Abstract
The human immunodeficiency virus (HIV) genome is AU rich, and this imparts a codon bias that is quite different from the one used by human genes. The codon usage is particularly marked for the gag, pol, and env genes. Interestingly, the expression of these genes is dependent on the presence of the Rev/Rev-responsive element (RRE) regulatory system, even in contexts other than the HIV genome. The Rev dependency has been explained in part by the presence of RNA instability sequences residing in these coding regions. The requirement for Rev also places a limitation on the development of HIV-based vectors, because of the requirement to provide an accessory factor. We have now synthesized a complete codon-optimized HIV-1 gag-pol gene. We show that expression levels are high and that expression is Rev independent. This effect is due to an increase in the amount of gag-pol mRNA. Provision of the RRE in cis did not lower protein or RNA levels or stimulate a Rev response. Furthermore we have used this synthetic gag-pol gene to produce HIV vectors that now lack all of the accessory proteins. These vectors should now be safer than murine leukemia virus-based vectors.
The expression of a gene in general, when the gene is introduced into mammalian cells, depends on various factors, including gene copy number, transcription control elements, site of chromosomal integration, mRNA stability, and translational efficiency.
Human immunodeficiency virus type 1 (HIV-1) gene expression is tightly regulated at multiple levels. Tight regulation may be beneficial for the virus to minimize its antigenic profile and maintain latent infection. Expression is controlled at the transcriptional level, mainly by the viral transactivator Tat. The binding of Tat to an RNA element, namely, TAR, which is found at the 5′ end of the HIV-1 transcript, stimulates transcription from the HIV-1 long terminal repeat.
At a posttranscriptional step, the binding of HIV-1 Rev to an RNA element, namely, the Rev-responsive element (RRE), found in all unspliced and singly spliced HIV-1 mRNAs, results in the nuclear export of the latter and the translation of Gag, Gag-Pol, Env, Vif, Vpr, and Vpu (51). Although Rev's main function seems to be the nuclear export of RRE-containing HIV-1 mRNAs, other roles have been suggested for this small protein. It has been proposed (56), although some argue against it (48), that Rev may act to stabilize these transcripts. One of the major factors that have been implicated in the instability of HIV-1 mRNAs is the high AU content that the virus exhibits (2, 36, 46, 47, 56–58). It has been shown for human genes that the existence of sequences such as AUUUA within highly unstable RNAs, such as c-myc or those of some cytokines, is associated with their instability (39, 53, 69). Stabilization of these transcripts occurs upon induction; e.g., the labile interleukin 6 transcript is stabilized by mitogen-activated protein kinase kinase kinase MEKK1. Rev may therefore be acting in a similar way. Several cis-acting repressive sequences, or instability sequences (INS), have been identified within HIV-1. These include sequences in the gag-pol (36, 56) and env genes (including the RRE) (9, 46). However, not all HIV-1 instability elements contain the AUUUA pentanucleotide, and some are not even AU rich (e.g., the RRE [56]). When the AU-rich INS within gag-pol were mutated, their inhibitory effects were eliminated, leading to increased protein production, which was due to an increase in steady-state mRNA levels (56).
Rev has also been proposed to enhance the polysomal association of RRE-containing RNAs and therefore to play a positive role in their translation (5, 15, 18). At the translational level, the unusual HIV-1 codon bias, which is markedly different from the one used by highly expressed human genes (33, 34, 59, 64), may also be affecting the expression of HIV genes, especially since the abundances of the various tRNA isoacceptor species correspond to the codon usages of highly expressed cellular genes (7, 65). The pausing of the ribosomes at rare codons may also lead to enhanced RNA turnover (reviewed by Hentze and Kulozik [27]). There is a plethora of examples in the literature of changes in codon usage having a significant effect on translation (24, 54, 72, 75). It has been demonstrated for HIV-1 env that converting the unfavorable HIV-1 codon bias to the one used by human genes results in enhanced translation of gp120 (24).
The HIV-1 genome is AU rich, and this correlates with the extreme codon bias that the virus exhibits, as well as with the stability of the HIV-1 transcripts. It has been shown that efficient expression of HIV proteins is restricted both by the adverse codon bias (24) and by instability sequences (see, e.g., reference 56). Efficient HIV-1 protein expression may be desirable in the context of HIV-1-based vectors. Recently HIV-1 has been used to develop a new class of gene transfer vectors that have utility in a variety of gene therapy applications principally because they exploit the ability of lentiviruses to stably transfer genes to the chromosomes of nondividing cells (for reviews, see references 43 and 66). Initially the vector system contained all of the accessory genes and the transferred genome contained large residual regions of the HIV genome (8, 44, 45). It has been possible to delete most of the viral genome in the transfer vector and to produce vectors without any of the accessory genes except rev (17, 31). In order to move these vectors into clinical application, however, it is necessary to make further refinements to remove the possibilities for homologous recombination between components both during vector production and in the patient. Efficient packaging of an HIV-based vector requires sequences in the amino-terminal-coding region of gag and in the 5′ untranslated region (UTR) (50). Although it has been reported that as little as 40 nucleotides (nt) of the gag coding sequence are sufficient (14), the retention of any region of gag will maintain homology with the gag-pol expression cassette that is used to produce the vector. In addition, the vector genome and the gag-pol expression cassette both contain the RRE, providing another region of homology.
We have now constructed a completely synthetic HIV-1 gag-pol gene (SYNGP) where the nucleotide sequence has been altered in the majority of codons to retain the primary amino acid sequence but to exploit the favored codon usage of human cells. The altered sequence contains no substantial regions of homology with any naturally occurring HIV-1 gag-pol sequence and has no potential for recombination with gag sequences in the transfer vector genome. In addition, the expression of SYNGP is entirely Rev independent, so the RRE can be removed from the gag-pol expression cassette thus removing the second region with recombination potential. We have characterized the expression of SYNGP with respect to the role of Rev and INS, and we have used the SYNGP expression cassette to produce a safer HIV-1-based vector that may have utility for delivering therapeutic genes for the treatment of AIDS.
MATERIALS AND METHODS
Cell lines.
293T cells (16) and HeLa cells (41) were maintained in Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum and supplemented with l-glutamine and antibiotics (penicillin-streptomycin). The 293T cells were obtained from D. Baltimore (Rockefeller University).
HIV-1 proviral clones.
Proviral clones pWI3 (30) and pNL4-3 (1) were used in this study.
Construction of the codon-optimized gene.
The codon-optimized gag-pol gene was constructed by annealing a series of short overlapping oligonucleotides (approximately 30- to 40-mers; 9 nt of overlap). Oligonucleotides were purchased from R&D Systems Europe Ltd. (Abingdon, United Kingdom). Codon optimization was performed using the sequence of the HXB2 strain (accession no. K03455) (20). A fragment from base 1222 from the beginning of gag until the end of gag (base 1503) was not optimized in order to maintain the frameshift site and the overlap between the gag and pol reading frames. This was from clone pNL4-3. When reference to base numbers within the gag-pol gene base is made, 1 is the A of the gag ATG, which corresponds to base 790 from the beginning of the HXB2 sequence. When reference to sequences outside gag-pol is made, the numbers refer to bases from the beginning of the HXB2 sequence, where base 1 corresponds to the beginning of the 5′ long terminal repeat. Some deviations from optimization were made in order to introduce convenient restriction sites. The gene was cloned into the vector pCIneo (Promega) in the EcoRI-NotI sites. The resulting plasmid was named pSYNGP. The RRE sequence (bases 7769 to 8021 of the HXB2 sequence) was amplified by PCR from the pWI3 proviral clone with primers bearing the NotI restriction site and was subsequently cloned into the NotI site of pSYNGP. The resulting plasmids were named pSYNGP-RRE (RRE in the correct orientation) and pSYNGP-ERR (RRE in the reverse orientation).
