Abstract
Simple Summary
Conifers are vital for both ecological and economic reasons, offering valuable insights into land plant evolution. Molecular phylogenetics plays a significant role in studying evolution, but research on conifers using large-scale data from multiple nuclear genes has been limited. Target enrichment sequencing has emerged as a crucial method in phylogenomic studies. However, a specific bait set for conifers is missing. The REMcon probe set targets around 100 single-copy nuclear loci for family- and species-level phylogenetic studies of conifers. High target recovery and read coverage were observed for the REMcon when tested on 69 species, including conifers and other gymnosperm taxa. Phylogenetic analysis based on the DNA sequences generated from REMcon recovered the existing understanding of conifer relationships. The REMcon bait set will be beneficial in generating large-scale nuclear data consistently for any conifer lineage.
Abstract
Conifers are an ecologically and economically important seed plant group that can provide significant insights into the evolution of land plants. Molecular phylogenetics has developed as an important approach in evolutionary studies, although there have been relatively few studies of conifers that employ large-scale data sourced from multiple nuclear genes. Target enrichment sequencing (target capture, exon capture, or Hyb-Seq) has developed as a key approach in modern phylogenomic studies. However, until now, there has been no bait set that specifically targets the entire conifer clade. REMcon is a target sequence capture probe set intended for family- and species-level phylogenetic studies of conifers that target c. 100 single-copy nuclear loci. We tested the REMcon probe set using 69 species, including 44 conifer genera across six families and four other gymnosperm taxa, to evaluate the efficiency of target capture to efficiently generate comparable DNA sequence data across conifers. The recovery of target loci was high, with, on average, 94% of the targeted regions recovered across samples with high read coverage. A phylogenetic analysis of these data produced a well-supported topology that is consistent with the current understanding of relationships among conifers. The REMcon bait set will be useful in generating relatively large-scale nuclear data sets consistently for any conifer lineage.
Keywords: conifers, gymnosperms, target capture sequencing, probe design, phylogenomics
1. Introduction
Conifers are the largest extant group among gymnosperms, with more than 722 species in 72 genera and 7 families, e.g., Araucariaceae, Cupressaceae, Pinaceae, Podocarpaceae, Sciadopityaceae, Cephalotaxaceae, and Taxaceae [1,2,3]. They are of great economic, ecological, and evolutionary significance, comprising approximately 39% of the world’s forests, and have a fossil record spanning more than 300 million years [4,5,6,7]. The complete understanding of conifer diversity, trait evolutions, genetic structure, and evolutionary history is still poorly explored [3,5,6]. Molecular phylogenetic studies play an important role in understanding the mode and tempo of evolution amongst conifers, but to date, most studies have applied a limited range of markers, principally a small number of chloroplast loci plus nuclear ribosomal DNA regions typically generated by direct amplicon sequencing (e.g., [5,8,9,10,11,12]). These studies have leveraged DNA direct amplicon sequencing data generated from these loci for phylogenetic and evolutionary analyses. Further complicating the study of molecular evolution in this major land plant lineage is the large genome size and overall complexity of conifer genomes [13,14], leaving a notable gap in the exploration of conifers using large-scale data. Target enrichment by hybridization capture (e.g., hyb-seq; Weitemier et al. 2014) [15] provides an efficient and cost-effective approach for generating DNA sequence data for a large number of single and low-copy nuclear gene regions across multiple samples.
Target enrichment approaches (File S1) have become the method of choice for many systematics, phylogenetic, and evolutionary studies in plants [16,17,18,19,20,21], in part fostered by the availability of ‘universal’ probe sets that can recover a common set of genes across broad evolutionary timescales. These include angiosperm-specific probe sets that target hundreds of nuclear genes [17,22] and one recently developed to enrich more than 400 nuclear genes across flagellate land plants [18]. While there are bait kits developed specifically for specific conifer families (e.g., Pinaceae; [23]), we are not aware of a conifer-specific bait set that targets the entire clade.
Here we present a new molecular toolkit (REMcon) which, based upon published transcriptomic (The 1000 Plant Transcriptomes Initiative, 1KP; [24]) and genomic data [25,26], uses RNA baits to target approximately 100 low-copy nuclear genes across conifers. In the present study, we demonstrate the universality and application of these new molecular tools for reconstructing phylogenetic relationships among conifers based on a broad sample of gymnosperm taxa. The approach typically recovers conservative coding regions plus more variable non-coding regions that flank the exons and has application for evolutionary analyses among closely related species, as we will demonstrate in an upcoming study of the Podocarpus ‘Australis’ clade (Khan et al., in prep) of family Podocarpaceae [6,27,28].
2. Materials and Methods
2.1. Probe Design
Target enrichment probes were designed using genes selected from Duarte et al. (2010) [29], who identified a set of orthologous low-copy nuclear genes shared across angiosperms (Arabidopsis, Populus, Vitis and Oryza). For each of the selected genes, we extracted the putatively orthologous coding sequence (CDS) from the spruce (Picea abies, Pinaceae: https://plantgenie.org/, accessed on 4 April 2021) and western red cedar (Thuja plicata; https://phytozome-next.jgi.doe.gov/info/Tplicata_v3_1, accessed on 14 May 2021) genomes. These were used to retrieve putatively homologous transcript sequences for conifers from 1KP (https://www.onekp.com, accessed on 22 April 2021) using the China National GenBank (https://db.cngb.org, accessed on 12 April 2021) BLAST portal and the following settings: Discontiguous Mega-Blast, expect value = 10, maximum target sequences = 1000, selected organisms = Pinidae (taxid: 3313).
