Abstract
Hepatitis C virus (HCV) is a major human pathogen causing chronic liver disease. We have recently found that the large extracellular loop (LEL) of human CD81 binds HCV. This finding prompted us to assess the structure-function features of HCV-CD81 interaction by using recombinant E2 protein and a recombinant soluble form of CD81 LEL. We have found that HCV-E2 binds CD81 LEL with a Kd of 1.8 nM; CD81 can mediate attachment of E2 on hepatocytes; engagement of CD81 mediates internalization of only 30% of CD81 molecules even after 12 h; and the four cysteines of CD81 LEL form two disulfide bridges, the integrity of which is necessary for CD81-HCV interaction. Altogether our data suggest that neutralizing antibodies aimed at interfering with HCV binding to human cells should have an affinity higher than 10−9 M, that HCV binding to hepatocytes may not entirely depend on CD81, that CD81 is an attachment receptor with poor capacity to mediate virus entry, and that reducing environments do not favor CD81-HCV interaction. These studies provide a better understanding of the CD81-HCV interaction and should thus help to elucidate the viral life cycle and to develop new strategies aimed at interfering with HCV binding to human cells.
Hepatitis C virus (HCV) is a positive-strand RNA virus belonging to the Flaviviridiae family (7). It has been estimated that 170 million people worldwide are chronically infected with HCV (18). Generally, HCV infection becomes chronic and may have very serious outcomes such as hepatitis, cirrhosis, and hepatocarcinoma. Although HCV was identified molecularly more than a decade ago (5), the virus has not been isolated nor have reliable in vitro systems for viral propagation been described, reverse transcription-PCR (RT-PCR) being the only way to detect HCV. Recently, we have shown that a bona fide HCV particle, i.e., HCV RNA associated with envelope, specifically binds human CD81 as demonstrated by quantitative PCR (14).
CD81 is a membrane-associated protein belonging to the family of tetraspanins (10). Like all tetraspanins, CD81 is organized in four highly hydrophobic transmembrane domains, which force the protein to traverse the membrane four times, creating two hydrophilic domains, a small one and a large one, protruding out of the cells. We have found that the large extracellular loop (LEL) of CD81 is sufficient to bind HCV via interaction with the major virus envelope protein E2 (14). Remarkably, chimpanzee sera containing antienvelope antibodies, which are capable of preventing HCV infection in vivo, inhibit the binding of HCV to CD81 in vitro (14, 16), supporting the idea that CD81 represents a cellular receptor for the virus.
In this work we have studied the HCV-CD81 interaction in more detail. First, we determined the affinity constant for binding of soluble CD81 LEL and monomeric HCV E2 by using highly purified recombinant LEL and E2 proteins. Second, we assessed the binding of recombinant E2 on fresh hepatocytes and hepatocarcinoma cell lines. Third, we quantitated the ability of cell surface-associated CD81 to mediate internalization of bound ligand. Finally, since CD81, like all tetraspanins, carries four cysteines in the large extracellular loop, we have investigated the role of disulfide bridging in E2 binding by using both genetic and biochemical approaches.
MATERIALS AND METHODS
Cloning and expression of CD81 LEL.
For the expression of CD81 LEL as a glutathione S-transferase (GST) fusion, LEL was amplified using Vent DNA polymerase (New England Biolabs), plasmid pCDM8-CD81 as template, and the primers 5′-CAAAAGGAATTCTATTTGTCATCAACAAGGACCAGATCGCCAAGG-3′ and 5′-CCCCAAGCTTTCAATGATGATGATGATGATGCAGCTTCCCGGAGAAG-3′. Both the amplified product and plasmid pGEX-KG (American Type Culture Collection) were cleaved with EcoRI and HindIII (Boehringer, Mannheim, Germany) and ligated. Plasmid pGST-LEL was obtained from the transformation of Escherichia coli TG1 (17) with the ligase mixture.
Purification of CD81 LEL.
E. coli TG1(pGST-LEL) cells were induced for 3 h and disrupted with a French press (Spectronic Instruments, Rochester, N.Y.). The protein solution was loaded onto a glutathione-Sepharose 4B column (Pharmacia Biotech, Uppsala, Sweden), and the retained proteins were eluted with 50 mM Tris-HCl–10 mM glutathione (pH 8.0). The eluted proteins were digested at 25°C for 7 h with thrombin (Pharmacia) at a protein/enzyme ratio of 100:1 (wt/wt) and applied again on the glutathione-Sepharose 4B column. The nonretained material was loaded onto a Ni2+-chelating Sepharose FF column (Pharmacia); the LEL domain was eluted with 20 mM sodium phosphate buffer–500 mM imidazole (pH 7.8) and finally loaded onto a Superdex 75 High Load column (Pharmacia). The total protein concentration was evaluated by the Bradford method (2).