Plasmid constructs for the study of the effects of INS on RNA and protein levels.
Briefly, the HIV-1 gag sequence or parts of it (nt 1 to 625 or 625 to 1503) were amplified by PCR from HIV-1 proviral clone pWI3. An ATG and a frameshift gag mutant were also constructed by PCR from the same clone. β-Galactosidase (β-Gal; encoded by lacZ) was excised from plasmid pCMV-βGal (Clontech), and parts of lacZ (i.e., nt 1 to 1093 or 72 to 1093) were excised from plasmid pH6Z (see next section). All fragments were blunt ended and were inserted into pSYNGP or pSYNGP-RRE at either a blunted EcoRI or NotI site (upstream or downstream of the codon-optimized gag-pol gene, respectively).
Vector genome constructs.
pH6nZ was derived from pH4Z (31) by the addition of a single nucleotide to place an extra guanine residue that was missing from pH4Z at the 5′ end of the vector genome transcript to optimize reverse transcription. In addition the gene coding for β-Gal (lacZ) was replaced by a gene encoding a nucleus-localizing β-Gal. (We are grateful to Enca Martin-Rendon and Said Ismail for providing pH6nZ.) In order to make genome constructs lacking the gene encoding Rev, the following modifications were made: (i) a 1.8-kb PstI-PstI fragment was removed from pH6nZ, resulting in plasmid pH6.1nZ, and (ii) an EcoNI (filled)-SphI fragment was replaced with a SpeI (filled)-SphI fragment from the same plasmid (pH6nZ), resulting in plasmid pH6.2nZ. In both cases sequences within gag (nt 1 to 625) were retained, as they have been shown to play a role in packaging (50). rev, RRE, and any other residual env sequences were removed. pH6.2nZ further contains the env splice acceptor, whereas pH6.1nZ does not. A series of vectors encompassing further gag deletions with or without a mutant major splice donor (SD) (GT-to-CA mutation) were also derived from pH6Z. These were made by PCR with primers bearing NarI (5′ primers) and SpeI (3′ primers) sites. The PCR products were inserted into pH6Z at the NarI-SpeI sites. The resulting vectors were named pHS1nZ (containing HIV-1 sequences up to gag 40), pHS2nZ (containing HIV-1 sequences up to gag 260), pHS3nZ (containing HIV-1 sequences up to gag 360), pHS4nZ (containing HIV-1 sequences up to gag 625), pHS5nZ (same as pHS1nZ but with a mutant SD), pHS6nZ (same as pHS2nZ but with a mutant SD), pHS7nZ (same as pHS3nZ but with a mutant SD), and pHS8nZ (same as pHS4nZ but with a mutant SD).
In addition, the RRE sequence (nt 7769 to 8021 of the HXB2 sequence) was inserted in the SpeI (filled) site of pH6.1nZ, pHS1nZ, pHS3nZ, and pHS7nZ, resulting in plasmids pH6.1nZR, pHS1nZR, pHS3nZR, and pHS7nZR, respectively.
Transient transfections, transductions, and determination of viral titers.
These operations were performed as previously described (31, 60). Briefly, 293T cells were seeded on 6-cm-diameter dishes and 24 h later they were transiently transfected by overnight calcium phosphate treatment. The medium was replaced 12 h posttransfection, and unless otherwise stated supernatants were harvested 48 h posttransfection and filtered (through 0.22- or 0.45-μm-pore-size filters), and titers were determined by transduction of 293T cells. For this reason, supernatant at appropriate dilutions of the original stock was added to 293T cells (plated onto 6- or 12-well plates 24 h prior to transduction). Polybrene (8 μg/ml; Sigma) was added to each well, and 48 h posttransduction viral titers were determined by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining.
Luminescent β-Gal assays.
These assays were performed on total cell extracts using a luminescent β-Gal reporter system (Clontech). Untransfected 293T cells were used as the negative control, and 293T cells transfected with pCMV-βgal (Clontech) were used as the positive control.
RNA analysis.
Total or cytoplasmic RNA was extracted from 293T cells by using the RNeasy minikit (Qiagen) 36 to 48 h posttransfection. Five to 10 μg of RNA was subjected to Northern blot analysis as previously described (55). A probe complementary to bases 1222 to 1503 of the gag-pol gene was amplified by PCR from the HIV-1 pNL4-3 proviral clone and was used to detect both the codon-optimized and wild-type gag-pol mRNAs. A second probe, complementary to nt 1510 to 2290 of the codon-optimized gene, was also amplified by PCR from plasmid pSYNGP and was used to detect the codon-optimized genes only. A 732-bp fragment complementary to all vector genomes used in this study was prepared by an SpeI-AvrII digestion of pH6nZ. A probe specific for ubiquitin (Clontech) was used to normalize the results. All probes were labeled by random labeling (Stratagene) with [α-32P]dCTP (Amersham). The results were quantitated by using a Storm PhosphorImager (Molecular Dynamics).
Protein analysis.
Total cell lysates were prepared from 293T cells 48 h posttransfection (unless otherwise stated) with an alkaline lysis buffer. For extraction of proteins from cell supernatants the supernatant was first passed through a 0.22-μm-pore-size filter and the vector particles were collected by centrifugation of 1 ml of supernatant at 21,000 × g for 30 min. Pellets were washed with phosphate-buffered saline and then resuspended in a small volume (2 to 10 μl) of lysis buffer. Equal protein amounts were separated on a sodium dodecyl sulfate–10 to 12% (vol/vol) polyacrylamide gel. Proteins were transferred to nitrocellulose membranes, which were probed sequentially with a 1:500 dilution of HIV-1-positive human serum (AIDS Reagent Project; ADP508, panel E) and a 1:1,000 dilution of horseradish peroxidase-labeled anti-human immunoglobulin G (IgG) (Sigma; A0176). Proteins were visualized using the ECL or ECL-plus Western blotting detection reagent (Amersham). To verify equal protein loading, membranes were stripped and reprobed with a 1:1,000 dilution of antiactin antibody (Sigma; A2066), followed by a 1:2,000 dilution or horseradish peroxidase-labeled anti-rabbit IgG (Vector Laboratories; PI-1000).
RESULTS
Construction of a codon-optimized HIV-1 gag-pol gene.