The sequences retrieved from the BLAST search for each target gene were downloaded and made into a BLAST database in Geneious (Kearse et al. 2012; https://www.geneious.com, accessed on 1 August 2021) [30]. We queried each BLAST database using the Thuja plicata gene family member with exon annotations manually added and the following settings: Discontiguous Mega-Blast, expect value = 10, maximum target sequences = 1200, results = Hit Table, retrieve = Matching Region with Annotation. We then extracted all sequences matching one or more exon annotations in P. abies with the caveat that the exon was >180 bp in length to allow for bait tiling. The extracted sequences were clustered using CD-HIT-EST ([31,32,33]; http://weizhong-lab.ucsd.edu/cdhit_suite, accessed on 4 August 2021) with a sequence identity cut-off fraction of 0.88 (see [34,35]) and a length similarity fraction of 0.2, and one representative sequence (the longest) per cluster was selected. A total of 1,124 representative sequences (mean length 1051 nucleotides, range 181–4416 nt) covering exons from 100 putative low-copy nuclear genes (Table 1; Table S1) were used for bait design with 120-nt baits and ~2× flexible tiling density for a total of 17,982 baits (see baits-Spruce [14853] for the nucleotide sequences of the baits). Bait design and synthesis were performed by Daicel Arbor Biosciences (formerly MYcroarray; Ann Arbor, MI, USA) in the generation of the myBaits Custom DNA-Seq kit™ Ann Arbor, MI, USA used for target enrichment-based next-generation sequencing.
Table 1.
Targeted nuclear gene regions and the length of the probe sequences.
| S# | Picea abies Gene Name | Arabidopsis thaliana Putative Homolog | Length of the Probe Sequences |
|---|---|---|---|
| 1 | MA_10437158 | AT5G06430 | 195 |
| 2 | MA_10437143 | AT1G12370 | 480 |
| 3 | MA_10437077 | AT5G02250 | 621 |
| 4 | MA_10437070 | AT5G10920 | 720 |
| 5 | MA_10436603 | AT1G03750 | 945 |
| 6 | MA_10436489 | AT4G37510 | 878 |
| 7 | MA_10435966 | AT4G38890 | 822 |
| 8 | MA_10435879 | AT2G33630 | 613 |
| 9 | MA_10435851 | AT5G04520 | 510 |
| 10 | MA_10435433 | AT2G44760 | 426 |
| 11 | MA_10435005 | AT2G40570 | 787 |
| 12 | MA_10434812 | AT1G36310 | 1088 |
| 13 | MA_10434753 | AT1G49380 | 539 |
| 14 | MA_10433768 | AT2G31955 | 942 |
| 15 | MA_10433107 | AT5G64150 | 825 |
| 16 | MA_10432498 | AT1G74640 | 453 |
| 17 | MA_10431375 | AT2G24830 | 321 |
| 18 | MA_10430781 | AT4G35910 | 432 |
| 19 | MA_10429426 | AT1G30070 | 240 |
| 20 | MA_10428930 | AT1G15390 | 256 |
| 21 | MA_10428614 | AT2G34640 | 259 |
| 22 | MA_10428345 | AT1G57770.1 | 315 |
| 23 | MA_10428134 | AT2G04560 | 273 |
| 24 | MA_10427767 | AT1G21370 | 291 |
| 25 | MA_10427729 | AT5G67530 | 1224 |
| 26 | MA_10427590 | AT1G17160 | 480 |
| 27 | MA_10427203 | AT2G36740 | 543 |
| 28 | MA_10426631 | AT4G36390 | 1533 |
| 29 | MA_10426581 | AT2G33450 | 231 |
| 30 | MA_10426376 | AT2G38270 | 504 |
| 31 | MA_9578808 | AT4G18372 | 387 |
| 32 | MA_9514062 | AT5G20220 | 315 |
| 33 | MA_9503281 | AT1G48175 | 257 |
| 34 | MA_8815984 | AT2G346401 | 693 |
| 35 | MA_8715484 | AT4G38020 | 501 |
| 36 | MA_8687206 | AT4G26980 | 408 |
| 37 | MA_8286794 | AT3G17170 | 342 |
| 38 | MA_8140147 | AT2G28605 | 480 |
| 39 | MA_7890741 | AT2G44660 | 783 |
| 40 | MA_5587080 | AT4G20060 | 447 |
| 41 | MA_957334 | AT1G05055 | 462 |
| 42 | MA_945784 | AT5G06410 | 380 |
| 43 | MA_939779 | AT4G27390 | 468 |
| 44 | MA_938037 | AT5G49570 | 580 |
| 45 | MA_894439_ | AT2G30100 | 1306 |
| 46 | MA_824260 | AT4G28020 | 441 |
| 47 | MA_762004 | AT1G28560 | 675 |
| 48 | MA_759516 | AT5G08720 | 461 |
| 49 | MA_749379 | AT4G11980 | 201 |
| 50 | MA_587488 | AT4G01040 | 377 |
| 51 | MA_546546 | AT4G17760 | 252 |
| 52 | MA_537299 | AT5G54840 | 264 |
| 53 | MA_458270 | AT5G06830 | 690 |
| 54 | MA_388031 | AT2G20330 | 486 |
| 55 | MA_341112 | AT5G11980 | 276 |
| 56 | MA_332596 | AT2G34460 | 333 |
| 57 | MA_314789 | AT1G56345.1 | 603 |
| 58 | MA_261436 | AT4G33030 | 1290 |
| 59 | MA_253636 | AT3G51050 | 768 |
| 60 | MA_225872 | AT5G14260 | 456 |
| 61 | MA_224167 | AT2G20790 | 900 |
| 62 | MA_199851 | AT3G01660 | 350 |
| 63 | MA_196209 | AT4G36530 | 273 |
| 64 | MA_187402 | AT4G31460 | 471 |
| 65 | MA_173127 | AT4G28740 | 548 |
| 66 | MA_159115 | AT2G27600 | 1191 |
| 67 | MA_159115 | AT4G27600 | 1056 |
| 68 | MA_127668 | AT3G15290 | 465 |
| 69 | MA_123340 | AT2G19870 | 1137 |
| 70 | MA_121485 | AT1G02410 | 749 |
| 71 | MA_121026 | AT1G08460 | 570 |
| 72 | MA_106933 | AT2G266801 | 636 |
| 73 | MA_104872 | AT3G26580 | 507 |
| 74 | MA_99242 | AT4G29070 | 412 |
| 75 | MA_98424 | AT1G07130 | 558 |
| 76 | MA_95157 | AT5G09820 | 292 |
| 77 | MA_83545 | AT5G65860 | 514 |
| 78 | MA_78599 | AT2G40760 | 252 |
| 79 | MA_73742 | AT2G21840 | 939 |
| 80 | MA_73742 | AT1G21840 | 939 |
| 81 | MA_67861 | AT2G26680 | 369 |
| 82 | MA_66902 | AT2G36145 | 234 |
| 83 | MA_66902 | AT2G34145 | 234 |
| 84 | MA_63465 | AT3G24315 | 290 |
| 85 | MA_61548 | AT1G65030 | 681 |
| 86 | MA_55048 | AT5G19130 | 858 |
| 87 | MA_43083 | AT5G48330 | 717 |
| 88 | MA_41847 | AT3G03790 | 303 |
| 89 | MA_35149 | AT3G02300 | 431 |
| 90 | MA_34295 | AT1G43580 | 378 |
| 91 | MA_30194 | AT5G16210 | 369 |