Preparation of HCV E2 envelope protein.
The HCV E2 protein used throughout this study was a clinical-grade batch prepared by Chiron Co. (Emeryville, Calif.). Briefly, the protein was prepared from a CHO cell line stably transfected with plasmid pCMVa120 (4) in which the E2 sequence from amino acids 383 to 715 was fused to the tissue plasminogen activator leader sequence. After cell disruption and debris removal by microfiltration (30-kDa cutoff; Millipore), the protein was purified by three subsequent chromatographic steps; lectin affinity chromatography, hydroxyapatite chromatography, and ion-exchange chromatography.
Affinity study of CD81-HCV E2 interaction.
E2 binding to CD81 was studied in 150 mM NaCl–10 mM Tris-HCl (pH 7.4) at either 25 or 37°C. The quenching of the intrinsic tryptophan fluorescence of E2 was monitored as a function of the LEL concentration in a SPEX-Fluoromax spectrofluorometer. The protein emission spectra were collected between 300 and 450 nm, using an excitation wavelength of 280 nm. The titrations were carried out at 347 nm by acquiring the fluorescence intensity at LEL concentrations ranging from 0 to 400 nM. The fluorescence intensity was corrected for the contribution of buffer and for protein dilution as already described (1, 6). To compensate the decrease in fluorescence due to the repeated exposure of the sample to a high-intensity light beam, all measurements were corrected with a control experiment where E2 was titrated with buffer alone. The data were analyzed according to the model proposed by Lohman and Bujalowski (11) and Eftink (6) for single-site tight-binding systems.
NOB assay.
The neutralization of binding (NOB) assay was performed as previously described (16).
Immunohistochemistry.
Cryostat sections from three different liver samples from patients with chronic hepatitis B were air dried, acetone fixed, and treated for 30 min with heat-inactivated normal human AB serum diluted 1:10 in Tris-HCl buffer–0.05 M NaCl (pH 7.8). Sections were then incubated with either recombinant E2 protein (2 μg) or anti-CD81 monoclonal antibody (MAb) (JS/81; 0.5 μg; Pharmingen) in Tris-buffered saline at room temperature for 1 h. Sections incubated with the E2 protein were further incubated with 1 μg of an anti-E2 MAb (291A2) in Tris-buffered saline. All sections were then incubated with rabbit anti-mouse serum (1/30 dilution; Dako, Glostrup, Denmark) followed by alkaline phosphatase–anti-alkaline phosphatase complex (1/50 dilution) (Dako) for 30 min at room temperature. In some experiments, E2 was preincubated with 1 μg of recombinant LEL before being used to stain liver sections. Naphthol As-BI phosphate (Sigma) was used as the substrate, and New Fuxin (Merk, Darmstadt, Germany) was used as the chromogen.
Assessment of CD81 down-modulation.
Human B cells were incubated at 5 × 105 cell per ml for 4, 8, 12, or 24 h at 37°C in the presence of purified anti-CD81 MAb JS/81 (5 μg/ml), washed twice, incubated for 30 min at 4°C with the same purified MAb (5 μg/ml), washed, and finally incubated with phycoerythrin (PE)-labeled goat anti-mouse serum. Cells were then washed twice, and mean fluorescence intensity (MFI) was determined on a FACS-Calibur (Becton Dickinson). In some experiments, the purified anti-CD81 MAb was cross-linked by a goat anti-mouse serum.
Analysis of LEL by HPLC.
Purified LEL was analyzed by high-pressure liquid chromatography (HPLC) using a uRPc C2/C18 column (Pharmacia). Protein elution was carried out in buffer A (0.06% trifluoroacetic acid in water) and different concentrations of buffer B (0.06% trifluoroacetic acid in 90% acetonitrile–10% water) as follows: 20 min in 2% buffer B, 120 min in 2 to 63% buffer B, and 20 min in 63 to 95% buffer B. Absorbance was measured at 214 nm.
Digestion of LEL with Lys-C protease and peptide analysis.
