 0.5 to 1 50% tissue culture infective doses (TCID50) per cell and stored at −80°C.
Primers for RT-PCR and sequencing.
Primers used for the amplification and sequencing of naturally occurring CVB3 5′ nontranslated and capsid coding regions are listed in Table 1. The B3-CO/Sac I primer was designed to incorporate the SacI restriction site (nt 751) into the CVB3/CO VP4 coding sequence and substituted 742G→A, 748G→A, and 751A→T without altering the amino acid sequence. Primers B3-1464, B3-1990, and B3-3324 used for RT-PCR and sequencing were designed based on published CVB3 sequences (5, 21, 22, 46). Primers B3-1226CC, B3-2689, and B3-2729 were designed based on the consensus sequence of multiple clinical CVB3 isolates (J. Romero, unpublished data). Primers B3-AS/Spe I, AS/20(3324)S, and 20/ApaL I were designed to incorporate the entire CVB3/AS capsid coding region in the CVB3/20 background using overlapping PCR products (see below).
TABLE 1.
Primers used for the amplification and sequencing of CVB3
| Name | Sequence | Locationa | Orientationb |
|---|---|---|---|
| T1 | 5′-TCACTATAGGTTAAAACAGC-3′ | 1 | S |
| JRp64 | 5′-ACGGTACCTTTGTGCGCCTGTTTTA-3′ | 63 | S |
| JRp577 | 5′-TGGCTGCTTATGGTGACAAT-3′ | 582 | S |
| B3-CO/Sac I | 5′-GATGGGAGCTCAAGTGTCG-3′ | 742 | S |
| JRpATG | 5′-TGAACTCGAGGGTA-3′ | 756 | A |
| B3-1226-CC | 5′-GGGCAAAACATGCAATATCACTACTTRGG-3′ | 1226 | S |
| B3-1464 | 5′-GTGTATAAYGCAGGCATGGG-3′ | 1464 | S |
| 3′PUFF | 5′-ATGTTATCCATAGGTACACT-3′ | 1593 | A |
| B3-1990 | 5′-GGCTTTCCACTGCAACCRGG-3′ | 1990 | S |
| ID3 | 5′-CACGCCACGTTAAGAACTACC-3′ | 2601 | S |
| B3-2689 | 5′-ACTCGGTGAAGTAAACACATGC-3′ | 2689 | A |
| B3-2729 | 5′-ATCACCCATTCRGCATACCG-3′ | 2729 | A |
| B3-AS/Spe I | 5′-GTTATAACTAGTACTCAACAGCC-3′ | 2806 | S |
| AS/20(3324)S | 5′-GGCGCTTTTGGACAACAATCAGGGGCAG-3′ | 3296 | S |
| B3-3324 | 5′-ACTGCCCCTGATTGTTGTCC-3′ | 3324 | A |
| 20/ApaL I | 5′-CCCGTTGTGCACTGACATCTGG-3′ | 3477 | A |
Numbered from the 5′ terminus relative to the CVB3/20 genome (46).
S, sense; A, antisense.
Extraction and RT-PCR of clinical CVB3 isolates.
The CVB3/CO and CVB3/AS RNA genome was extracted from 100 μl of a previously aliquoted virus stock preparation using the IsoQuick (ORCA Research Inc., Bothel, Wash.) guanidinium isothiocyanate kit as specified by the manufacturer. The precipitated nucleic acid was washed in 70% ethanol and dried.
High-fidelity RT-PCR was carried out in duplicate essentially as described previously (36, 37). Following RT-PCR, duplicate reactions were pooled, loaded in 1 to 3% low-melting-point agarose gels (Agarose SF; Amresco, Solon, Ohio) containing ethidium bromide (0.5 μg/ml), and electrophoresed at 80 to 120 V in 1× Tris-acetate-EDTA) buffer for approximately 1 h. Appropriate-sized DNA bands were identified, excised from the gel, purified, and resuspended in sterile H2O. The purified amplification products were stored at −20°C until needed.
Sequencing of RT-PCR products and cloned amplicons.
Direct sequencing of RT-PCR products and clones was performed using the ThermoSequenase (Amersham, Cleveland, Ohio) terminator cycle sequencing protocol according to the manufacturer's instructions.
Samples were electrophoresed through 8% Page Plus (Amresco) gels in 0.6× glycerol tolerant buffer (1.78 M Tris-HCl, 0.57 M taurine, 0.01 M EDTA · Na2 · H2O) (U.S. Biochemical, Cleveland, Ohio) for periods of 1.5 to 6.5 h at 85-W constant power. Gels were transferred to Whatman paper and dried. Autoradiography was performed by exposure to film (Kodak/IBI, Rochester, N.Y.) for 12 to 36 h. Sequence analysis was performed with the MacVector software (version 6.0) (Kodak/IBI) and the Wisconsin Sequence Analysis Package (version 10.0) (Genetics Computer Group, Inc., Madison, Wis.).
Construction and generation of recombinant viruses.
RT-PCR products generated from CVB3/CO and CVB3/AS RNA templates were blunt-end cloned into the SmaI site of pCR-Script SK(+) (Stratagene, La Jolla, Calif.) as instructed by the manufacturer. Each clone was sequenced entirely to verify its fidelity to the directly sequenced RT-PCR products. All infectious cDNA genomic constructs were verified by restriction endonuclease mapping and sequence analysis. The cDNA copy of the cardiovirulent CVB3/20 genome in the modified plasmid pSVN (5), and the cloned 5′NTR and capsid coding sequences of the noncardiovirulent CVB3/CO and cardiovirulent CVB3/AS clinical isolates were used to generate full-length infectious intratypic chimeras.
The complete capsid coding region (nt 743 to 3296 relative to the CVB3/20 genome [46]) of CVB3/CO was constructed using overlapping subcloned fragments spanning the primer pairs B3-3324–B3-1464 and B3-2689–B3-CO/Sac I using the AlwNI (nt 2153) restriction site. The capsid coding region of CVB3/20 was replaced with that of CVB3/CO by exchange of the homologous SacI (nt 751)-to-AflIII (nt 3109) fragment which excludes only a single deduced amino acid difference between these strains (CVB3/20 R1264→CVB3/CO Q).