| 92 | MA_29076 | AT3G57910 | 513 |
| 93 | MA_26068 | AT2G37560 | 414 |
| 94 | MA_25177 | AT1G07970 | 472 |
| 95 | MA_24252 | AT4G24090 | 600 |
| 96 | MA_19954 | AT2G02590 | 414 |
| 97 | MA_11407 | AT3G47860 | 312 |
| 98 | MA_10909 | AT2G04270 | 318 |
| 99 | MA_6888 | AT3G24080 | 2286 |
| 100 | MA_4586 | AT2G22650 | 303 |
2.2. Taxon Selection
A total of 44 conifer genera representing six families, three species of Cycadales, and Gingko biloba (69 taxa) were included to evaluate the efficiency of target capture across conifers and more widely among gymnosperms. Most plant specimens were freshly collected from the living collections held at the Botanic Gardens of South Australia and dried in silica gel, and some were sampled from preserved specimens held at the State Herbarium of South Australia (Table S2).
2.3. DNA Extraction, Library Preparation, Hybrid Capture and Sequencing
For DNA extractions, about 15 mg of silica gel dried leaf material per sample was used, and homogenized in a Omni ruptor (Omni International, Kennesaw, GA, USA) using ceramic beads. DNA was extracted using the Qiagen Plant Mini kit, QIAGEN, Germantown, MD, USA and normalized 2 ng/uL before proceeding to library preparation, which follows the steps outlined in Waycott et al. [36] for their nuclear bait set. Hybrid capture was performed following the manual provided by myBaits with a hybridization temperature of 65 °C and 150 bp paired-end sequencing was performed at the Australian Genome Research Facility (AGRF), Melbourne, Australia on an Illumina NovaSeq S1 flow cell.
2.4. Bioinformatics Analyses
High-throughput paired-end reads were de-multiplexed and quality trimmed (using a Phred score threshold of 20) using CLC Genomics Workbench (v. 20; https://www.qiagenbioinformatics.com/, accessed on 22 April 2022). The Sequence Capture Processor pipeline SECAPR v 2.2.3: Andermann et al. [37], http://antonellilab.github.io/seqcap_processor/, accessed on 24 April 2022) was used to generate nuclear DNA sequence data sets from the trimmed reads. First, the reads from each sample were assembled de novo using SPAdes [38] with default kmer values. Contigs matching the reference Thuja plicata reference sequences (i.e., annotated exons, see above) were extracted from the de novo assemblies with LASTZ v. 1.04 [39] using a target length of 0.5 and a similarity fraction of 0.75 (i.e., 50% of the contig has to overlap with the target gene and be no less than 75% similar). The ‘keep paralogs’ flag was activated and deactivated to assess the extent of paralogy in the data. SECAPR identifies paralogs as multiple overlapping contigs matching a target sequence, keeping the longest contig if the ‘keep paralogs’ flag is activated. The extracted contigs were aligned per locus to produce multiple sequence alignments (MSA) using MAFFT [40]. The aligned contigs were subsequently used for a reference-based assembly using the BWA read mapper v.0.7.16a-r1181 [41], and the ‘sample specific’ flag, i.e., each sample is extracted from the alignment and mapped separately. Consensus sequences per sample from subsequent read mappings were again aligned using MAFFT to produce MSAs for each targeted gene region. The approach developed by Yang and Smith [42] and modified with containerization for target capture data (Jackson et al. [43]; https://github.com/chrisjackson-pellicle/Yang-and-Smith-paralogy-resolution, accessed on 29 April 2022) was used to resolve groups of orthologous sequences (orthology inference) from targeted gene regions. Following various filtering steps, the approach uses phylogenetic tree-based methods and the pruning of duplicated taxa from rooted phylogenies to resolve orthologous groups of sequences. In this study, de novo contigs for each sample from the SECAPR pipeline (above) were first imported into Geneious Prime (v. 2022.0.1; (https://www.geneious.com, accessed on 24 May 2022) and made into a Blast database. This database was queried using the extracted contigs from an outgroup (Ginkgo biloba) matching the targeted gene regions in P. abies. The contigs from Ginkgo were annotated with the CDS from P. abies, and the coding region(s) were queried against the Blast database using blast-n with a maximum expected value of 1e-10 and maximum hits set to 1000. The Blast output was filtered using a minimum coverage fraction (query coverage of at least 0.4) to remove poorly aligned and short sequence fragments [42]. The resulting contigs were then used as input into the Yang-and-Smith-paralogy-resolution pipeline [43]. We used the monophyletic outgroups (MO) method to identify ortholog groups using reference genes from Gingko biloba as the outgroup. For downstream analyses, we retained alignments with >10 individuals in order to reduce the influence of missing data in tree inference. The aligned ortholog groups were concatenated, and a phylogeny was generated using IQ-tree 2 [44] using model finder [45] to estimate the optimum partitioning scheme and partition-specific nucleotide substitution model (MFP+MERGE flag activated) and 1000 ultrafast bootstrap [46] replicates to assess branch support.