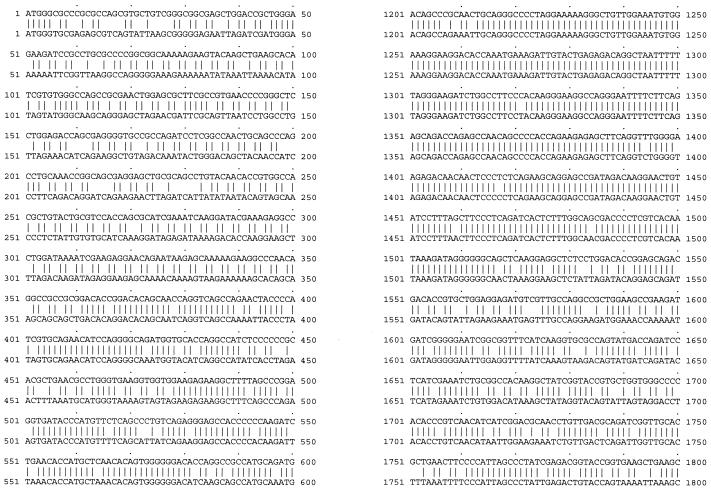
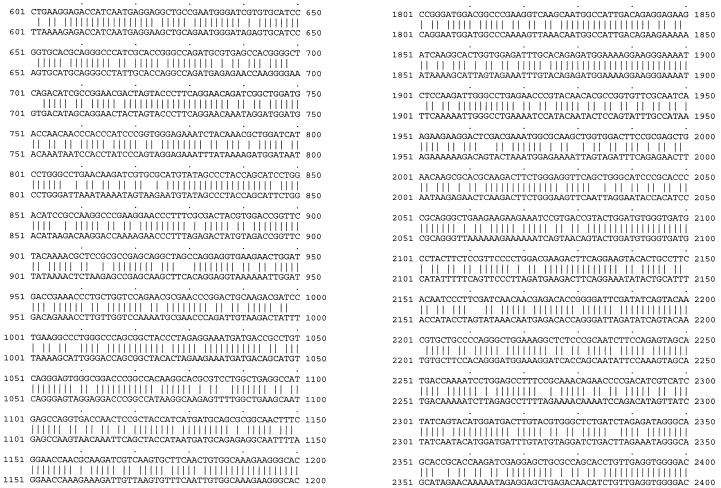
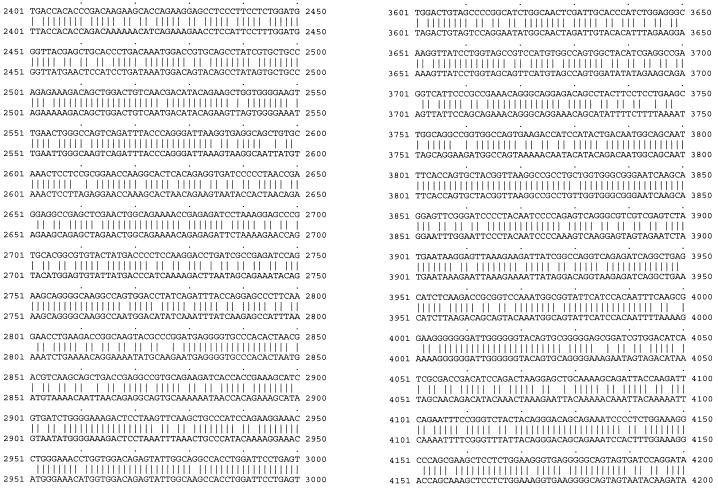
To investigate whether the poor expression and Rev dependence of HIV-1 gag-pol were due to the HIV codon bias, we constructed a codon-optimized HIV-1 gag-pol gene with codons frequently used by highly expressed human genes. We based the codon optimization on the sequence of the HIV-1 HXB2 strain. The codon-optimized gene was synthesized by annealing oligonucleotides (approximately 30- to 40-mers) corresponding to both strands of the coding sequence, with a 25% overlap (9 nt). The Kozak consensus sequence for optimal translation initiation (32) was also included. The only sequence not optimized was that encompassing the frameshift site. The −1 ribosomal frameshift (occurring at the sequence TTTTTTA, 1,296 bases downstream of the gag ATG) brings the overlapping out-of-phase gag and pol genes into translational phase (29, 68), thus allowing expression of the Gag-Pol polyprotein. Sequences downstream of the frameshift site that form secondary structures also play a role in facilitating the event (by stalling the ribosomes and allowing the “slippage” to occur) (10, 49). Consequently, a 281-bp fragment spanning the frameshift site and the overlapping region of the two reading frames (gag and gag-pol) was retained. Some deviations from optimal codon usage were made in order to accommodate convenient restriction sites. The final codon usage, shown in Table 1, now resembles that of highly expressed human genes and is quite different from that of the wild-type HIV-1 gag-pol. The codon-optimized gag-pol gene was cloned into the mammalian expression vector pCIneo (Promega). The resulting plasmid was named pSYNGP. Sequencing the gene in both strands verified the absence of any mistakes. A sequence comparison between the codon-optimized and wild-type HIV gag-pol sequences is shown in Fig. 1.
TABLE 1.
Codon usage in human genes, wild-type HIV-1 gag-pol, and codon-optimized HIV-1 gag-pol
| Amino acid | 1st 2 bases | 3rd base | Codon usage (%) ina:
|
||
|---|---|---|---|---|---|
| MH | WT | CO | |||
| Ala | GC | A | 13 | 46 | 8 |
| C | 53 | 19 | 65 | ||
| G | 17 | 11 | 8 | ||
| T | 17 | 24 | 19 | ||
| Arg | AG | A | 10 | 58 | 10 |
| G | 18 | 29 | 11 | ||
| CG | A | 6 | 6 | 0 | |
| C | 37 | 0 | 61 | ||
| G | 21 | 6 | 10 | ||
| T | 7 | 0 | 5 | ||
| Asn | AA | C | 78 | 29 | 71 |
| T | 22 | 71 | 29 | ||
| Asp | GA | C | 75 | 64 | 70 |
| T | 25 | 36 | 30 | ||
| Cys | TG | C | 68 | 10 | 70 |
| T | 32 | 90 | 30 | ||
| Gln | CA | A | 12 | 53 | 21 |
| G | 88 | 47 | 79 | ||
| Glu | GA | A | 25 | 65 | 38 |
| G | 75 | 35 | 62 | ||
| Gly | GG | A | 14 | 53 | 21 |
| C | 50 | 21 | 55 | ||
| G | 24 | 24 | 24 | ||
| T | 12 | 3 | 0 | ||
| His | CA | C | 79 | 30 | 90 |
| T | 21 | 70 | 10 | ||
| Ile | AT | A | 5 | 58 | 8 |
| C | 18 | 19 | 92 | ||
| T | 77 | 23 | 0 | ||
| Leu | CT | A | 3 | 14 | 3 |
| C | 26 | 8 | 17 | ||
| G | 58 | 14 | 70 | ||
| T | 5 | 11 | 6 | ||
| TT | A | 2 | 42 | 6 | |
| G | 6 | 11 | 0 | ||
| Lys | AA | A | 18 | 58 | 28 |
| G | 82 | 42 | 72 | ||
| Phe | TT | C | 80 | 45 | 45 |
| T | 20 | 55 | 55 | ||
| Pro | CC | A | 16 | 52 | 24 |
| C | 48 | 15 | 39 | ||
| G | 17 | 3 | 21 | ||
| T | 19 | 30 | 15 | ||
| Ser | AG | C | 34 | 35 | 55 |
| T | 10 | 10 | 3 | ||
| TC | A | 5 | 38 | 17 | |
| C | 28 | 10 | 14 | ||
| G | 9 | 3 | 7 | ||
| T | 13 | 3 | 3 | ||
| Thr | AC | A | 14 | 45 | 16 |
| C | 57 | 29 | 52 | ||
| G | 15 | 0 | 19 | ||
| T | 14 | 26 | 13 | ||
| Tyr | TA | C | 74 | 20 | 80 |
| T | 26 | 80 | 20 | ||
| Val | GT | A | 5 | 56 | 4 |
| C | 25 | 8 | 20 | ||
| G | 64 | 24 | 76 | ||
| T | 7 | 12 | 0 | ||
MH, human genes; WT, wild-type HIV-1 gag-pol; CO, codon-optimized HIV-1 gag-pol. For each amino acid, the usage for the most frequently used codon is in boldface.