LEL (10 nmol) in 100 mM Tris-HCl (pH 6.8) was digested at 37°C for 16 h with Lys-C endoprotease (Boehringer) at an enzyme/LEL ratio of 1:200 (wt/wt). The peptides were analyzed by HPLC using the elution conditions described above. The peptides were also analyzed under reducing conditions [10 min at 65°C in 20% of 100 mM Tris(2-carboxyethyl)phosphine (TCEP)–850 mM citric acid]. The reduced peptides were separated on the same uRP C2/C18 column in buffers A and B, applying a 2 to 65% buffer B gradient for 70 min with a flow rate of 40 μl/min. Automated Edman degradation of LEL and its derived peptides was performed with a Beckman model LF 3000 sequencer.
RESULTS
Affinity of interaction between CD81 LEL and HCV EC2.
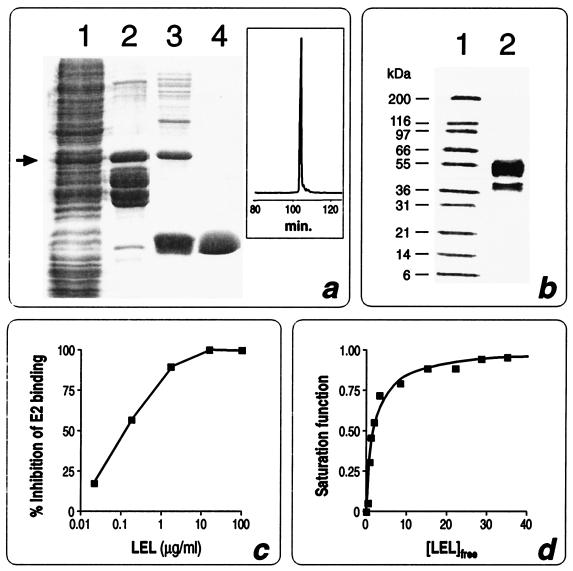
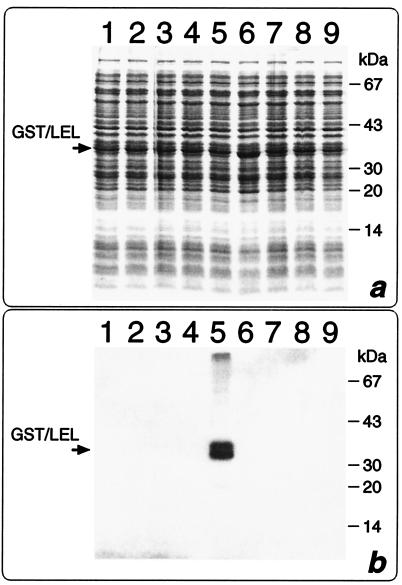
We have previously shown that when fused to the C-terminal end of thioredoxin (Trx) LEL is expressed in E. coli in a soluble form and the fusion product binds the recombinant HCV E2 protein (14). To determine the affinity constant of LEL-E2 interaction, the LEL domain was purified in a “standalone” configuration. Since the cleavage and purification of LEL from the Trx-LEL fusion was quite inefficient, we produced a second construct in which LEL, carrying a six-histidine tail at its C terminus, was fused to GST. Although almost 90% of the fusion, named GST-LEL, was proteolytically cleaved inside the cells at its C terminus by cellular proteases (Fig. 1a, lanes 1 and 2), approximately 3 mg of intact fusion per liter could be recovered from the cell extract. This was accomplished by using a four-step purification procedure. First, the crude extract was subjected to a glutathione affinity column (Fig. 1a, lane 2), which allowed the separation of the protein species carrying the GST moiety. The affinity column step was repeated after the thrombin hydrolysis, which turned out to be very efficient and specific, to remove the GST protein. Third, taking advantage of the histidine tail located at its C terminus, LEL was purified by immobilized metal affinity chromatography, allowing separation of the full-length LEL from the partially degraded products (Fig. 1a, lane 3). Finally, the protein was purified by gel filtration chromatography, obtaining a product which appeared to be more than 98% pure, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and reverse-phase HPLC (RP-HPLC) (Fig. 1a, lane 4 and boxed chromatogram). The purified domain was subjected to five cycles of N-terminal sequencing, obtaining the expected sequence GSPIS (see Fig. 5a). Interestingly, the gel filtration chromatography indicated that the apparent molecular mass of LEL was 36 kDa, suggesting that the molecule forms a trimer under the experimental conditions used. Since in an SDS-polyacrylamide gel the protein moves with the expected molecular mass of 12 kDa in both the presence and the absence of reducing agents (data not shown), the trimer is held together by noncovalent interactions.