3. Results and Discussion
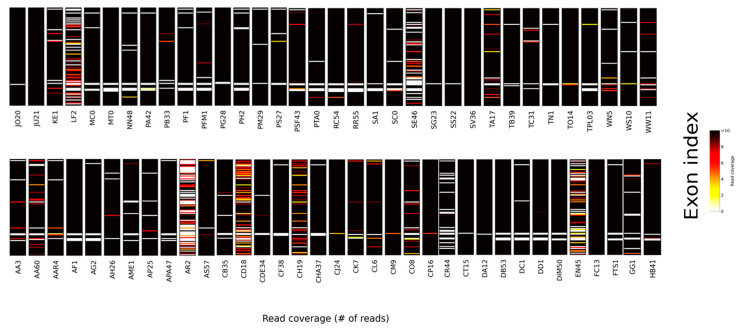
The retrieval of target loci across the conifers was high, with, on average, 94% of the targeted gene regions recovered per sample (range 53–100), and 27 loci were recovered across all samples (Table 2). For the recovered loci, read coverage (read depth/position, averaged across loci) was also high, averaging c.146 across the included taxa with a maximum of 622 in Chamaecyparis pisifera and a minimum of c. 6 in Agathis robusta (Figure 1—coverage heatmap). In general, the recovery of genes across the six conifer families was relatively consistent, and with the exception of Araucariaceae, the mean number of loci captured per family exceeds 90 (Table 2). The lower mean value for Araucariaceae (87 loci) is influenced by the poor recovery for Agathis robusta (57 loci), which is likely a consequence of DNA quality and/or issues with the library construction, given that target recovery amongst close relatives (e.g., Agathis microstachya, 94 loci) was high (Figure 1, Table S2). There was a lower recovery of target genes for the non-coniferous gymnosperm species (c. 80% of genes recovered across the four samples), although this is not unexpected given that these were not specifically targeted in the probe design. Furthermore, the identity of the target sequences (here, sourced from Thuja) could influence locus recovery with increasing evolutionary distance, and an approach similar to McLay et al. [47] may be valuable in increasing target locus recovery from specific lineages.
Table 2.
Recovery of targeted gene regions, including potentially paralogous loci, across conifer families and four non-conifer gymnosperms. Locus recovery is averaged across samples (n), and the minimum and maximum recovery per sample is indicated.
| Families | N | Average Locus Recovery | Min | Max |
|---|---|---|---|---|
| Araucariaceae | 5 | 85 | 53 | 97 |
| Cupressaceae | 22 | 98 | 89 | 100 |
| Pinaceae | 11 | 96 | 95 | 90 |
| Podocarpaceae | 26 | 93 | 76 | 97 |
| Sciadopityaceae | 1 | 95 | - | - |
| Taxaceae | 3 | 97 | 96 | 97 |
| Non conifer Gymnosperms | 4 | 81 | 75 | 92 |
Figure 1.
Heatmap showing gene recovery and read depth across samples. The gene recovery includes samples that were flagged as potentially paralogous using the SECAPR pipeline. Sample abbreviations as in Table S1.