The complete capsid coding region of CVB3/AS was generated using overlapping subcloned fragments spanning the primer pairs JRp577–3′PUFF, B3-1226-CC–B3-2729, and B3-3324–B3-1990, using the NsiI (nt 1291) and XmnI (nt 2649) restriction sites. To generate a full-length chimera encompassing the complete capsid sequence of CVB3/AS, PCR fragments containing CVB3/AS sequences from nt 2806 to 3304, which included the SpeI (nt 2813) site, and CVB3/20 sequences from nt 3305 to 3477, which included the ApaLI (nt 3466) site, were amplified, overlapped, and cloned by the method of Ho et al. (17) with the following modifications. Briefly, a CVB3/AS sequence specific PCR fragment spanning nt 2806 to 3324 (primer pair B3-AS/Spe I–B3-3324) and a CVB3/20 sequence-specific PCR fragment spanning nt 3296 to 3477 [primer pair AS/20(3324)S–20/ApaL I] were used as template to generate the overlap PCR product described above. The entire capsid coding region of CVB3/20 was replaced with that of CVB3/AS by exchange of the SacI (nt 751)-to-SpeI (nt 2813) fragment of the CVB3/AS capsid clone plus the SpeI-to-ApaLI (nt 3466) fragment of the AS/20 overlap PCR product clone.
The 5′NTR of CVB3/CO was amplified using the primer pair JRp64-JRpATG (36). The 5′NTR of CVB3/AS was amplified using the primer pair T1-JRpATG. Both 5′NTRs were cloned as described previously and used to replace the homologous region of CVB3/20 between nt 69 and 751, using the conserved KpnI and SacI restriction sites, respectively.
Virus was generated from cDNA genomes by electroporation of 20 μg of plasmid DNA into approximately 2 × 106 HeLa cells. Cells were incubated at 37°C and 5% CO2 for 3 days or until cytopathic effect (CPE) was observed and then were thrice frozen and thawed. Viral progeny were clarified by centrifugation at 3,000 × g, and 500 μl of supernatant was added to 106 HeLa cells and incubated until complete CPE was observed.
Viral titer was determined using 96-well plates (Becton Dickinson, Franklin Lakes, N.J.) with 103 HeLa cells/well. Samples were serially diluted to 10−8, added to the appropriate wells, incubated at 37°C and 5% CO2 for 5 days, and scored for the presence of complete CPE. TCID50 calculations were determined according to the formula of Cunningham (8).
Viral replication in cell culture.
HeLa monolayers of 2 × 105 cells in 35-mm2 plates (Corning, Acton, Mass.) were washed once with phosphate-buffered saline, infected at an MOI of 10 TCID50/cell in a volume of 500 μl, and allowed to adsorb for 30 min at room temperature. Plates were then washed twice with phosphate-buffered saline to remove unbound virus, refed with 2 ml of supplemented MEM, incubated at 37°C and 5% CO2, and, at the given intervals, removed and frozen. Plates were thrice frozen and thawed, and 1 ml was transferred to a microcentrifuge tube and clarified by centrifugation. Viral titer was determined as described above.
Determination of viral cardiovirulence phenotype in mice.
Groups of five juvenile (21 to 25 days of age) C3H/HeJ male mice (Jackson Laboratory, Bar Harbor, Maine) were inoculated intraperitoneally with 2 × 105 to 5 × 105 TCID50 of virus in 0.1 ml of unsupplemented MEM or with medium alone (negative control) as described previously (5, 46, 48). Groups of mice were maintained in separate microisolators in a ventilated containment facility. Ten days postinoculation (dpi), mice were sacrificed and hearts were excised (48). One half of each heart was fixed in buffered formalin, embedded in paraffin, and sectioned. Three to six heart sections (6 μm) were stained with hemotoxylin and eosin and examined by light microscopy for evidence of myocarditis. The myocarditis lesion score of each group was determined by the method of Knowlton et al. (22) according to the following scale: 0, no myocarditis; 1, 1 to 10 lesions per section; 2, 11 to 20 lesions per section; 3, 21 to 40 lesions per section; and 4, widespread and confluent inflammation.
Approximately one quarter of each heart was weighed and homogenized in 400 μl of supplemented MEM using a Dounce homogenizer (Fisher Scientific, Pittsburgh, Pa.). Following centrifugation (12,000 × g) to clarify the remaining cellular debris, 200 μl of the supernatant was used to determine cardiac viral titers. Titers from homogenized heart were performed (see above) and expressed as log TCID50 per gram of heart tissue. RT-PCR was performed with 100 μl from each of two homogenized cardiac specimens from each group exhibiting inflammatory lesions; these reaction products were directly cycle sequenced (see above) to verify the infecting genotype.
Nucleotide sequence accession numbers.
GenBank accession numbers of the 5′NTRs and capsid protein coding sequences are AF169665 and AF169666 for CVB3/CO and AF169670 and AF169671 for CVB3/AS.
RESULTS
Nucleotide analysis of the 5′NTR.
There are currently five full-length infectious CVB3 cDNA clones for which complete genomic sequence data are available. These include four cardiovirulent strains (19, 22, 23, 46) and one artificially attenuated noncardiovirulent strain (5). Nucleotide sequence analysis indicates that all five are highly related (37). Several of these CVB3 strains have been used to examine the viral genetics of cardiovirulence, revealing major determinants in the 5′NTR (24, 48) and capsid coding region (22).
The 5′NTR nucleotide sequences of the noncardiovirulent CVB3/CO and cardiovirulent CVB3/AS strains were determined and compared to the full-length CVB3 cDNA clones to examine genotypic diversity and the significance of the 5′NTR in the determination of CVB3 cardiovirulence phenotype. The 5′NTR sequences of multiple clinical CVB3 strains including CO and AS have been examined (6, 37). The CVB3/AS 5′NTR spanning nt 87 to 742 (relative to CVB3/20 genome) displayed a significantly higher nucleotide identity (90.9 to 92.3%) with the CVB3/20 (46), CVB3/0 (5), CVB3/Nancy (21), CVB3/H3 (22), and CVB3/M (23) strains than did the noncardiovirulent CVB3/CO 5′NTR (83.9 to 84.3%). Among themselves, CVB3/AS and CO exhibited 82.9% nucleotide identity within the 5′NTR.