FIG. 1.
Purification and affinity of interaction between CD81 LEL and HCV E2. (a) SDS-PAGE analysis of different purification steps of CD81 LEL. Lane 1, total cell extract; lane 2, protein elution after the first glutathione-Sepharose 4B column; lane 3, LEL preparation after thrombin cleavage and Ni2+-chelating sepharose FF column; lane 4, LEL after Superdex 75 HL column. The RP-HPLC chromatogram of the same material as in lane 4 is boxed. The arrow indicates the position of the GST-LEL fusion. (b) SDS-PAGE analysis of purified HCV E2. Lane 1, molecular weight standards; lane 2, purified E2. Western blot analysis, N-terminal sequencing, and amino acid analysis have shown that the lower band (42 kDa) is a degradation product of the major 55-kDa species, most likely processed at amino acid 663 (data not shown). (c) NOB assay to determine inhibition of E2 binding to Molt4 cells by increasing concentration of LEL. (d) E2 saturation as function of free LEL concentration. Experiments were carried out at three different concentrations of E2 ranging from 20 to 80 nM. The continuous lane was obtained by nonlinear least-squares fitting of experimental data with binding parameter Kd = 1.8 nM at 25°C.
FIG. 5.
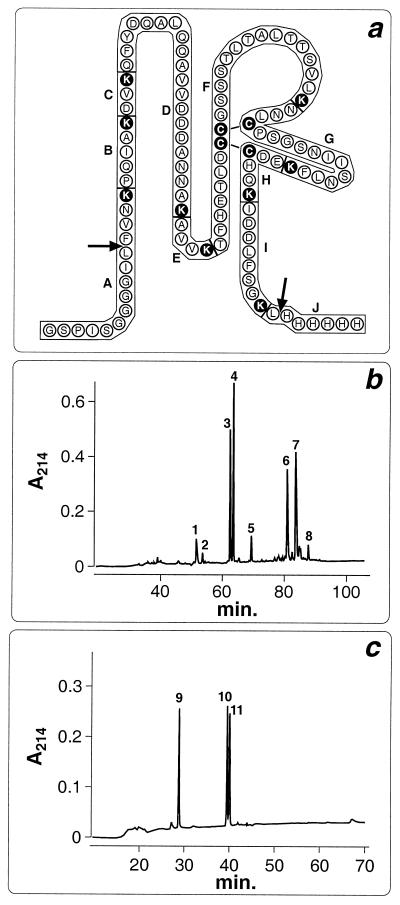
Identification of disulfides in CD81 LEL by Lys-C protease digestion and HPLC analysis. (a) Amino acid sequence of purified LEL. Black circles highlight lysines and cysteines. Arrows indicate beginning and end of CD81 LEL. Additional amino acids at the N terminus belong to the joining region between GST and LEL; at the C terminus is the hystidine tail. Letters (A to J) indicate the peptide fragments generated by protease digestion. (b) HPLC analysis of LEL after Lys-C protease digestion. LEL was digested with Lys-C protease under nonreducing conditions, and the generated fragments were separated by RP-HPLC. Each peak was purified and subjected to N-terminal sequencing. Peak 1, fragment I; peak 2, no sequence; peak 3, fragment A; peak 4, fragment D; peak 5, no sequence; peak 6, fragments F, G, and H; peak 7, fragments F, G, and H-I; peak 8, fragments F and G-H. (c) RP-FPLC analysis of peak 7. Peak 7 was purified, reduced with TCEP, and analyzed by RP-HPLC. Peak 9, fragment H-I; peak 10, fragment F; peak 11, fragment G.
The quality of the purified LEL was also assessed by determining the concentration of protein sufficient to inhibit 50% of binding of recombinant HCV E2 protein to Molt4 cells, using the NOB assay previously described (16). We found that 0.2 μg/ml was the LEL concentration sufficient to obtain 50% inhibition of E2 binding to human cells (Fig. 1c), indicating that, on a molar basis, the soluble LEL domain is a 5- to 10-fold-better inhibitor than the Trx-LEL fusion (14).
For the NOB assay, a highly purified, recombinant E2 preparation was used. As shown in Fig. 1b, the E2 preparation appeared to consist of two molecular species, a major one migrating on a reducing SDS-polyacrylamide gel as a 55-kDa protein and a second one with an apparent molecular mass of 42 kDa. Western blot analysis, N-terminal sequencing, and amino acid analysis revealed that the 42-kDa species represents a C-terminal degradation product of the 55-kDa protein, the protein being most likely processed at amino acid 663 of the E2 sequence (data not shown). When the 42-kDa form was purified at analytical scale by gel filtration chromatography, it was still able to bind CD81 (data not shown); therefore, no further attempts were made to purify the 55-kDa form.