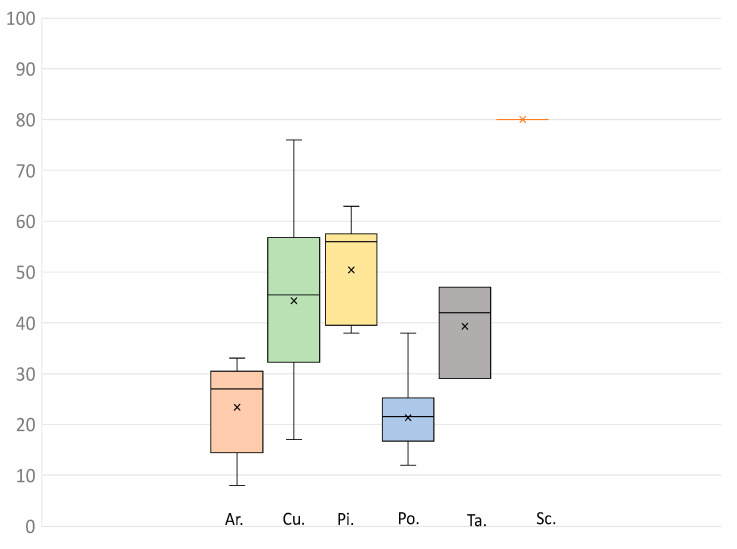
Overall, there was a large number of putatively paralogous gene copies recovered at most loci (c. 36% of loci/sample, averaged across all samples) but this was highly variable among taxa (Prumnopitys andina, Podocarpaceae: c. 13% of recovered loci; Sciadopitys verticillata, Sciadopityaceae: 80% of recovered loci) (Figure 2; Table S2). The extent of paralogy might reflect the generally large genome size of conifers, which is also highly variable, with at least an order of magnitude difference between the smallest and the largest conifer genomes [14]. Polyploidy is a major driver of genome size evolution amongst angiosperms, although until recently [48,49], this phenomenon was thought to be relatively rare among conifers (e.g., [13,50,51]). In addition, conifer genome size evolution has been attributed to other factors, such as a high copy number of long transposable elements (e.g., [25]). The distribution of paralogs in our data supports the view that genome size, per se, is only partly related to the frequency of duplicated genes. For instance, Podocarpaceae has the smallest average genome size [14] and the smallest proportion of paralogs in our data. On the other hand, Pinaceae generally have large genomes, and we found a large proportion of putative paralogs among samples from this family. However, the relatively large genome size among Araucariaceae is not strongly associated with a high number of paralogous genes in our data (Figure 2), suggesting that factors other than gene duplication (e.g., transposable elements, larger introns, and abundant pseudogenes; [13,25,48] are also important drivers of genome size evolution.
Figure 2.
Box and whisker plots showing the recovery of paralogs (number of paralogous genes/sample) by family. Abbreviations: Ar., Araucariaceae; Cu., Cupressaceae; Pi., Pinaceae; Po., Podocarpaceae; Ta., Taxaceae; Sc., Sciadopityaceae. X = Mean, Middle horizontal bar = Median, and the lower bounds of the box are the 75 and 25 quartiles.
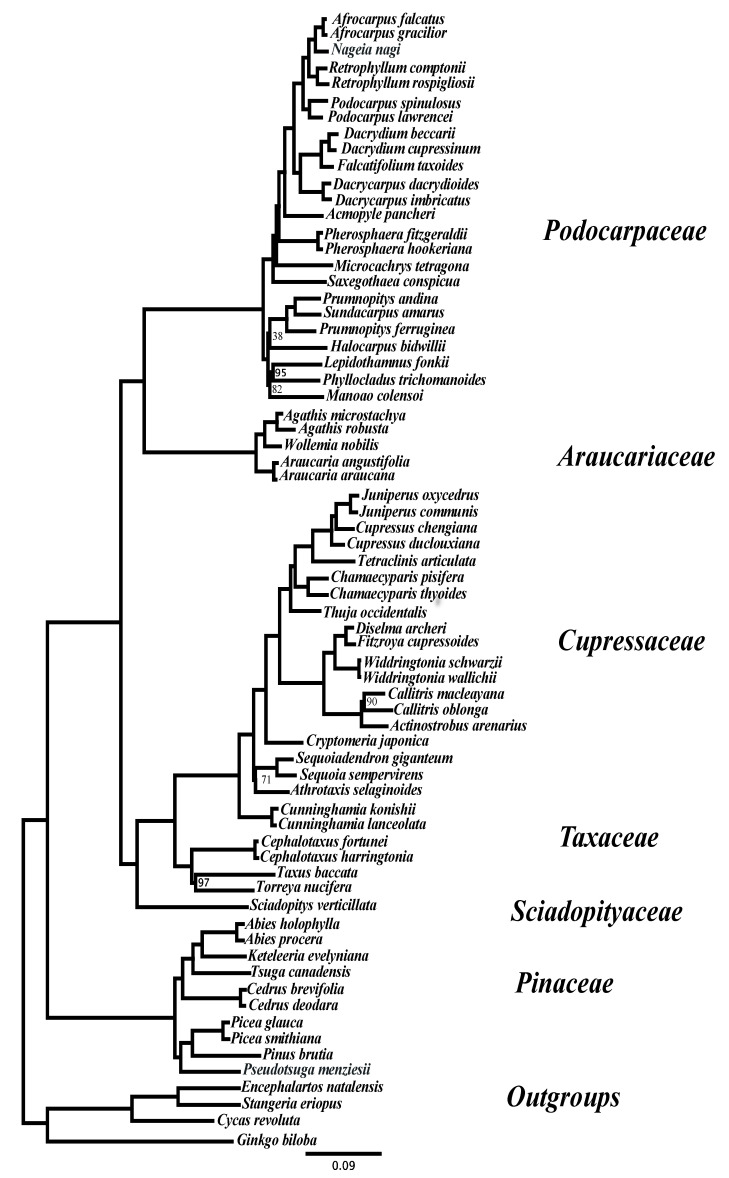
Of the 100 targeted gene regions, 90 were recovered for Gingko, and these were included in the paralogy resolution analyses. Orthology inference recovered 98 MO ortholog groups and 95 with more than 10 samples included, which were retained for phylogenetic inference. The concatenated length of the 95 loci was an average of c. 48,770 bp with an aligned length of c. 74,179 bp and approximately 34% missing values. The average aligned length of the individual loci was c. 780 bp and ranged from 440–1691 bp. The concatenated alignment includes 47,469 (c. 64%) variable positions, of which 36,350 (c. 49%) are parsimony informative and 44,818 (c. 60%) variable and 34,411 (c. 46%) parsimony informative characters within the conifer clade. The maximum likelihood topology inferred from these data is shown in Figure 3. Of the 65 clades recovered, only 7 have a bootstrap support value < 100, and of these, only 3 received less than 80% support (Figure 3). The inferred topology is generally in agreement with our current understanding of conifer relationships (e.g., [2,3,5]), while the poorly supported nodes are associated with short branches and may be inherently difficult to resolve (e.g., [52,53,54]). For example, the relationships within the Prumnopityoid clade of Podocarpaceae, and in particular the placement of Halocarpus, were found to be unstable in the recent analyses of Chen et al. [55] using a large transcriptome data set of c. 1000 nuclear and c. 40 chloroplast gene regions and is poorly resolved here (Figure 3).