Sequence identities differed significantly among the three CVB3 strains used in this study in upstream and downstream sequences of the 5′NTR comprised by nt 88 to 181 and nt 452 to 742, respectively. CVB3/CO displayed 61.6 and 62.6% nucleotide identity with the myocarditic CVB3/AS and CVB/20 strains, respectively, in the upstream region. For the same region, CVB3/AS and CVB3/20 were 85.1% identical among themselves. For sequences preceding and up to the coding start site (nt 452 to 742), CVB3/20 and CVB3/AS were 89.7% identical. CVB3/CO was only 80.3 and 82.1% identical to CVB3/AS and CVB3/20, respectively, in this same region. Analysis of the intervening sequences of the 5′NTR revealed that all three CVB3 shared between 91.8 and 97.0% nucleotide identity.
Nucleotide and amino acid analysis of the capsid coding region.
The capsid nucleotide and deduced amino acid sequences of CVB3/CO and CVB3/AS were examined and compared to the homologous region of the full-length CVB3 cDNA clones to assess differences which might affect the cardiovirulence phenotype. Nucleotide analysis of the entire P1 coding region showed that CVB3/CO and CVB3/AS exhibited 79.5% nucleotide identity among themselves. The CVB3/20 strain shared 79.6 and 87.5% nucleotide identity with CVB3/CO and CVB3/AS, respectively. In fact, capsid nucleotide sequence analysis indicated that CVB3/AS and all full-length sequenced CVB3 strains clustered within the same genotypic group, while CVB3/CO fell into a separate genotype. Comparison of the deduced capsid amino acid sequences of CVB3/AS and CVB3/CO with those of the full-length sequenced CVB3 genomes showed that all strains shared greater than 97% identity (Table 2). The vast majority of nucleotide differences occurred at second- and third-base codon positions leaving the amino acid sequence unaffected.
TABLE 2.
Capsid amino acid differences among CVB3 isolates
| Protein | Amino acid positiona | Amino acid at that position in strainb:
|
||||||
|---|---|---|---|---|---|---|---|---|
| AS | CO | 20 | N | 0 | H3 | M | ||
| VP4 | 4016 | G | G | R | R | G | G | G |
| 4018 | S | N | N | N | N | N | N | |
| 4030 | I | I | I | I | V | I | I | |
| 4042 | N | T | N | N | N | N | N | |
| 4047 | A | A | T | T | T | T | T | |
| 4051 | G | G | G | G | G | S | S | |
| VP2 | 2013 | V | V | V | A | A | V | V |
| 2045 | S | N | S | S | S | S | S | |
| 2108 | I | I | V | V | V | I | I | |
| 2138* | D | D | D | D | D | N | N | |
| 2144* | A | E | A | A | A | A | A | |
| 2151* | S | A | S | T | S | S | S | |
| 2171 | V | A | V | V | V | V | V | |
| 2179 | V | I | V | V | V | V | V | |
| 2245 | V | V | V | V | V | I | I | |
| VP3 | 3046 | I | I | I | I | I | I | V |
| 3058* | I | I | V | V | V | V | V | |
| 3062* | I | V | V | V | V | V | V | |
| 3078* | S | T | S | S | S | S | S | |
| 3155 | V | V | V | I | V | V | V | |
| 3178 | Y | Y | F | F | Y | Y | Y | |
| 3234* | Q | Q | Q | Q | E | Q | Q | |
| 3237* | F | L | F | F | F | F | F | |
| VP1 | 1007 | V | I | I | I | I | I | I |
| 1045 | S | G | G | G | G | S | S | |
| 1064 | V | I | I | I | I | I | I | |
| 1080* | E | E | K | K | E | E | E | |
| 1084 | A | S | A | A | A | A | A | |
| 1085* | K | N | K | K | K | K | K | |
| 1092 | I | I | L | L | L | I | I | |
| 1094 | P | T | P | P | P | P | P | |
| 1098 | A | V | A | A | A | A | A | |
| 1110 | V | M | V | V | V | V | V | |
| 1180 | I | I | I | I | I | V | V | |
| 1200* | S | A | S | S | S | S | S | |
| 1223* | T | A | A | A | A | A | A | |
| 1264* | Q | Q | R | Q | Q | Q | Q | |
Cardiovirulence phenotype and cardiac viral titers of chimeric CVB3.
The full-length infectious clone of CVB3/20 has been characterized and used previously to examine determinants of cardiovirulence (48). Typically, at 10 dpi, CVB3/20 induces widespread inflammatory lesions with significant necrosis and calcification in the hearts of C3H/HeJ mice (48). Infectious CVB3/20 was readily detectable in murine hearts at 10 dpi (48). The cardiovirulence phenotypes of CVB3/AS and CVB3/CO have been defined previously in C3H/HeJ and CD-1 mice (6, 14, 47). CVB3/AS was cardiovirulent for adolescent (4- to 6-week-old) male and female CD-1 mice, with detectable virus in the murine myocardium at 7 dpi (14). CVB3/AS was also found to induce inflammatory lesions and muscle damage in hearts of 3- to 4-week-old male C3H/HeJ mice 10 dpi (6). No myocarditic lesions were observed following inoculation of CVB3/CO in male or female CD-1 or C3H/HeJ mice (6, 47).
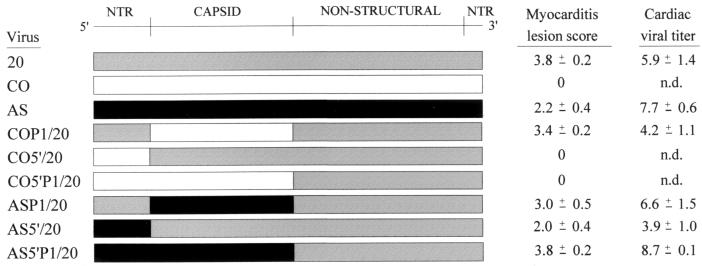
Chimeric CVB3 (Fig. 1) viruses were generated and inoculated into juvenile C3H/HeJ male mice to examine whether the 5′ NTR and/or capsid coding region contained sequences that significantly influenced CVB3-induced inflammatory heart disease. To assess the role of the capsid proteins of CVB3 strains in determining the cardiovirulent phenotype, the capsid coding region of the noncardiovirulent CVB3/CO strain was used to replace the homologous region of the full-length infectious clone of the cardiovirulent CVB3/20 strain. The resultant CVB3 chimera (COP1/20) retained the cardiovirulence phenotype of CVB3/20 in mice (Fig. 2F). Similarly, the myocarditic phenotype of CVB3/20 was not altered when the capsid coding region was exchanged for that of the cardiovirulent CVB3/AS strain (ASP1/20) (Fig. 2E). Virus was detectable in murine hearts 10 days after inoculation of strains COP1/20 and ASP1/20 (Fig. 1).