Having demonstrated the high quality of the LEL preparation, we used the purified material to establish the affinity constant of E2-LEL interaction. Since LEL binding to E2 affects the E2 intrinsic fluorescence intensity (most likely due to tryptophan emission quenching or conformational changes), by plotting the fluorescence intensity of E2 as a function of LEL concentration (Fig. 1d) and using the single-site-binding equation described by Eftink (6), Kd values of 1.8 nM at 25°C and 9.1 nM at 37°C were determined. These data are in good agreement with preliminary calculations made during our studies of E2-whole cell interaction, where an affinity of 10 nM was estimated (16).
HCV envelope binding to liver cells.
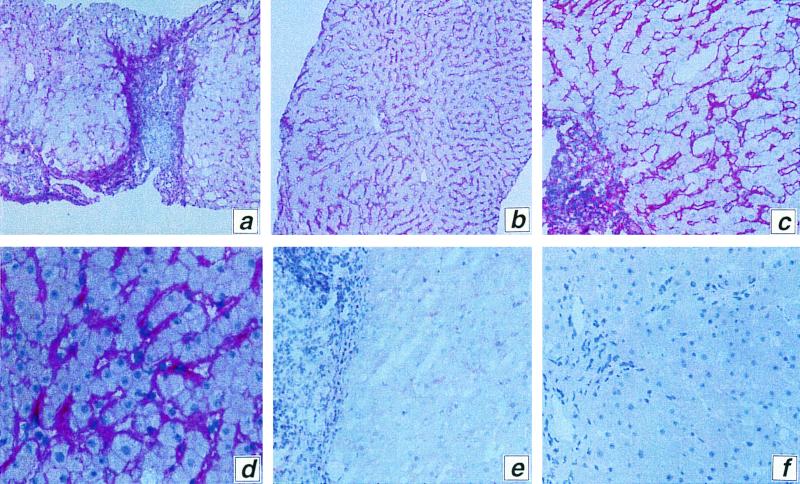
Given the high affinity of CD81-E2 interaction, we used recombinant E2 as if it were an antibody to CD81. First, we demonstrated that an anti-CD81 antibody can stain liver sections (data not shown). Second, the representative staining from a liver biopsy (Fig. 2a to d) shows that HCV-E2 binds all cell lineages (endothelial cells, Kupffer cells, hepatocytes, and lymphocytes) present in liver samples, though with different intensities. Moreover, Fig. 2e shows that preincubation of HCV E2 with recombinant CD81 LEL almost completely blocked the binding of E2. These experiments confirm that CD81 is a widely expressed molecule and suggest that HCV can attach to various target cells in the liver.
FIG. 2.
E2 binding to liver tissue. (a to d) Representative liver sections stained with E2 and anti-E2 MAb; (e) as above except that E2 was preincubated with soluble CD81 LEL domain; (f) negative control stained only with anti-E2 MAb. Magnifications: a and b, ×100; c, e, and f, ×250; d, ×400.
Interestingly, we observed that some subclones of the hepatocarcinoma cell line HepG2, which do not express CD81 and are negative for CD81 mRNA by RT-PCR, retain a low but detectable capacity to bind HCV E2 (data not shown). It is therefore likely that other molecules contribute to HCV binding to hepatocytes.
Down-modulation of cell surface CD81.