FIG. 1.
Sequence comparison between the wild-type HIV gag-pol sequence (pGP-RRE3; bottom) and the codon-optimized gag-pol sequence (pSYNGP; top).
In order to assess Rev (in)dependence, the HIV-1 RRE was amplified by PCR from HIV-1 HXB2 and inserted in both orientations downstream of the codon-optimized gag-pol gene, resulting in plasmids pSYNGP-RRE and pSYNGP-ERR.
Expression of gag-pol gene products and vector particle production.
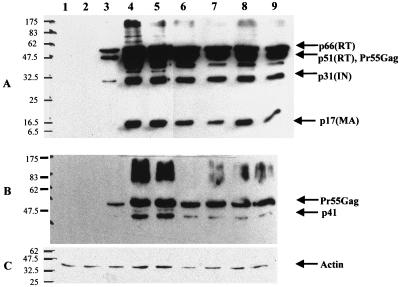
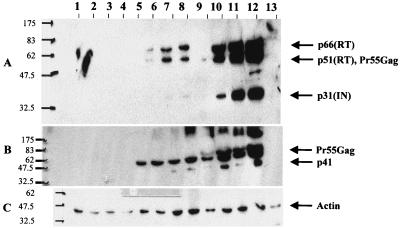
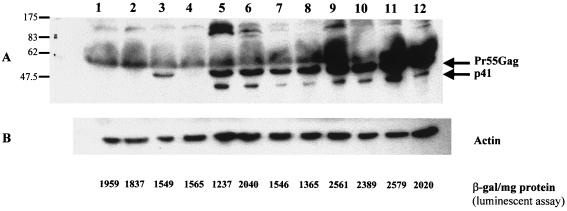
The wild-type gag-pol (pGP-RRE3) (31) and codon-optimized expression vectors (pSYNGP, pSYNGP-RRE, and pSYNGP-ERR) were transiently transfected into 293T cells. Transfections were performed in the presence or absence of Rev expression vector pCMV-Rev (19) in order to assess the Rev dependence of expression. Western blot analysis was performed on cell lysates and supernatants to assess protein production. The results are shown in Fig. 2. As expected (25), expression of the wild-type gene is observed only when Rev is provided in trans (lanes 2 and 3). In contrast, when the codon-optimized gag-pol was used, there was high-level expression in both the presence and absence of Rev (lanes 4 and 5), indicating that in this system there was no requirement for Rev. Protein levels were higher for the codon-optimized gene than for the wild-type gag-pol (compare lanes 4 to 9 with lane 3). The difference was more evident in the cell supernatants (approximately 10-fold-higher protein levels for the codon-optimized gene than for the wild-type one; quantitated by using a PhosphorImager) than in the cell lysates.
FIG. 2.
Protein expression and particle formation are high and Rev independent. The gag-pol expression plasmids (5 μg) were transfected into 293T cells in the presence or absence of Rev (pCMV-Rev; 1 μg), and protein levels were determined 48 h posttransfection in culture supernatants (A) and cell lysates (B). HIV-1-positive human serum was used to detect the Gag-Pol proteins. The blots were reprobed with an antiactin antibody, as an internal control (C). The protein marker (New England Biolabs) sizes (in kilodaltons) are at the left. Lanes: 1, mock-transfected 293T cells; 2, pGP-RRE3; 3, pGP-RRE3 plus pCMV-Rev; 4, pSYNGP; 5, pSYNGP plus pCMV-Rev; 6, pSYNGP-RRE; 7, pSYNGP-RRE plus pCMV-Rev; 8, pSYNGP-ERR; 9, pSYNGP-ERR plus pCMV-Rev.
In previous studies where the RRE has been included in gag-pol expression vectors that had been engineered to remove INS, inclusion of the RRE led to a decrease in protein levels, which was restored by providing Rev in trans (56). In our hands, the presence of the RRE in the fully codon-optimized gag-pol mRNA did not affect protein levels and the provision of Rev in trans did not further enhance expression (lanes 6 and 7).
In order to compare translation rates between the wild-type and codon-optimized gene, protein production from the wild-type and codon-optimized expression vectors was determined at several time intervals after transfection into 293T cells. Protein production and particle formation were determined by Western blot analysis, and the results are shown in Fig. 3. Protein production and particle formation were 10-fold higher for the codon-optimized gag-pol at all time points.
FIG. 3.
Translation rates of the wild-type and codon-optimized genes. 293T cells were transfected with 2 μg of pGP-RRE3 (with or without pCMV-Rev [1 μg]) or 2 μg of pSYNGP. Protein samples from culture supernatants (A) and cell extracts (B) were analyzed by Western blotting 12, 25, 37, and 48 h posttransfection. HIV-1-positive human serum was used to detect Gag-Pol proteins (A and B), and an antiactin antibody was used as an internal control (C). The protein marker sizes (kilodaltons) at the left. A PhosphorImager was used for quantification of the results. Lanes: 1, pGP-RRE3 at 12 h; 2, pGP-RRE3 at 25 h; 3, pGP-RRE3 at 37 h; 4, pGP-RRE3 at 48 h; 5, pGP-RRE3 plus pCMV-Rev at 12 h; 6, pGP-RRE3 plus pCMV-Rev at 25 h; 7, pGP-RRE3 plus pCMV-Rev at 37 h; 8, pGP-RRE3 plus pCMV-Rev at 48 h; 9, pSYNGP at 12 h; 10, pSYNGP at 25 h; 11, pSYNGP at 37 h; 12, pSYNGP at 48 h; 13, mock-transfected 293T cells.
To further determine whether the enhanced expression that was observed with the codon-optimized gene was due to better translation or due to effects on the RNA, RNA analysis was carried out.
Codon optimization increases the mRNA levels of HIV-1 gag-pol.
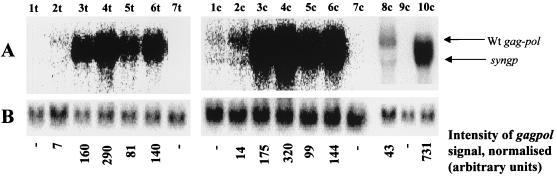
Northern blot analysis was performed to quantify the effect of codon optimization on RNA levels. Total and cytoplasmic RNA was isolated for this purpose. Correct fractionation was verified by staining the agarose gel. In the total cellular fractions the 47S rRNA precursor could be clearly seen, whereas it was absent from the cytoplasmic fractions (data not shown). The blots were hybridized with a probe specific for both the codon-optimized and wild-type gag-pol mRNAs (a 281-nt probe complementary to the overlapping region between the gag and pol reading frames; see Materials and Methods). Reprobing with a probe specific for human ubiquitin was performed in order to assess differences in the amounts of RNA loaded on the gel. The results are shown in Fig. 4. As expected (38), Rev stimulates the cytoplasmic accumulation of wild-type gag-pol mRNA (lanes 1c and 2c). RNA levels were 10- to 20-fold higher for the codon-optimized gene than for the wild-type one, both in total and cytoplasmic fractions (compare lanes 3t and 2t, 3c and 2c, and 10c and 8c). The RRE sequence did not significantly destabilize the codon-optimized RNAs since RNA levels were similar for codon-optimized RNAs whether or not they contained the RRE sequence (compare lanes 3 and 5). Rev did not markedly enhance cytoplasmic accumulation of the codon-optimized gag-pol mRNAs, even when they contained the RRE sequence (differences in RNA levels were less than twofold; compare lanes 3 and 4 or 5 and 6).