FIG. 1.
Schematic representation, myocarditis lesion score and cardiac viral titer of CVB3 and intratypic chimeras. Myocarditis lesion scoring (22) (see Materials and Methods) and cardiac viral titers (log of TCID50 per gram of heart tissue) are given as mean ± standard error of the mean. n.d., no virus titer detected.
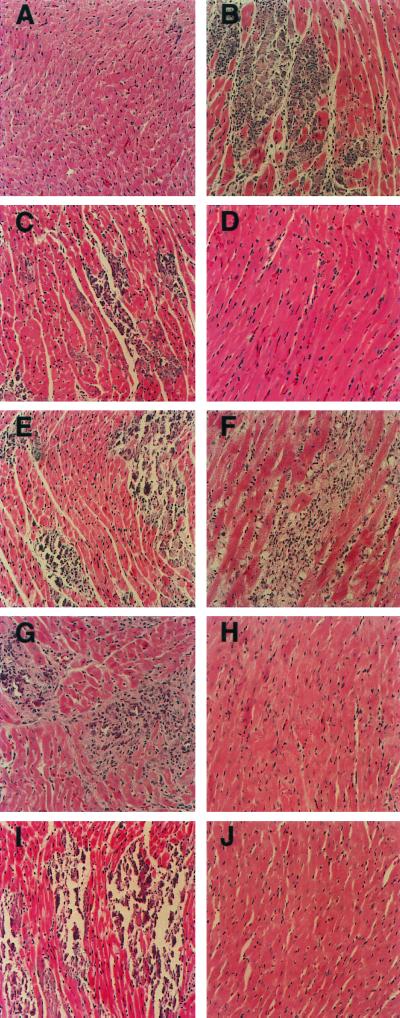
FIG. 2.
Representative histology of murine myocardium following inoculation of CVB3 and chimeric viruses. Juvenile male C3H/HeJ mice were inoculated with virus, and at 10-dpi hearts were recovered, sectioned, and stained as described in Materials and Methods. Shown are murine myocardium of negative control (A), typical myocarditis seen following inoculation of positive control CVB3/20 (B), murine myocardium following inoculation of CVB3/AS (C) and CVB3/CO (D), intratypic capsid chimeras ASP1/20 (E) and COP1/20 (F), intratypic 5′NTR chimeras AS5′/20 (G) and CO5′/20 (H), and intratypic 5′NTR/capsid chimeras AS5′P1/20 (I) and CO5′P1/20 (J). Imaged at ×200 magnification.
The 5′NTRs of CVB3/CO and CVB3/AS were then examined to determine their effects, alone and in combination with their corresponding capsid coding regions, on the cardiovirulence phenotype. The 5′NTR (spanning nt 88 to 742) of CVB3/20 was replaced with that from CVB3/CO. Examination of the murine myocardium following inoculation of the resultant chimera (CO5′/20) revealed no evidence of myocarditis (Fig. 2H). Similarly, when the CVB3/CO 5′NTR and capsid coding region were combined to replace the homologous regions of CVB3/20 (CO5′P1/20), no myocarditic lesions were found (Fig. 2J). Ten days following inoculation of CO5′/20 or CO5′P1/20, detectable virus was not recovered from murine hearts by infectious assay (Fig. 1). In contrast, when the CVB3/AS 5′NTR (nt 71 to 742) alone or in combination with the AS capsid coding region was used to replace the homologous regions of CVB3/20 (AS5′/20 and AS5′P1/20, respectively), inflammatory lesions and significant necrosis were observed (Fig. 2G and I). Cardiac viral titers were obtained following inoculation of AS5′/20 and AS5′P1/20 (Fig. 1).
RT-PCR amplification and direct cycle sequencing of viral RNA from homogenized cardiac specimens from two separate animals of each group of mice exhibiting inflammatory heart lesions was performed. Sequence analysis of nt 630 to 850 (relative to the CVB3/20 genome) demonstrated that the infecting genotype was as had been cloned in this region and that no mutations had occurred in or around the SacI restriction site (nt 751) utilized for homologous exchange of the 5′ NTR and/or capsid coding region (data not shown).
Growth kinetics of parental and recombinant viruses.
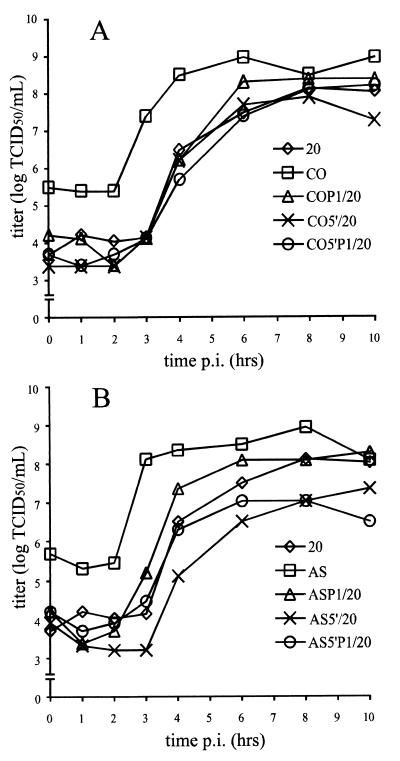
The single-step growth characteristics of the chimeric CVB3 were examined in HeLa cells and compared to those of the parental CVB3/20, CVB3/CO, and CVB3/AS strains (Fig. 3). All parental and recombinant viruses replicated efficiently in HeLa cell cultures. All strains exhibited logarithmic growth by 3 to 4 h postinfection, and maximum titers were achieved after 6 to 8 h of incubation. While differences were observed in the duration of the lag phase (∼1 h) and overall virus yield (∼1 log TCID50/ml), these did not appear to correlate with differences in myocarditic phenotype. The data indicated that homologous exchange of CVB3 5′ NTR and capsid coding region between various strains resulted in viable progeny with similar replication phenotypes in HeLa cells.