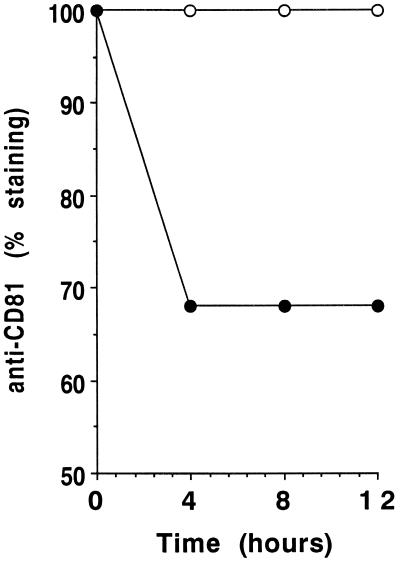
It is known that virus attachment to a cell surface molecule per se does not necessarily lead to uptake and infection. We therefore assessed the ability of cellular CD81 to internalize bound ligands by following down-modulation of CD81 using an anti-CD81 antibody, JS/81, which binds CD81 on the same region as recombinant HCV E2 (14). Human B cells were incubated with a purified anti-CD81 antibody for different times at 37°C. Cells were then washed and reincubated at 4°C with the same purified anti-CD81 antibody, washed, and incubated with a PE-labeled goat anti-mouse serum to assess cell surface expression of CD81 by flow cytometry. Figure 3 shows that even after 12 h of incubation at 37°C, only 30% of cell surface CD81 had been down-modulated. In contrast, molecules such as CD71 or CD4 (this B-cell line is CD4 positive) are very efficiently down-modulated by interaction with specific antibodies, as about 50 to 80% of the molecules disappeared from the cell surface in 30 to 60 min (data not shown). These experiments demonstrate that CD81 is a surface molecule with relatively poor ability to internalize ligands and suggest that it may serve as an HCV attachment receptor rather than as a receptor for virus entry.
FIG. 3.
Down-modulation of CD81. Human B cells were preincubated for 4, 8, or 12 h at 37°C in the presence of purified anti-CD81 MAb (closed circles) or purified anti-HLA class I MAb as a control (open circles), washed, and incubated for 30 min at 4°C with the same anti-CD81 MAb. Cells were then washed, incubated with a PE-labeled goat anti-mouse serum, and washed again; then MFI determined by flow cytometry. MFI values in the presence of preincubation with anti-HLA MAb (positive control), in the presence of preincubation with anti-CD81 MAb (experimental values), and in the absence of MAb (negative control) were measured, and specific down-modulation of CD81 was determined as [(positive control MFI − experimental MFI)/(positive control MFI − negative control MFI)] × 100.
Role of disulfides in E2-LEL interaction.
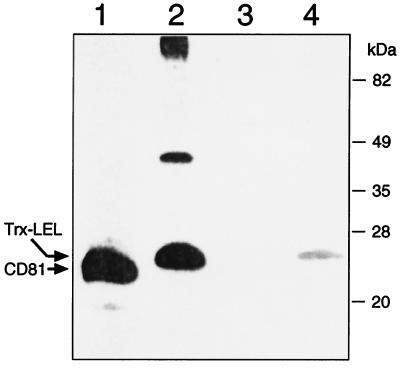
Like all tetraspanins, CD81 carries four cysteines which are located in the LEL domain (positions 156, 157, 175, and 190 [see Fig. 5a]). The presence of disulfides in CD81 and their importance in E2 binding are suggested by our findings that E2 does not detect CD81 on Western blots if CD81 is resolved by SDS-PAGE under reducing conditions (Fig. 4, lanes 1 and 3). Similar results (Fig. 4, lanes 2 and 4) are obtained with the purified Trx-LEL fusion (14).
FIG. 4.
Role of CD81 disulfides in E2 binding. Membrane proteins from Molt4 cells (lanes 1 and 3) and Trx-LEL fusion protein (lanes 2 and 4) were prepared as described previously (14) and loaded onto a 15% polyacrylamide gel in the presence (lanes 3 and 4) or absence (lanes 1 and 2) of 10 mM dithiothreitol. The proteins were transferred to a polyvinylidene difluoride membrane and treated with recombinant E2 (1 μg/ml) and anti-E2 MAb (291A2) (14). E2 interaction with CD81 and Trx-LEL was detected with peroxidase-conjugated goat anti-mouse antibodies.
To assess whether all four cysteines were engaged in the formation of disulfide bridges, the free thiol groups of LEL were titrated by Ellman's method (15). From this analysis, it was calculated that less than 2% of total LEL was in a reduced state, whereas the denatured and reduced LEL preparation gave a total amount of -SH groups of 42 nmol, in excellent agreement with the amount of protein (10 nmol) determined by the Bradford method (2). Therefore, we conclude that all four cysteines of the CD81 LEL are engaged in disulfide bond formation.
To confirm the presence of two disulfides, we digested the purified LEL domain with the protease Lys-C and separated the peptide fragments by RP-HPLC, obtaining eight major peaks (Fig. 5b). These peaks were collected and subjected to N-terminal sequencing. The sequence analysis revealed that peaks 6 and 7 contained the proteolytic fragments (F, G, and H in peak 6; F, G, and H-I in peak 7) carrying the four cysteines of LEL (Fig. 5a). These fragments could not be resolved under any other elution regimen tested (data not shown), suggesting that they were held together by covalent interactions. To demonstrate this, peak 7 was collected, reduced, and applied to the RP-HPLC column. As shown in Fig. 5C, three peaks were generated which corresponded to fragments F, G, and H-I of LEL, as determined by sequence analysis.