FIG. 4.
gag-pol mRNA levels in total and cytoplasmic fractions. Total and cytoplasmic RNA was extracted from 293T cells 36 h after transfection with 5 μg of the gag-pol expression plasmid (with or without pCMV-Rev [1 μg]), and mRNA levels were estimated by Northern blot analysis. A probe complementary to nt 1222 to 1503 of both the wild-type (Wt) and codon-optimized gene was used. (A) Band corresponding to HIV-1 gag-pol. The sizes of the mRNAs are 4.4 kb for the codon-optimized gene and 6 kb for the wild-type gene. (B) Band corresponding to human ubiquitin (internal control for normalization of results). Quantification was performed using a PhosphorImager. Lanes (c, cytoplasmic fraction; t, total RNA fraction): 1, pGP-RRE3; 2, pGP-RRE3 plus pCMV-Rev; 3, pSYNGP; 4, pSYNGP plus pCMV-Rev; 5, pSYNGP-RRE; 6, pSYNGP-RRE plus pCMV-Rev; 7, mock-transfected 293T cells; 8, pGP-RRE3 plus pCMV-Rev; 9, mock-transfected 293T cells; 10, pSYNGP.
It appeared from a comparison of Fig. 2 and 4 that all of the increase in protein expression from SYNGP could be accounted for by the increase in RNA levels. In order to investigate whether this was due to saturating levels of RNA in the cell, we transfected 0.1, 1, and 10 μg of the wild-type or codon-optimized expression vectors into 293T cells and compared protein production. In all cases protein production was 10-fold higher for the codon-optimized gene for the same amount of transfected DNA, while the increase in protein levels was proportional to the amount of transfected DNA for each individual gene (data not shown). It seems likely, therefore, that the enhanced expression of the codon-optimized gene can be mainly attributed to the enhanced RNA levels present in the cytoplasm and not to increased translation. However, the molecular basis of this finding was not identified.
The effect of HIV-1 gag INS on the codon-optimized gene is position dependent.
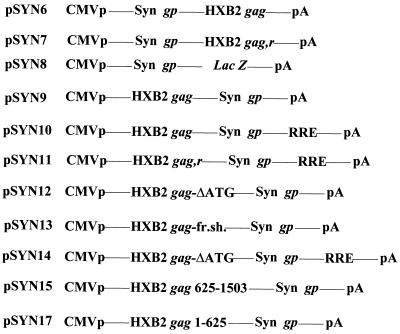
It has previously been demonstrated that insertion of wild-type HIV-1 gag sequences downstream of other RNAs, e.g., HIV-1 tat (58), HIV-1 gag (56), or chloramphenicol acetyltransferase gene (36) can lead to a dramatic decrease in steady-state mRNA levels, presumably as a result of the INS. In other cases, e.g., for β-globin (42), it was shown that the effect was splice site dependent. Cellular AU-rich elements that are found in the 3′ UTR of labile mRNAs may confer mRNA destabilization by inducing cytoplasmic deadenylation of the transcripts (71). To test whether HIV-1 gag INS would destabilize the codon-optimized RNA, the HIV-1 wild-type gag from the HXB2 strain was inserted downstream of the codon-optimized gag-pol gene (pSYN6). As controls the wild-type HIV-1 gag in the reverse orientation (as INS have been shown to act in an orientation-dependent manner [36]) (pSYN7) and lacZ (in the correct orientation) (pSYN8) were also inserted in the same site. Contrary to our expectation, as shown in Fig. 5, the wild-type HIV-1 gag sequence did not appear to significantly affect RNA or protein levels of the codon-optimized gene. We further constructed another series of plasmids where the wild-type HIV-1 gag in the sense or reverse orientation, subfragments of gag (nt 1 to 625 or 625 to 1503), the wild-type HIV-1 gag without the ATG or with a frameshift mutation 25 bases downstream of the ATG, or the first 1,093 bases of lacZ with or without the ATG were inserted upstream of the codon-optimized HIV-1 gag-pol gene in pSYNGP and/or pSYNGP-RRE (pSYN9-pSYN22; Fig. 6). Northern blot analysis showed that insertion of the wild-type HIV-1 gag gene upstream of the codon-optimized HIV-1 gag-pol (pSYN9, pSYN10) led to diminished RNA levels in the presence or absence of Rev/RRE (Fig. 7A, lanes 1 to 4, and 7B, lanes 1 and 12). The effect was not dependent on translation, as insertion of a wild-type HIV-1 gag lacking the ATG or with a frameshift mutation (pSYN12, pSYN13, and pSYN14) also diminished RNA levels (Fig. 7B, lanes 1 to 7). Western blot analysis verified that there was no HIV-1 Gag translation product for pSYN12-14 (data not shown). However, it is possible that, as the wild-type HIV-1 gag exhibits such an adverse codon usage, it may act as a nontranslatable long 5′ leader for SYNGP, and if this is the case, then the ATG mutation should not have any effects.
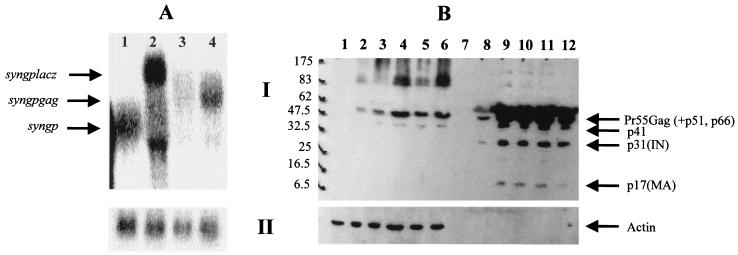
FIG. 5.
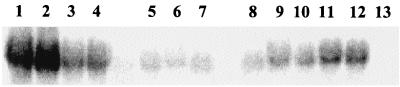
Effect of the insertion of the wild-type gag sequence downstream of the codon-optimized gene on RNA and protein levels. The wild-type gag sequence was inserted downstream of the codon-optimized gene in both orientations (NotI site), resulting in plasmids pSYN6 (correct orientation; Fig. 6) and pSYN7 (reverse orientation; Fig. 6). The gene encoding β-Gal (lacZ) was also inserted in the same site and in the correct orientation (plasmid pSYN8; Fig. 6). 293T cells were transfected with 5 μg of each plasmid, and 48 h posttransfection mRNA and protein levels were determined as previously described by means of Northern and Western blot analyses, respectively. (A) Northern blot analysis of cytoplasmic RNA fractions. The blot was probed with a probe complementary to nt 1510 to 2290 of the codon-optimized gene (I) and was reprobed with a probe specific for human ubiquitin (II). Lanes: 1, pSYNGP; 2, pSYN8; 3, pSYN7; 4, pSYN6. (B) Western blot analysis. HIV-1-positive human serum was used to detect the Gag-Pol proteins (I), and an antiactin antibody was used as an internal control (II). Lanes with cell lysates: 1, mock-transfected 293T cells; 2, pGP-RRE3 plus pCMV-Rev; 3, pSYNGP; 4, pSYN6; 5, pSYN7; 6, pSYN8; lanes with supernatants: 7, mock-transfected 293T cells; 8, pGP-RRE3 plus pCMV-Rev; 9, pSYNGP; 10, pSYN6; 11, pSYN7; 12, pSYN8. The protein marker (New England Biolabs) sizes are at the left.