FIG. 3.
One-step growth curves of parental and chimeric viruses in HeLa cells. (A) Replication of cardiovirulent CVB3/20, noncardiovirulent CVB3/CO, and intratypic chimeras; (B) replication of cardiovirulent CVB3/20, CVB3/AS, and intratypic chimeras. Cells were inoculated as described in Materials and Methods and harvested by freezing at specific times shown. Virus titers were determined on HeLa cell monolayers. p.i., postinoculation.
DISCUSSION
To date, the vast majority of the studies of enteroviral determinants of cardio-, pancreo-, or neurovirulence have relied on phenotypically diverse virus strains recovered in the laboratory using cell culture techniques, passage in animal systems, or immunologic variability. Initial work in developing the live attenuated oral PV vaccines relied on some of these methods (39). Unfortunately, as exemplified by human immunodeficiency virus, prototypic laboratory strains are sometimes so phenotypically and genotypically distinct from those found in infected individuals that therapeutic or preventative measures effective against these laboratory strains do not always correlate with naturally circulating strains (27).
In this study, we used two phenotypically and genotypically distinct clinical CVB3 strains to map the natural genetics of cardiovirulence. This is the first report mapping the genetics of enterovirus virulence to use so-called wild-type strains, that is, clinical viral isolates that have low passage histories in cell culture. Using one such murine noncardiovirulent strain, CVB3/CO, we demonstrate here that the 5′NTR contains a site(s) that attenuates cardiovirulence. No inflammatory heart lesions were observed following inoculation of mice with any viral construct containing the CVB3/CO 5′NTR (Fig. 2H and J). In addition, infectious virus could not be recovered from the heart at 10 dpi (Fig. 1), similar to the amyocarditic CVB3/CO strain itself. Similar experiments using the 5′NTR of the cardiovirulent CVB3/AS clinical isolate revealed no significant change in cardiovirulence (Fig. 2G and I) or in recovery of virus from the heart (Fig. 1). The capsid coding regions of the CVB3/CO and CVB3/AS isolates used to replace the homologous region of the full-length infectious CVB3/20 cDNA did not alter the cardiovirulence phenotype of the resultant viruses (Fig. 2E and F). Additionally, infectious virus was readily detected in the murine myocardium at 10 dpi (Fig. 1). Thus, the capsid amino acid differences observed between the cardiovirulent CVB3/20 and noncardiovirulent CVB3/CO strains (Table 2) do not modify the myocarditic phenotype of CVB3/20. Although the fine mapping of the sites within the 5′NTR that contribute to cardiovirulence and the molecular mechanisms that determine this phenotype remain to be identified, our data support the hypothesis that sequences within the CVB3 5′NTR are the primary determinants of the myocarditic phenotype of CVB3 strains.
The severity of disease and cardiac viral titers observed in this study following inoculation of the parental CVB3/20, CVB3/AS, and CVB3/CO strains correlated to that seen in previous work (14, 47, 48). Typically, myocardial lesion scores obtained following inoculation of CVB3/AS were less than those seen with CVB3/20 (14, 47). However, this and other reports have documented that for these two strains the quantity of virus recovered from the myocardium did not correlate with the extent of inflammatory heart disease (Fig. 1) (14, 47). Gauntt et al. also demonstrated that the quantity of CVB3-induced myocardial lesions was not necessarily reflected by the corresponding cardiac viral titer (13). This holds true for the intratypic CVB3 chimeras in this study, in which no statistically significant correlation could be determined between the mean lesion score and cardiac viral titer (data not shown).
We have demonstrated that the CVB3 5′NTR contains the primary determinant(s) that attenuates cardiovirulence. However, we cannot discount the possibility that other viral genomic regions (e.g., capsid) may contain additional determinants that modulate the expression of the 5′NTR phenotype. Note the CVB3/AS5′P1/20 chimera in which inclusion of the CVB3/AS capsid augments the cardiovirulent phenotype of CVB3/AS5′/20 (Fig. 1). Similarly, although the primary determinants of PV neurovirulence localize to the 5′NTR (discussed below), variations within the coding region influence the full expression of this phenotype (2, 7, 25, 26, 50).
By comparison to the near atomic structure of the CVB3 virion (31), several capsid amino acid differences between the CVB3/AS and CVB3/CO isolates and sequenced full-length CVB3 strains could be mapped to positions exposed on the surface of the virion (Table 2). Surface differences in VP1 localized in and around the BC loop (residues 1081 to 1085), which is known to be an important neutralizing antigenic site for the PVs and rhinoviruses (28, 32, 40, 41). The VP2 surface differences were found in the prominent EF loop (residues 2129 to 2180). An exterior region of the EF loop known to contain neutralizing antigenic sites for the PVs and rhinoviruses (28, 32, 40, 41) was found to be identical among all CVB3 strains examined. Both CVB3/AS and CVB3/CO differed from the full-length sequenced CVB3 strains in the major surface protrusion of VP3 (residues 3058 to 3069). However, none of these capsid surface or other differences appear to affect the cardiovirulence phenotype of CVB3/20 when the capsid is replaced by that of either CVB3/CO or CVB3/AS.
The findings presented here are in contrast to those of Knowlton et al. (22). An attenuated antibody escape mutant derived from the cardiovirulent CVB3/H3 strain was used to examine cardiovirulence determinants. This strain was attenuated by a single Asn→Asp mutation at residue 165 in VP2. When the Asn→Asp165 change was introduced into the parental CVB3/H3 genome, cardiovirulence was severely attenuated; similarly, changing Asp→Asn165 in the antibody escape mutant restored cardiovirulence (22). In that system, the presence of Asn165 corresponded to a cardiovirulent phenotype. However, our analysis of multiple CVB3 isolates (including CVB3/AS and CVB3/CO) demonstrated that all strains encode Asn165 regardless of cardiovirulence phenotype (Romero, unpublished data). Based on these findings and the chimeric construct data presented here that show no impact on cardiovirulence due to the capsid proteins, the findings of Knowlton and colleagues can best be explained as the mapping of an attenuating site that is not found in circulating strains. Ramsingh et al. derived a pancreovirulent strain of CVB4 by repeated passage through mice with isolation from the pancreatic tissue (33). The sites that determine the pancreovirulent phenotype of this strain were then mapped using a series of chimeric constructs to a single amino acid change in each of the VP1 and VP4 capsid proteins (3, 34). While these data suggest that CVB4 murine pancreovirulence might naturally map to these sites, it remains necessary to confirm these findings in studies using murine pancreovirulent clinically isolated CVB4 strains.