Figure 3.
Maximum likelihood phylogeny estimated from the concatenated MO ortholog gene alignments. All branches have maximum bootstrap support unless indicated adjacent to the branch. Sample abbreviations as in Table S1.
4. Conclusions
In conclusion, we present a conifer-specific hybrid-capture bait set that has been shown to perform well in terms of the consistency of locus recovery across a broad range of gymnosperms, and these data can be applied to credibly resolve deep phylogenetic relationships within the conifer clade. As part of ongoing studies (Khan et al. in prep) [6,27,28], we have found the REMcon bait set to be similarly successful in resolving relationships among closely related species groups within Podocarpaceae. The REMcon bait set offers an efficient and relatively cost-effective approach that fills an important gap in conifer and gymnosperm phylogenomics. This hybrid-capture bait set has exciting future applications, including the resolution of complex phylogenetic relationships, population, and comparative genomics, providing valuable insights into the evolution and conservation of conifers and other gymnosperms.
Acknowledgments
We acknowledge the provision of plant material with permission from Adelaide Botanic Garden, Mount Lofty Botanical Garden South Australia, and the State Herbarium of South Australia.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biology13060361/s1, Table S1: Details of targeted genes; Table S2: Sample details; Table S3: Recovery of targeted gene regions across conifer familes and four non conifer gymnosperms. Locus recovery is averaged across samples (n) and the minimum and maximum recovery per seample is indicated. Figure S1: Paralogs/Family; File S1: HybCap76 NEBnext Podocarp03 half plate.
Author Contributions
Conceptualization, R.K., M.W. and E.B.; methodology, R.K., M.W., K.-j.v.D. and E.B.; software, E.B. and R.K.; validation, R.K., M.W., K.-j.v.D., R.S.H., J.L. and E.B.; formal analysis, E.B. and R.K.; investigation, E.B. and R.K.; resources, M.W. and R.S.H.; data curation, R.K and E.B.; writing—original draft preparation, R.K. and E.B.; writing—review and editing, M.W., K.-j.v.D., R.S.H., J.L. and E.B.; supervision, M.W. and R.S.H.; project administration, M.W. and R.S.H.; funding acquisition, M.W. and R.S.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the Botanic Gardens and State Herbarium collaborative student support 2020–2022; collections digitization support from the Board of the Botanic Gardens and State Herbarium (South Australia) 2021–2022; PhD Student support from the University of Adelaide, School of Biological Sciences 2020–2022; and a capacity building grant from The Environment Institute, the University of Adelaide 2021; Key Research Program of Frontier Sciences, CAS (ZDBS-LY-7001), the National Natural Science Foundation of China (42211540718), the Top-notch Young Talents Project of Yunnan Provincial ‘Ten Thousand Talents Program’ (YNWR-QNBJ-2018-146), the Xingdian Talent Support Program of Yunnan Province (XDYC-QNRC-2022-0068), and the CAS ‘Light of West China’ Program. Raees Khan was supported by the Postdoctoral International Exchange Program of the Office of China Postdoctoral Council, the Postdoctoral Targeted Funding, and the Postdoctoral Research Fund of Yunnan Province.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ran J.-H., Gao H., Wang X.-Q. Fast evolution of the retroprocessed mitochondrial rps3 gene in Conifer II and further evidence for the phylogeny of gymnosperms. Mol. Phylogenetics Evol. 2010;54:136–149. doi: 10.1016/j.ympev.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y., Ferguson D.K., Liu B., Mao K.S., Gao L.M., Zhang S.Z., Wan T., Rushforth K., Zhang Z.X. Recent advances on phylogenomics of gymnosperms and an updated classification. Plant Divers. 2022;44:340–350. doi: 10.1016/j.pld.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan R., Hill R.S., Liu J., Biffin E. Diversity, Distribution, Systematics and Conservation Status of Podocarpaceae. Plants. 2023;12:1171. doi: 10.3390/plants12051171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armenise L., Simeone M.C., Piredda R., Schirone B. Validation of DNA barcoding as an efficient tool for taxon identification and detection of species diversity in Italian conifers. Eur. J. For. Res. 2012;131:1337–1353. doi: 10.1007/s10342-012-0602-0. [DOI] [Google Scholar]
- 5.Leslie A.B., Beaulieu J., Holman G., Campbell C.S., Mei W., Raubeson L.R., Mathews S. An overview of extant conifer evolution from the perspective of the fossil record. Am. J. Bot. 2018;105:1531–1544. doi: 10.1002/ajb2.1143. [DOI] [PubMed] [Google Scholar]