To further test our hypothesis that the two disulfide bonds are required for HCV E2 binding to CD81, the four cysteines of LEL were replaced by site-directed mutagenesis with either alanine or serine, generating eight GST-LEL mutants. None of the mutated fusion proteins showed detectable E2 binding activity, as assessed by Western blot (Fig. 6) and NOB (data not shown) assays.
FIG. 6.
Inability of GST-LEL cysteine mutants to bind E2. Bacterial cells expressing the GST-LEL mutants were directly lysated in SDS-PAGE loading buffer, and the total proteins were resolved on an SDS–12.5% polyacrylamide gel. The proteins were either stained with Coomassie blue (A) or transferred to a polyvinylidene difluoride membrane and treated with E2 and anti-E2 291/A2 MAb as described in the legend to Fig. 4 (b). Lane 1, GST LEL Cys156→Ala; lane 2, GST-LEL Cys157→Ala; lane 3, GST-LEL Cys175→Ala; lane 4, GST LEL Cys190→Ala; lane 5, WT GST LEL; lane 6, GST LEL Cys156→Ser; lane 7, GST LEL Cys157→Ser; lane 8, GST LEL Cys175→Ser; lane 9, GST LEL Cys190→Ser.
DISCUSSION
We have recently shown that HCV binds specifically to the LEL of human CD81, suggesting that the latter may be a cellular receptor for HCV (14). Since the efficiency with which the virus interacts with its receptor represents an important factor in determining both the onset and the progression of viral infection, the first objective of this work was to determine the affinity of the interaction of the recombinant HCV envelope protein E2 with the soluble CD81 LEL domain. For this purpose, we first developed efficient expression and purification systems for the production of both E2 and CD81 LEL. Using the procedures here described, both proteins turned out to be of high enough quality for affinity studies. The recombinant E2 preparation contained two major protein species, one of them, representing approximately 25% of the total protein content, being a truncated form of the major protein species of 55 kDa. Altogether, the two protein species account for more than 95% of the total protein content in the final E2 preparation. Since the truncated form of E2 was still able to bind CD81 (data not shown), the E2 preparation was considered appropriate for affinity studies. Similarly, CD81 LEL was purified to a level higher than 95%. The purification procedure was complicated by the tendency of the E. coli cytoplasmic proteases to degrade the GST-LEL fusion, generating a series of degradation products in which the LEL domain was partially or totally truncated. Therefore, addition of the histidine tail to the C terminus of LEL turned out to be key to obtaining a homogeneous preparation of full-length LEL.
Using fluorescence analysis to monitor the E2-LEL interaction, we calculated that the binding affinities of the E2-LEL interaction are 1.8 nM at 25°C and 9.1 nM at 37°C. These affinity values are in the range of the 1 to 4 nM which was reported for the human immunodeficiency virus (HIV) gp120-CD4 interaction (12), whereas they are 2 to 3 logs higher than the values of 0.5 to 2 mM reported for the binding of rhinovirus to ICAM-1 (3). At present, it is not possible to measure the affinity of interaction between HCV and CD81 because HCV is not available as purified virus. However, we can extrapolate from the HIV case where CD4-gp120 interaction in solution occurs with an affinity similar to that of E2-CD81 (12). It has been demonstrated that at 37°C, the affinity of interaction of CD4 with HIV gp120 on the virion surface is indistinguishable from the affinity of CD4 for the equivalent concentration of soluble recombinant gp120 (13). Should the high affinity of E2-CD81 binding be representative of the envelope-receptor interaction for HCV in vivo, only antienvelope antibodies with affinity constants higher than 10−9 M should efficiently block virus attachment to human cells and therefore neutralize infection. Studies are in progress to assess whether the high rate of chronic HCV infections is related to the low titers of high-affinity antienvelope antibodies elicited by infection and whether spontaneous resolution of infection (8) is due to high-affinity antibodies. Finally, the affinity data are in good agreement with preliminary calculations made during our studies on E2 binding to cells (16). Thus, the recombinant E2 and CD81 LEL are suitable to establish a cell-free system for screening possible inhibitors of virus attachment to human cells.