In three studies to date, sites in the CVB3 5′NTR have been implicated as affecting the cardiovirulence phenotype. The single C→U change at position 234 within the 5′NTR that conferred a cardiovirulent phenotype to CVB3/0 (48) did not correlate with the cardiovirulence phenotypes of multiple CVB3 strains sequenced to date, in which position 234 was always U regardless of cardiovirulence phenotype (6, 37). This U→C234 mutation is, therefore, not found in nature and can be regarded as artificial. On the basis of 5′NTR sequence comparisons, Gauntt and Pallansch (14) suggested one or more sites might be involved in determining the myocarditic phenotype. Using a noncardiovirulent CVB3 strain obtained following multiple passages of the cardiovirulent CVB3/Nancy in human dermatofibroblasts, Zhang et al. demonstrated that a mutation found in the 5′NTR was not attenuating (53). Using clinical isolates for the construction of infectious intratypic chimeric cDNA genomes, we demonstrate that replacement of the 5′NTR of a cardiovirulent strain's genome with that from an amyocarditic strain ablates cardiovirulence. Reconstruction of the genome with the parental 5′NTR or the use of the 5′NTR from another cardiovirulent strain reestablishes the cardiovirulent phenotype. These data implicate the 5′NTR as containing one or more sites that determine the cardiovirulence phenotype.
In this work, we differentiate between so-called wild-type or clinically isolated CVB3 strains and strains that have been manipulated in the laboratory with a concomitant change in phenotype. The approach taken in this paper is the only one possible at present, for there exists no amyocarditic strain of CVB3 that has been cloned as an infectious cDNA copy. All infectious cDNA clones of CVB3 genomes are from strains that either are naturally cardiovirulent (19, 22, 23, 46) or, in the case of CVB3/0, can be made to be cardiovirulent if engineered to revert the C→U234 mutation (5, 6). In general, the nucleotide sequences of recent clinical CVB3 isolates differ significantly from the published full-length sequences in both the 5′ NTR (37) and capsid coding region (Romero, unpublished data). A recent epidemiologic investigation using CVB3 isolates from 1952 to 1997 demonstrated that the original CVB3/Nancy (1949) strain, from which most of the full-length infectious cDNAs and sequences have been generated, is no longer circulating in the general population (J. R. Romero, D. Schnurr, M. S. Oberste, S. S. Bradrick, C. Kirk, and C. Price, Sci. Program Abstr. 18th Annu. Meet. Am. Soc. Virol. 1999, abstr. W50-2, p. 148, 1999).
How might naturally occurring differences in the 5′NTR determine the cardiovirulence phenotype of a CVB population? To date, this question has been examined only in artificially attenuated virus strains or strains that do not circulate in nature. Enteroviruses include within the 5′NTR an internal ribosome entry site (IRES) that is required for efficient cap-independent translation of the viral coding region. Mutations within the IRES can seriously impair or abrogate translation (11). Attenuated Sabin strains of PV exhibit decreased efficiency of translation in cells of neural origin compared to wild-type, neurovirulent strains (42–44). This difference in translational efficiency was found to correspond to the major attenuating mutations at nt 480, 481, and 472 within the IRESs of PV1, PV2, and PV3, respectively (reviewed in reference 29). The Sabin PV vaccine strains, however, were artificially derived and not naturally occurring, attenuated strains. The attenuating mutations in the PV vaccine strains readily revert in passage in the human gut, largely reacquiring the virulent phenotype in the process (12). Similarly, following prolonged replication in scid mice, CVB3/0 nt 234C changes to U and the virus reverts to the cardiovirulent phenotype (48). While it is tempting to speculate that, like the case for the PVs, perturbation(s) in translational efficiency may be the mechanism for the attenuated cardiovirulence phenotype of CVB3/CO, alteration of viral replication could also be affected. As seen with the U→C234 transition in the 5′NTR of CVB3/0, replication of viral RNA is significantly decreased in murine cells of cardiac origin but not in HeLa cells (48). This would suggest that interaction between some specific cardiac cell factor(s) and the CVB3/0 5′NTR may be responsible. All parental and intratypic CVB3 studied here grew efficiently in HeLa cell cultures (Fig. 3). Although differences in lag-phase duration (∼1 h) and overall virus yield (∼1 log TCID50/ml) were observed, these variations did not correlate with the ability of CVB3 strains to induce inflammatory heart disease. It will be necessary to characterize the ability of these strains to replicate in cells of cardiac origin to determine if and how production of viable progeny is altered.
The extensive sequence diversity within specific regions of the 5′NTR among the CVB3 strains used in this study suggests that RNA elements necessary for efficient viral translation and/or replication in the murine myocardium may differ between amyocarditic and cardiovirulent isolates. CVB3/CO displayed only 61.6 and 62.6% nucleotide identity with CVB3/AS and CVB3/20, respectively, in a CVB3 5′NTR region predicted to form a single stem-loop (nt 88 to 181) (52). Among themselves, CVB3/AS and CVB3/20 were 85.1% identical in this region. Eukaryotic initiation factor 2α, pyrimidine tract-binding protein (PTB), and a complex of other HeLa cell proteins are known to bind to the homologous region of PV (10, 16). In addition, a major genomic determinant of CVB1 virulence was localized to within this portion of the 5′NTR (nt 117 to 161) (35). The region encompassed by nt 452 to 742 preceding and up to the true initiation codon also differed among CVB3 strains. Here CVB3/AS and CVB3/20 were 89.7% identical but shared only 80.3 and 82.1% identity, respectively, with CVB3/CO. Within this region of the 5′NTR are included the major attenuating sites of the Sabin PV strains (discussed above), multiple PV PTB binding sites (16), and the variable region observed among enteroviruses (45). Sequences within this region have been shown to be crucial for the expression of neurovirulence phenotype in PV (15). All three CVB3 strains displayed between 91.8 and 97.0% nucleotide identity within the region spanning nt 182 to 451. For the amyocarditic CVB3/CO strain, the significant comparative differences observed in the upstream and downstream 5′NTR sequences may affect viral RNA higher-order structure such that cardiac-specific protein-viral RNA interaction(s) is disrupted, preventing efficient viral infection.