FIG. 6.
Plasmids used to study the effect of HIV-1 gag INS on the codon-optimized gene. The backbone for all constructs was pCI-Neo. Syn gp, codon-optimized HIV-1 gag-pol gene; HXB2 gag, wild-type HIV-1 gag gene. HXB2 gag,r, wild-type HIV-1 gag gene in the reverse orientation; HXB2 gag-ΔATG, wild-type HIV-1 gag gene without the gag ATG; HXB2 gag-fr.sh., wild-type HIV-1 gag gene with a frameshift mutation; HXB2 gag 625-1503, nt 625 to 1503 of the wild-type HIV-1 gag gene; HXB2 gag 1-625, nt 1 to 625 of the wild-type HIV-1 gag gene.
FIG. 7.
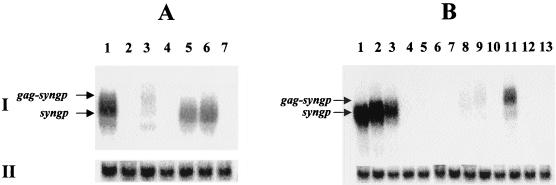
Insertion of HIV-1 gag upstream of the codon optimized gene affects cytoplasmic RNA levels. Cytoplasmic RNA was extracted 48 h after transfection of 293T cells (5 μg of each pSYN plasmid was used, and 1 μg of pCMV-Rev was cotransfected in some cases). The probe that was used was designed to be complementary to nt 1510 to 2290 of the codon-optimized gene (I). A probe specific for human ubiquitin was used as an internal control (II). (A) Lanes: 1, pSYNGP; 2, pSYN9; 3, pSYN10; 4, pSYN10 plus pCMV-Rev; 5, pSYN11; 6, pSYN11 plus pCMV-Rev; 7, pCMV-Rev. (B) Lanes: 1, pSYNGP; 2, pSYNGP-RRE; 3, pSYNGP-RRE plus pCMV-Rev; 4, pSYN12; 5, pSYN14; 6, pSYN14 plus pCMV-Rev; 7, pSYN13; 8, pSYN15; 9, pSYN17; 10, pGP-RRE3; 11, pSYN6; 12, pSYN9; 13, pCMV-Rev.
Insertion of smaller parts of the wild-type HIV-1 gag gene (pSYN15 and pSYN17) also led to a decrease in RNA levels (Fig. 7B, lanes 1 to 3 and 8 and 9) but not to levels as low as when the whole gag sequence was used (compare lanes 1 to 3, 4 to 7, and 8 and 9 in Fig. 7B). This indicates that the effect of INS is dependent on their sizes. Insertion of the wild-type HIV-1 gag in the reverse orientation (pSYN11) had no effect on RNA levels (Fig. 7A, lanes 1 and 5 and 6). However a splicing event seemed to take place in that case, as indicated by the size of the RNA (equal to the size of the codon-optimized gag-pol RNA) and by the translation product (Gag-Pol in equal amounts compared to that for pSYNGP, as verified by Western blot analysis; data not shown).
These data indicate, therefore, that wild-type HIV-1 gag INS act in a position- and size-dependent manner, probably irrespective of translation. However, we were unable at this stage to identify the mechanism underlying this effect. It should also be noted that the RRE was unable to rescue the destabilized RNAs through interaction with Rev. It has been shown (42) that the ability of the RRE to stimulate a Rev response could depend on the HIV sequence or other sequences surrounding the RRE, and this may explain why the Rev/RRE system was dysfunctional in our experiments. It would be interesting to investigate whether a Rev response would be observed if the RRE was to be placed, for example, between the wild-type and codon-optimized sequences.
The codon-optimized gag-pol gene does not use the exportin-1 nuclear export pathway.
Rev mediates the export of unspliced and singly spliced HIV-1 mRNAs via the nuclear export receptor exportin-1 (CRM1) (21, 22, 51, 62, 63). Leptomycin B (LMB) has been shown to inhibit leucine-rich nuclear export sequence (NES)-mediated nuclear export by disrupting the formation of the exportin-1–NES–RanGTP complex (48, 51). In particular, LMB inhibits nucleocytoplasmic translocation of Rev and Rev-dependent HIV mRNAs (70). To investigate whether exportin-1 mediates the export of the codon-optimized gag-pol constructs, the effect of LMB on protein production was tested. Western blot analysis was performed on cell lysates from cells transfected with the gag-pol constructs (with or without pCMV-Rev) and treated with LMB (7.5 nM; for 20 h, beginning 5 h posttransfection) or not treated. To confirm that LMB had no global effects on transport, the expression of β-Gal from the control plasmid pCMV-βGal was also measured. An actin internal control was used to account for protein variations between samples. The results are shown in Fig. 8. As expected (70), the wild-type gag-pol was not expressed in the presence of LMB (compare lanes 3 and 4), whereas LMB had no effect on protein production from the codon-optimized gag-pol, irrespective of the presence of the RRE in the transcript and the provision of Rev in trans (compare lanes 5 and 6, 7 and 8, 9 and 10, 11 and 12; also, compare lanes 5 and 6 to lanes 11 and 12). The resistance of the expression of the codon-optimized gag-pol to inhibition by LMB indicates that the exportin-1 pathway is not used and therefore an alternative export pathway must be used. This offers a possible explanation for the Rev-independent expression. The fact that the presence of a nonfunctional Rev-RRE interaction did not affect expression implies that the RRE does not necessarily act as an inhibitory (e.g., nuclear retention) signal per se, which is in agreement with previous observations (11, 42).
FIG. 8.
Nuclear export: the effect of LMB on protein production. 293T cells were transfected with 1 μg of pCMV-Rev and 3 μg of pGP-RRE3, pSYNGP, or pSYNGP-RRE (with or without pCMV-Rev [1 μg]). Transfections were done in duplicate. Five hours posttransfection the medium was replaced with fresh medium in the first set and with fresh medium containing 7.5 nM LMB in the second. Twenty hours later the cells were lysed and protein production was estimated by Western blot analysis. HIV-1-positive human serum was used to detect the Gag-Pol proteins (A), and an antiactin antibody was used as an internal control (B). Lanes: 1, pGP-RRE3; 2, pGP-RRE3 plus LMB; 3, pGP-RRE3 plus pCMV-Rev; 4, pGP-RRE3 plus pCMV-Rev plus LMB; 5, pSYNGP; 6, pSYNGP plus LMB; 7, pSYNGP plus pCMV-Rev; 8, pSYNGP plus pCMV-Rev plus LMB; 9, pSYNGP-RRE; 10, pSYNGP-RRE plus LMB; 11, pSYNGP-RRE plus pCMV-Rev; 12, pSYNGP-RRE plus pCMV-Rev plus LMB.