We have shown previously that CD81 is an attachment receptor for HCV (14), but we did not know whether it mediates virus entry in human cells. We therefore assessed the ability of CD81 to mediate internalization of bound ligands and found that even after 12 h only 30% of CD81 is down-modulated by engagement with antibodies. This is very inefficient compared to the uptake ability of CD4, the HIV receptor (9). However, it is possible that the inefficient internalization of CD81 might still be sufficient for HCV to enter human cells and establish productive infection. Alternatively, it may be that the role of CD81 is to concentrate virus particles at the cell surface for subsequent interaction with an entry receptor. With regard to this possibility, we have recently found that some hepatoma cell lines can bind HCV envelope in the absence of CD81, suggesting that other molecules exist on human cells which interact with HCV. We are currently analyzing the nature of this interaction.
Our data show that two disulfides are required in CD81 LEL to interact with HCV E2. The implication of this finding is that no association between CD81 and HCV should occur in a reducing environment such as the cytoplasmic milieu.
As for the topological organization of the disulfide bridges, the absence in the Lys-C digestion analysis of covalent binding between fragments G and H, together with the unlikely possibility of disulfide binding between two adjacent cysteines, allow us to exclude the partnering between Cys175 and Cys190 and Cys156 and Cys157. However, because of the presence of two adjacent cysteines, no conclusive information on the disulfide organization can be obtained. Recently, in the attempt to define the correct partnering of cysteines, a Lys-C protease cleavage site was introduced between Cys156 and Cys157 by inserting either a single lysine residue or the sequence Gly-Ser-Lys-Ser-Gly (data not shown). Although the two mutants were efficiently expressed in E. coli, none of them bound E2 (data not shown). While these experiments still do not help elucidate the structure of the disulfide bonds, they indicate that insertions between the adjacent cysteines impair LEL structure or interaction with E2. This implies that in searches for molecules which disturb attachment of HCV to CD81, the structural organization of the region around the four cysteines of CD81 must be highly preserved.
In conclusion, our data on the CD81-HCV interaction will help clarify the virus life cycle and develop new strategies aimed at interfering with HCV binding to human cells.
ACKNOWLEDGMENTS
We thank N. Valiante, P. Pileri, Y. Uematsu, and G. Del Giudice for critical reading of the manuscript and G. Corsi for artwork.
REFERENCES
- 1.Altekar W. Fluorescence of proteins in aqueous neutral salt solutions. II. Influence of monovalent cation chlorides, particularly cesium chloride. Biopolymers. 1997;16:369–386. doi: 10.1002/bip.1977.360160211. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Casasnovas J M, Springer T A. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 4.Chapman B S, Thayer R M, Vincent K A, Haigwood N L. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Eftink M R. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- 7.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 8.Ishii K, Rosa D, Watanabe Y, Katayama T, Harada H, Wyatt C, Kiyosawa K, Aizaki H, Matsuura Y, Houghton M, Abrignani S, Miyamura T. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology. 1998;28:1117–1120. doi: 10.1002/hep.510280429. [DOI] [PubMed] [Google Scholar]
- 9.Lanzavecchia A, Roosneck E, Gregory T, Berman P, Abrignani S. T cells can present antigens such as HIV gp120 targeted to their own surface molecules. Nature. 1988;334:530–532. doi: 10.1038/334530a0. [DOI] [PubMed] [Google Scholar]
- 10.Levy S, Todd S C, Maecker H T. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 11.Lohman T M, Bujalowski W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol. 1991;208:258–290. doi: 10.1016/0076-6879(91)08017-c. [DOI] [PubMed] [Google Scholar]
- 12.Moore J P. Simple methods for monitoring HIV-1 and HIV-2 gp120 binding to soluble CD4 by enzyme-linked immunosorbent assay: HIV-2 has a 25-fold lower affinity than HIV-1 for soluble CD4. AIDS. 1990;4:297–305. doi: 10.1097/00002030-199004000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Moore J P, McKeating J A, Norton W A, Sattentau Q J. Direct measurement of soluble CD4 binding to human immunodeficiency virus type 1 virions: gp120 dissociation and its implications for virus-cell binding and fusion reactions and their neutralization by soluble CD4. J Virol. 1991;65:1133–1140. doi: 10.1128/jvi.65.3.1133-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 15.Riddles P W, Blakeley R L, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 16.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q-L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.World Health Organization. Hepatitis C. Weekly Epidemiol Rec. 1997;72:65–69. [Google Scholar]