In summary, although it has been recognized since at least 1955 that the CVB can cause acute viral myocarditis (9), the genetics that naturally determine the viral cardiovirulence phenotype have remained unsolved. The data presented here are consistent with the hypothesis that the CVB3 5′NTR contains one or more primary genetic determinants that regulate the naturally occurring expression of cardiovirulence. Experiments to more finely map and identify the site(s) responsible within the CVB3 5′NTR are under way.
ACKNOWLEDGMENTS
This work was supported by grants from the Edna Ittner Foundation for Pediatric Research (J.R.R.) and the American Heart Association (S.T. and N.M.C.).
REFERENCES
- 1.Baboonian C, Davies M J, Booth J C, McKenna W J. Coxsackie B viruses and human heart disease. In: Tracy S, Chapman N M, Mahy B W J, editors. The coxsackie B viruses. Berlin, Germany: Springer-Verlag; 1997. pp. 31–52. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard M J, Lam D, Racaniello V R. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J Virol. 1995;69:4972–4978. doi: 10.1128/jvi.69.8.4972-4978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caggana M, Chen P, Ramsingh A. Identification of a single amino acid residue in the capsid protein of VP1 of coxsackievirus B4 that determines the virulent phenotype. J Virol. 1993;67:4797–4803. doi: 10.1128/jvi.67.8.4797-4803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameron-Wilson C L, Pandolfino Y A, Zhang H Y, Archard L C. Nucleotide sequence of an attenuated mutant of coxsackievirus B3 compared with the cardiovirulent wildtype: assessment of candidate mutations by analysis of a revertant to cardiovirulence. Clin Diagn Virol. 1998;9:99–105. doi: 10.1016/s0928-0197(98)00008-7. [DOI] [PubMed] [Google Scholar]
- 5.Chapman N M, Tu Z, Tracy S, Gauntt C J. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3 strain: its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch Virol. 1994;135:115–130. doi: 10.1007/BF01309769. [DOI] [PubMed] [Google Scholar]
- 6.Chapman N M, Romero J R, Pallansch M A, Tracy S. Sites other than nucleotide 234 determine cardiovirulence in natural isolates of coxsackievirus B3. J Med Virol. 1997;52:258–261. [PubMed] [Google Scholar]
- 7.Couderc T, Guedo N, Calvez V, Pelletier I, Hogle J, Colbere-Garapin F, Blondel B. Substitutions in the capsids of poliovirus mutants selected in human neuroblastoma cells confer on the Mahoney type 1 strain a phenotype neurovirulent in mice. J Virol. 1994;68:8386–8391. doi: 10.1128/jvi.68.12.8386-8391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham C H. Quantal and enumerative titration of virus in cell cultures. In: Kruse P F Jr, Patterson M K Jr, editors. Tissue culture methods and application. New York, N.Y: Academic Press, Inc.; 1973. pp. 527–532. [Google Scholar]
- 9.Dalldorf G. The coxsackie viruses. Annu Rev Microbiol. 1955;9:277–296. doi: 10.1146/annurev.mi.09.100155.001425. [DOI] [PubMed] [Google Scholar]
- 10.Del Angel R M, Papavassiliou A G, Fernandez-Tomas C, Silverstein S J, Racaniello V R. Cell proteins bind to multiple sites within the 5′ untranslated region of poliovirus RNA. Proc Natl Acad Sci USA. 1989;86:8299–8303. doi: 10.1073/pnas.86.21.8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 12.Evans D M, Dunn G, Minor P D, Schild G C, Cann A J, Stanway G, Almond J W, Currey K, Maizel J V. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature. 1985;314:548–550. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 13.Gauntt C J, Gomez P T, Duffey P S, Grant J A, Trent D W, Witherspoon S M, Paque R E. Characterization and myocarditic capabilities of coxsackievirus B3 variants in selected mouse strains. J Virol. 1984;52:598–605. doi: 10.1128/jvi.52.2.598-605.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauntt C J, Pallansch M A. Coxsackievirus B3 clinical isolates and murine myocarditis. Virus Res. 1996;41:89–99. doi: 10.1016/0168-1702(95)01250-8. [DOI] [PubMed] [Google Scholar]
- 15.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellen C U T, Pestova T V, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 19.Kandolf R, Hofschneider P H. Molecular cloning of the genome of cardiotropic coxsackie B3 virus: full-length reverse-transcribed recombinant cDNA generates infectious virus in mammalian cells. Proc Natl Acad Sci USA. 1985;82:4818–4822. doi: 10.1073/pnas.82.14.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan M H, Klein S W, McPhee J, Harper R G. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev Infect Dis. 1983;5:1019–1032. doi: 10.1093/clinids/5.6.1019. [DOI] [PubMed] [Google Scholar]
- 21.Klump W M, Bergmann I, Muller B C, Ameis D, Kandolf R. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J Virol. 1990;64:1573–1583. doi: 10.1128/jvi.64.4.1573-1583.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knowlton K U, Jeon E, Berkley N, Wessely R, Huber S. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J Virol. 1996;70:7811–7818. doi: 10.1128/jvi.70.11.7811-7818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C, Maull E, Chapman N, Tracy S, Wood J, Gauntt C. Generation of an infectious cDNA of a highly cardiovirulent coxsackievirus B3 (CVB3m) and comparison to other infectious CVB3 cDNAs. Virus Res. 1997;50:225–235. doi: 10.1016/s0168-1702(97)00059-2. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Maull E, Chapman N, Tracy S, Gauntt C. Genomic regions of coxsackievirus B3 associated with cardiovirulence. J Med Virol. 1997;52:341–347. [PubMed] [Google Scholar]
- 25.Lu H, Yang C, Murdin A D, Klein M H, Harber J J, Kew O M, Wimmer E. Mouse neurovirulence determinants of poliovirus type 1 strain LS-a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J Virol. 1994;68:7505–7515. doi: 10.1128/jvi.68.11.7507-7515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacAdam A J, Pollard S R, Ferguson G, Skuce R, Wood D, Almond J W, Minor P D. Genetic basis of attenuation of the Sabin type 2 vaccine strain of poliovirus in primates. Virology. 1993;192:18–26. doi: 10.1006/viro.1993.1003. [DOI] [PubMed] [Google Scholar]