The efficiency of vector production using the codon-optimized gag-pol gene.
To determine the effects of the codon-optimized gag-pol on vector production, we used an HIV vector genome, pH6nZ (see Materials and Methods), and the vesicular stomatitis virus glycoprotein (VSV-G) envelope expression plasmid pHCMVG (73), in combination with either pSYNGP, pSYNGP-RRE, pSYNGP-ERR, or pGP-RRE3, as a source for the gag-pol in a plasmid ratio of 2:1:2 in a three-plasmid cotransfection of 293T cells (31). Whole-cell extracts and culture supernatants were evaluated by Western blot analysis for the presence of the gag and gag-pol gene products. Particle production was, as expected (Fig. 2), 5- to 10-fold higher for the codon-optimized genes than for the wild type (data not shown).
To determine the effects of the codon-optimized gag-pol gene on vector titers, several ratios of the vector components were used. The results are summarized in Table 2. Whereas the gag-pol was the limiting component in the system (as determined by the drop in titers observed with the wild-type gene), titers were 10-fold higher for the codon-optimized vectors. This is in agreement with the higher protein production observed for these vectors but suggests that under normal conditions of vector production Gag-Pol is saturating and the codon optimization gives no maximum-yield advantage.
TABLE 2.
Vector titersa
| Amt of pH6NZ (μg) | Ratio of gag-pol expression plasmid to pH6NZ (μg:μg) | Titer (TU/ml of virus stock) for:
|
|||
|---|---|---|---|---|---|
| pGP-RRE3 | pSYNGP | pSYNGP- RRE | pSYNGP- ERR | ||
| 5 | 1:1 | 6.0 × 105 | 5.0 × 105 | 4.0 × 105 | 3.0 × 105 |
| 12.5 | 1:2.5 | 8.0 × 105 | 6.5 × 105 | ||
| 10 | 1:10 | 3.0 × 105 | 4.0 × 105 | ||
| 5 | 1:20 | 3.5 × 105 | 2.5 × 105 | ||
| 12.5 | 1:50 | 3.5 × 104 | 3.0 × 105 | 3.0 × 105 | 3.0 × 105 |
| 18 | 1:180 | 2.5 × 104 | 3.0 × 105 | ||
Viral stocks were generated by cotransfection of each gag-pol expression plasmid, pH6nZ (vector genome plasmid), and pHCMVG (VSV envelope expression plasmid; 2.5 μg for each transfection) on 293T cells. Titers were measured on 293T cells by counting the number of blue colonies following X-Gal staining 48 h after transduction. Experiments were performed at least twice, and the variation between experiments was less than 15%.
Construction of an HIV-1-based vector system that lacks all the accessory proteins.
Previously, several HIV-1-based vector systems that lack all accessory proteins but Rev were reported (31, 43). We wished to investigate whether the codon-optimized gene would permit the construction of an HIV-1-based vector system that lacks all accessory proteins. We initially deleted rev/RRE and any residual env sequences but kept the first 625 nt of gag, as they have been shown to play a role in efficient packaging (50). Two vector genome constructs were made, pH6.1nZ (retaining only HIV sequences up to nt 625 of gag) and pH6.2nZ (same as pH6.1nZ but also retaining the env splice acceptor). These were derived from a conventional HIV vector genome that contains RRE and that expresses Rev (pH6nZ). Our three-plasmid vector system now expressed only HIV-1 Gag-Pol and the VSV-G envelope protein. Vector particle titers were determined as described in the previous section. A 2:2:1 ratio of the vector genome (pH6Z, pH6.1nZ, or pH6.2nZ) to the Gag-Pol expression vector (pGP-RRE3 or pSYNGP) to pHCMV-G was used. Transfections were performed in the presence or absence of pCMV-Rev, as Gag-Pol expression was still Rev dependent for the wild-type gene. The results are summarized in Table 3 and indicate that an HIV vector could be produced in the total absence of Rev but that maximum titers were 20-fold lower than could be achieved in the presence of Rev. As Gag-Pol expression should be the same for pSYNGP with pH6nZ, pH6.1nZ, or pH6.2nZ (since it is Rev independent), as well as for pGP-RRE3 when Rev is provided in trans, we suspected that the vector genome retained a requirement for Rev and was therefore limiting the titers. To confirm this, Northern blot analysis was performed on cytoplasmic RNA prepared from cells transfected with pH6nZ or pH6.1nZ in the presence or absence of pCMV-Rev. As can be seen in Fig. 9, lanes 1 to 4, the levels of cytoplasmic RNA derived from pH6nZ were 5- to 10-fold higher than those obtained with pH6.1nZ (compare lanes 1 and 2 to lanes 3 and 4). These data support the notion that RNA produced from the vector genome requires the Rev/RRE system to ensure high cytoplasmic levels. This may be due to inefficient nuclear export of the RNA, as INS residing within gag were still present.
TABLE 3.
Vector titers from genomes with or without Rev/RREa
| Gag-Pol expression vector | Vector genome | Rev expression/ plasmid | Titer (TU/ml of viral stock ± SD) |
|---|---|---|---|
| pGP-RRE3 | pH6nZ | Yes/pH6Z | (8.0 ± 0.2) × 105 |
| pGP-RRE3 | pH6.1nZ | No | (2.1 ± 0.3) × 103 |
| pGP-RRE3 | pH6.1nZ | Yes/pCMV-Rev | (4.1 ± 0.3) × 104 |
| pGP-RRE3 | pH6.2nZ | No | (1.7 ± 0.4) × 103 |
| pGP-RRE3 | pH6.2nZ | Yes/pCMV-Rev | (5.6 ± 0.3) × 104 |
| pSYNGP | pH6nZ | Yes/pH6Z | (7.8 ± 0.2) × 105 |
| pSYNGP | pH6.1nZ | No | (5.4 ± 0.3) × 104 |
| pSYNGP | pH6.1nZ | Yes/pCMV-Rev | (5.2 ± 0.4) × 104 |
| pSYNGP | pH6.2nZ | No | (5.0 ± 0.2) × 104 |
| pSYNGP | pH6.2nZ | Yes/pCMV-Rev | (5.5 ± 0.3) × 104 |
The retroviral vector was generated as described in Materials and Methods. Titers were determined in 293T cells.
FIG. 9.
Cytoplasmic RNA levels of the vector genomes. 293T cells were transfected with 10 μg of each vector genome. Cytoplasmic RNA was extracted 48 h posttransfection, and 20 μg of RNA was used from each sample for Northern blot analysis. The 700-bp probe was designed to hybridize to all vector genome RNAs (see Materials and Methods). Lanes: 1, pH6nZ; 2, pH6nZ plus pCMV-Rev; 3, pH6.1nZ; 4, pH6.1nZ plus pCMV-Rev; 5, pHS1nZ; 6, pHS2nZ; 7, pHS3nZ; 8, pHS4nZ; 9, pHS5nZ; 10, pHS6nZ; 11, pHS7nZ; 12, pHS8nZ; 13, pCMV-Rev.