- 6.Khan R., Hill R.S., Dörken V.M., Biffin E. Detailed seed cone morpho-anatomy of the Prumnopityoid clade: An insight into the origin and evolution of Podocarpaceae seed cones. Ann. Bot. 2022;130:637–655. doi: 10.1093/aob/mcac097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan R., Hill R.S., Dörken V.M., Biffin E. Detailed Seed Cone Morpho-Anatomy Provides New Insights into Seed Cone Origin and Evolution of Podocarpaceae; Podocarpoid and Dacrydioid Clades. Plants. 2023;12:3903. doi: 10.3390/plants12223903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelch D.G. Phylogeny of Podocarpaceae: Comparison of evidence from morphology and 18S rDNA. Am. J. Bot. 1998;85:986–996. doi: 10.2307/2446365. [DOI] [PubMed] [Google Scholar]
- 9.Conran J.G., Wood G.M., Martin P.G., Dowd J.M., Quinn C.J., Gadek P.A., Price R.A. Generic relationships within and between the gymnosperm families Podocarpaceae and Phyllocladaceae based on an analysis of the chloroplast gene rbcL. Aust. J. Bot. 2000;48:715–724. doi: 10.1071/BT99062. [DOI] [Google Scholar]
- 10.Sinclair W., Mill R., Gardner M., Woltz P., Jaffré T., Preston J., Hollingsworth M., Ponge A., Möller M. Evolutionary relationships of the New Caledonian heterotrophic conifer, Parasitaxus usta (Podocarpaceae), inferred from chloroplast trn LF intron/spacer and nuclear rDNA ITS2 sequences. Plant Syst. Evol. 2002;233:79–104. doi: 10.1007/s00606-002-0199-8. [DOI] [Google Scholar]
- 11.Knopf P., Schulz C., Little D.P., Stützel T., Stevenson D.W. Relationships within Podocarpaceae based on DNA sequence, anatomical, morphological, and biogeographical data. Cladistics. 2012;28:271–299. doi: 10.1111/j.1096-0031.2011.00381.x. [DOI] [PubMed] [Google Scholar]
- 12.Little D.P., Knopf P., Schulz C. DNA barcode identification of Podocarpaceae—The second largest conifer family. PLoS ONE. 2013;8:e81008. doi: 10.1371/journal.pone.0081008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahuja M.R., Neale D.B. Evolution of genome size in conifers. Silvae Genet. 2005;54:126–137. doi: 10.1515/sg-2005-0020. [DOI] [Google Scholar]
- 14.Zonneveld B.J.M. Conifer genome sizes of 172 species, covering 64 of 67 genera, range from 8 to 72 picogram. Nord. J. Bot. 2012;30:490–502. doi: 10.1111/j.1756-1051.2012.01516.x. [DOI] [Google Scholar]
- 15.Weitemier K., Straub S.C., Cronn R.C., Fishbein M., Schmickl R., McDonnell A., Liston A. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2014;2:1400042. doi: 10.3732/apps.1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vatanparast M., Powell A., Doyle J.J., Egan A.N. Targeting legume loci: A comparison of three methods for target enrichment bait design in Leguminosae phylogenomics. Appl. Plant Sci. 2018;6:e1036. doi: 10.1002/aps3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson M.G., Pokorny L., Dodsworth S., Botigue L.R., Cowan R.S., Devault A., Eiserhardt W.L., Epitawalage N., Forest F., Kim J.T., et al. A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol. 2019;68:594–606. doi: 10.1093/sysbio/syy086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breinholt J.W., Carey S.B., Tiley G.P., Davis E.C., Endara L., McDaniel S.F., Neves L.G., Sessa E.B., von Konrat M., Chantanaorrapint S., et al. A target enrichment probe set for resolving the flagellate land plant tree of life. Appl. Plant Sci. 2021;9:e11406. doi: 10.1002/aps3.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah T., Schneider J.V., Zizka G., Maurin O., Baker W., Forest F., Brewer G.E., Savolainen V., Darbyshire I., Larridon I. Joining forces in Ochnaceae phylogenomics: A tale of two targeted sequencing probe kits. Am. J. Bot. 2021;108:1201–1216. doi: 10.1002/ajb2.1682. [DOI] [PubMed] [Google Scholar]
- 20.Baker W., Dodsworth S., Forest F., Graham S., Johnson M., McDonnell A., Pokorny L., Tate J.A., Wicke S., Wickett N. Exploring Angiosperms353: An open, community toolkit for collaborative phylogenomic research on flowering plants. Am. J. Bot. 2021;108:1059–1065. doi: 10.1002/ajb2.1703. [DOI] [PubMed] [Google Scholar]
- 21.Zuntini A.R., Carruthers T., Maurin O., Bailey P.C., Leempoel K., Brewer G.E., Epitawalage N., Françoso E., Gallego-Paramo B., Baker W.J., et al. Phylogenomics and the rise of the angiosperms. Nature. 2024. Online ahead of print . [DOI] [PMC free article] [PubMed]
- 22.Léveillé-Bourret É., Starr J.R., Ford B.A., Moriarty Lemmon E., Lemmon A.R. Resolving rapid radiations within angiosperm families using anchored phylogenomics. Syst. Biol. 2018;67:94–112. doi: 10.1093/sysbio/syx050. [DOI] [PubMed] [Google Scholar]
- 23.Montes J.R., Peláez P., Willyard A., Moreno-Letelier A., Piñero D., Gernandt D.S. Phylogenetics of Pinus subsection Cembroides Engelm. (Pinaceae) inferred from low-copy nuclear gene sequences. Syst. Bot. 2019;44:501–518. doi: 10.1600/036364419X15620113920563. [DOI] [Google Scholar]
- 24.Leebens-Mack J.H., Barker M.S., Carpenter E.J., Deyholos M.K., Gitzendanner M.A., Graham S.W., Grosse I., Li Z., Melkonian M., Mirarab S., et al. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nystedt B., Street N.R., Wetterbom A., Zuccolo A., Lin Y.C., Scofield D.G., Vezzi F., Delhomme N., Giacomello S., Jansson S., et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 26.Shalev T.J., El-Dien O.G., Yuen M.M., Shengqiang S., Jackman S.D., Warren R.L., Coombe L., van der Merwe L., Stewart A., Bohlmann J., et al. The western redcedar genome reveals low genetic diversity in a self-compatible conifer. Genome Res. 2022;32:1952–1964. doi: 10.1101/gr.276358.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan R., Hill R.S. Reproductive and leaf morpho-anatomy of the Australian alpine podocarp and comparison with the Australis subclade. Bot. Lett. 2022;169:237–249. doi: 10.1080/23818107.2022.2042381. [DOI] [Google Scholar]