- 27.Mascola J R, Snyder S W, Weislow O S, Belay S F, Belshe R B, Schwartz D H, Clements M L, Dolin R, Graham B S, Gorse G J, Keefer M C, McElrath M J, Walker M C, Wagner K F, McNeil J G, McCutchan F E, Burke D S. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 28.Minor P D, Ferguson M, Evans D M, Almond J W, Icenogle J P. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J Gen Virol. 1986;67:1283–1291. doi: 10.1099/0022-1317-67-7-1283. [DOI] [PubMed] [Google Scholar]
- 29.Minor P D. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065–3077. doi: 10.1099/0022-1317-73-12-3065. [DOI] [PubMed] [Google Scholar]
- 30.Modlin J F. Update on enterovirus infections in infants and children. Adv Pediatr Infect Dis. 1997;12:155–180. [PubMed] [Google Scholar]
- 31.Muckelbauer J K, Kremer M, Minor I, Diana G, Dutko F J, Groarke J, Pevear D C, Rossman M G. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure. 1995;3:653–667. doi: 10.1016/s0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 32.Page G S, Mosser A G, Hogle J M, Filman D J, Reuckert R R, Chow M. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J Virol. 1988;63:1781–1794. doi: 10.1128/jvi.62.5.1781-1794.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramsingh A, Slack J, Silkworth J, Hixson A. Severity of disease induced by a pancreatropic coxsackie B4 virus correlates with the H-2Kq locus of the major histocompatibility complex. Virus Res. 1989;14:347–358. doi: 10.1016/0168-1702(89)90027-0. [DOI] [PubMed] [Google Scholar]
- 34.Ramsingh A, Collins D N. A point mutation in the VP4 coding sequence of coxsackievirus B4 influences virulence. J Virol. 1995;69:7278–7281. doi: 10.1128/jvi.69.11.7278-7281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinehart J E, Gomez R M, Roos R P. Molecular determinants for virulence in coxsackievirus B1 infection. J Virol. 1997;71:3986–3991. doi: 10.1128/jvi.71.5.3986-3991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero J R, Rotbart H A. Sequence analysis of the downstream 5′ nontranslated region of seven echoviruses with different neurovirulence phenotypes. J Virol. 1995;69:1370–1375. doi: 10.1128/jvi.69.2.1370-1375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero J R, Price C, Dunn J J. Genetic divergence among the group B coxsackieviruses. In: Tracy S, Chapman N M, Mahy B W J, editors. The coxsackie B viruses. Berlin, Germany: Springer-Verlag; 1997. pp. 97–152. [DOI] [PubMed] [Google Scholar]
- 38.Rossman M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kramer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 39.Sabin A B. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J Infect Dis. 1985;151:420–436. doi: 10.1093/infdis/151.3.420. [DOI] [PubMed] [Google Scholar]
- 40.Sherry B, Reuckert R R. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J Virol. 1985;53:137–143. doi: 10.1128/jvi.53.1.137-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherry B, Mosser A G, Colonno R J, Reuckert R R. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J Virol. 1986;57:246–257. doi: 10.1128/jvi.57.1.246-257.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svitkin Y V, Maslova S V, Agol V I. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology. 1985;147:243–252. doi: 10.1016/0042-6822(85)90127-8. [DOI] [PubMed] [Google Scholar]
- 43.Svitkin Y V, Pestova T V, Maslova S V, Agol V I. Point mutations modify the response of poliovirus RNA to a translation or initiation factor: a comparison of neurovirulence and attenuated strains. Virology. 1988;166:394–404. doi: 10.1016/0042-6822(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 44.Svitkin Y V, Cammack N, Minor P D, Almond J W. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472-U. Virology. 1990;175:103–109. doi: 10.1016/0042-6822(90)90190-3. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes: implication for genetic relationship, gene function, and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 46.Tracy S, Chapman N M, Tu Z. Coxsackievirus B3 from an infectious cDNA is cardiovirulent in mice. Arch Virol. 1992;122:398–409. doi: 10.1007/BF01317202. [DOI] [PubMed] [Google Scholar]
- 47.Tracy S M, Gauntt C J. Phenotypic and genotypic differences among naturally occurring coxsackie virus B3 variants. Eur Heart J. 1987;8(Suppl. J):445–448. [Google Scholar]
- 48.Tu Z, Chapman N M, Hufnagel G, Tracy S, Romero J R, Barry W H, Zhao L, Currey K, Shapiro B. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J Virol. 1995;69:4607–4618. doi: 10.1128/jvi.69.8.4607-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Houten N, Bouchard P E, Moraska A, Huber S A. Selection of an attenuated coxsackievirus B3 variant using a monoclonal antibody reactive to myocyte antigen. J Virol. 1991;65:1286–1290. doi: 10.1128/jvi.65.3.1286-1290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westrop G D, Wareham K A, Evans D M A, Dunn G, Minor P D, Magrath D I, Taffs F, Marsden S, Skinner M A, Schild G C, Almond J W. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J Virol. 1989;63:1338–1344. doi: 10.1128/jvi.63.3.1338-1344.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimmer E, Hellen C U, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 52.Zell R, Sidigi K, Henke A, Schmidt-Brauns J, Hoey E, Martin S, Stelzner A. Functional features of the bovine enterovirus 5′-non-translated region. J Gen Virol. 1999;80:2299–2309. doi: 10.1099/0022-1317-80-9-2299. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H Y, Yousef G E, Cunningham L, Blake N W, OuYang X, Bayston T A, Kandolf R, Archard L C. Attenuation of a reactivated cardiovirulent coxsackievirus B3: the 5′-nontranslated region does not contain major attenuation determinants. J Med Virol. 1993;41:129–137. doi: 10.1002/jmv.1890410208. [DOI] [PubMed] [Google Scholar]