- 28.Khan R., Hill R.S. Morpho-anatomical affinities and evolutionary relationships of three paleoendemic podocarp genera based on seed cone traits. Ann. Bot. 2021;128:887–902. doi: 10.1093/aob/mcab113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duarte J.M., Wall P.K., Edger P.P., Landherr L.L., Ma H., Pires P.K., Leebens-Mack J., dePamphilis C.W. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 2010;10:1–18. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., Jaroszewski L., Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 32.Li W., Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y., Niu B., Gao Y., Fu L., Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock-Hanser B.L., Frey A., Leslie M.S., Dutton P.H., Archer F.I., Morin P.A. Targeted multiplex next-generation sequencing: Advances in techniques of mitochondrial and nuclear DNA sequencing for population genomics. Mol. Ecol. Resour. 2013;13:254–268. doi: 10.1111/1755-0998.12059. [DOI] [PubMed] [Google Scholar]
- 35.Hugall A.F., O’Hara T.D., Hunjan S., Nilsen R., Moussalli A. An exon-capture system for the entire class Ophiuroidea. Mol. Biol. Evol. 2015;33:281–294. doi: 10.1093/molbev/msv216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waycott M., van Dijk J.K., Biffin E. A hybrid capture RNA bait set for resolving genetic and evolutionary relationships in angiosperms from deep phylogeny to intraspecific lineage hybridization. bioRxiv. 2022 doi: 10.1101/2021.09.06.456727. [DOI] [Google Scholar]
- 37.Andermann T., Cano Á., Zizka A., Bacon C., Antonelli A. SECAPR—A bioinformatics pipeline for the rapid and user-friendly processing of targeted enriched Illumina sequences, from raw reads to alignments. PeerJ. 2018;6:e5175. doi: 10.7717/peerj.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris R.S. Ph.D. Thesis. The Pennsylvania State University; State College, PA, USA: 2007. Improved Pairwise Alignment of Genomic DNA. [Google Scholar]
- 40.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Smith S.A. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: Improving accuracy and matrix occupancy for phylogenomics. Mol. Biol. Evol. 2014;31:3081–3092. doi: 10.1093/molbev/msu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson C., McLay T., Schmidt-Lebuhn A.N. hybpiper-nf and paragone-nf: Containerization and additional options for target capture assembly and paralog resolution. Appl. Plant Sci. 2023;11:e11532. doi: 10.1002/aps3.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A., Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S., Minh B.Q., Wong T.K., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang D.T., Chernomor O., Von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLay T.G., Birch J.L., Gunn B.F., Ning W., Tate J.A., Nauheimer L., Joyce E.M., Simpson L., Schmidt-Lebuhn A.N., Baker W.J., et al. New targets acquired: Improving locus recovery from the Angiosperms353 probe set. Appl. Plant Sci. 2021;9:e11420. doi: 10.1002/aps3.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z., Baniaga A.E., Sessa E.B., Scascitelli M., Graham S.W., Rieseberg L.H., Barker M.S. Early genome duplications in conifers and other seed plants. Sci. Adv. 2015;1:e1501084. doi: 10.1126/sciadv.1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stull G.W., Qu X.J., Parins-Fukuchi C., Yang Y.Y., Yang J.B., Yang Z.Y., Hu Y., Ma H., Soltis P.S., Soltis D.E., et al. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nat. Plants. 2021;7:1015–1025. doi: 10.1038/s41477-021-00964-4. [DOI] [PubMed] [Google Scholar]
- 50.Murray B.G. Nuclear DNA amounts in gymnosperms. Ann. Bot. 1998;82((Suppl. 1)):3–15. doi: 10.1006/anbo.1998.0764. [DOI] [Google Scholar]
- 51.Kinlaw C.S., Neale D.B. Complex gene families in pine genomes. Trends Plant Sci. 1997;2:356–359. doi: 10.1016/S1360-1385(97)84624-9. [DOI] [Google Scholar]
- 52.Philippe H., Brinkmann H., Lavrov D.V., Littlewood D.T.J., Manuel M., Wörheide G., Baurain D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitfield J.B., Lockhart P.J. Deciphering ancient rapid radiations. Trends Ecol. Evol. 2007;22:258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Mongiardino Koch N. Phylogenomic subsampling and the search for phylogenetically reliable loci. Mol. Biol. Evol. 2021;38:4025–4038. doi: 10.1093/molbev/msab151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L., Jin W.T., Liu X.Q., Wang X.Q. New insights into the phylogeny and evolution of Podocarpaceae inferred from transcriptomic data. Mol. Phylogenetics Evol. 2022;166:107341. doi: 10.1016/j.ympev.2021.107341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article or Supplementary Materials.