Further deletions in the gag sequences of the vector genome might therefore be necessary to restore titers. To date efficient packaging has been reported to require 360 (17) or 255 (14) nt of gag in vectors that still retain env sequences, or about 40 nt of gag in a particular combination of splice donor (SD) mutation and gag and env deletions (12, 14). In an attempt to remove the requirement for Rev/RRE in our vector genome without compromising efficient packaging, we constructed a series of vectors derived from pH6nZ containing progressively larger deletions of HIV-1 sequences (only sequences upstream and within gag were retained) with or without a mutant major SD (GT-to-CA mutation). Vector particle titers were determined as described before, and the results are summarized in Table 4. As can be seen, deletion of up to nt 360 in gag (vector pHS3nZ) resulted in an increase in titers (compared to those with pH6.1nZ or pH6.2nZ) and only a fivefold decrease (titers were 1.3 × 105 to 1.7 × 105 TU/ml) compared to those with pH6nZ. Further deletions resulted in titers lower than those with pHS3nZ and similar to those with pH6.1nZ. In addition, the SD mutation did not have a positive effect on vector titers, and for pHS3nZ it resulted in a 10-fold decrease in titers (compare titers for pHS3nZ and pHS7nZ in Table 4). Northern blot analysis of cytoplasmic RNA (Fig. 9, lanes 1 and 5 to 12) showed that RNA levels were indeed higher for pH6nZ, which could account for the maximum titers observed with this vector. RNA levels were equal for pHS1nZ (lane 5), pHS2nZ (lane 6), and pHS3nZ (lane 7), whereas titers were five- to eightfold higher for pHS3nZ. It is possible that further deletions (a larger deletion than that found in pHS3nZ) in gag might result in less-efficient packaging (because for HIV-1 the packaging signal extends into gag) and therefore that even though all three vectors produce similar amounts of RNA only pHS3nZ retains maximum packaging efficiency. It is also interesting that the SD mutation resulted in increased RNA levels in the cytoplasm (compare lanes 6 and 10, 7 and 11, or 8 and 12 in Fig. 9) but equal or decreased titers (Table 4). The GT dinucleotide that was mutated is in the stem of SL2 of the packaging signal (26). It has been reported that SL2 might not be very important for HIV-1 RNA encapsidation (26, 40), whereas SL3 is of great importance (35). The folding of the wild-type and SD mutant vector sequences with the RNAdraw software program revealed that the mutation alters significantly the secondary structure of the RNA and not only that of SL2. It is likely, therefore, that although the SD mutation enhances cytoplasmic RNA levels it does not increase titers because it alters the secondary structure of the packaging signal.
TABLE 4.
Vector titers for the pHS series of vector genomesa
| Vector genome | gag length (nt) | SD mutation | Titer (TU/ml of viral stock ± SD) for:
|
||
|---|---|---|---|---|---|
| pSYNGP | pGP-RRE3 | pGP-RRE + Revb | |||
| pH6nZ | 625 | No | (6.7 ± 0.7) × 105 | Not done | Not done |
| pH6.1nZ | 625 | No | (3.2 ± 0.1) × 104 | Not done | Not done |
| pH6.2nZ | 527 | No | (2.7 ± 0.3) × 104 | Not done | Not done |
| pHS1nZ | 40 | No | (2.1 ± 0.4) × 104 | (2.1 ± 0.9) × 103 | (1.4 ± 0.3) × 104 |
| pHS2nZ | 260 | No | (1.5 ± 0.3) × 104 | (4.2 ± 0.6) × 103 | (3.0 ± 0.5) × 104 |
| pHS3nZ | 360 | No | (1.3 ± 0.3) × 105 | (6.5 ± 0.7) × 103 | (1.7 ± 0.4) × 105 |
| pHS4nZ | 625 | No | (1.0 ± 0.9) × 104 | (1.2 ± 0.7) × 103 | (1.0 ± 0.3) × 104 |
| pHS5nZ | 40 | Yes | (2.0 ± 0.5) × 104 | (6.0 ± 0.8) × 102 | (2.3 ± 0.5) × 104 |
| pHS6nZ | 260 | Yes | (1.0 ± 0.2) × 104 | (8.1 ± 0.6) × 102 | (1.0 ± 0.4) × 104 |
| pHS7nZ | 360 | Yes | 1.9 × 104 | (2.4 ± 0.9) × 103 | (4.2 ± 0.8) × 104 |
| pHS8nZ | 625 | Yes | (8.0 ± 1.0) × 103 | (1.0 ± 0.5) × 103 | (4.0 ± 0.9) × 103 |
The retroviral vector was generated as described in Materials and Methods. Titers were determined in 293T cells.
Rev was provided from pCMV-Rev. Note that pH6nZ expresses Rev and contains the RRE. None of the other genomes express Rev or contain the RRE. Expression from pSYNGP is Rev independent, whereas it is Rev dependent for pGP-RRE3.
To investigate whether the titer differences that were observed with the vectors lacking the Rev coding sequence were indeed due to the Rev dependence of the genomes, the RRE sequence (nt 7769 to 8021 of the HXB2 sequence) was inserted in the SpeI site (downstream of the gag sequence and just upstream of the internal cytomegalovirus promoter) of pH6.1nZ, pHS1nZ, pHS3nZ, and pHS7nZ, resulting in plasmids pH6.1nZR, pHS1nZR, pHS3nZR, and pHS7nZR, respectively. Vector particle titers were determined with pSYNGP and pHCMVG in the presence or absence of Rev (pCMV-Rev) as before, and the results are summarized in Table 5. In the absence of Rev titers were further compromised for pH6.1nZR (7-fold compared to those for pH6.1nZ), pHS3nZR (6-fold compared to those for pHS3nZ), and pHS7nZR (2.5-fold compared to those for pHS7nZ). This was expected, as the RRE also acts as an INS (9) and so it would be expected to confer Rev dependence. In the presence of Rev titers were restored to the maximum titers observed for pH6nZ in the case of pHS3nZR (5 × 105 TU/ml) and pH6.1nZR (2 × 105 TU/ml). Titers were not restored for pHS7nZR in the presence of Rev. This supports the hypothesis that the SD mutation in pHS7nZ affects the structure of the packaging signal and thus the packaging ability of this vector genome, because in this case Rev may be able to stimulate vector genome RNA levels, as for pHS3nZR and pH6.1nZR, but it cannot affect the secondary structure of the packaging signal. For vector pHS1nZ inclusion of the RRE did not lead to a decrease in titers. This could be due to the fact that pHS1nZ contains only 40 nt of gag sequences and therefore even with the RRE the sizes of INS are not larger than the sizes for pHS2nZ that give equal titers to pHS1nZ. Rev was able to partially restore titers for pHS1nZR (10-fold increase compared to those for pHS1nZ and 8-fold lower than those for pH6nZ) but not fully as for pHS3nZ. This is also in agreement with the hypothesis that 40 nt of HIV-1 gag sequences might not be sufficient for efficient vector RNA packaging, and this could account for the incomplete restoration in titers observed with pHS1nZR in the presence of Rev.
TABLE 5.
Vector titers for the pHS series of vector genomes in the presence or absence of Rev/RREa
| Vector genome | Titer (TU/ml) for:
|
|
|---|---|---|
| pSYNGP | PSYNGP + Revb | |
| pH6nZ | 6.5 × 105 | 6.9 × 105 |
| pH6.1nZ | 1.5 × 104 | 1.7 × 104 |
| pH6.1nZR | 2.3 × 103 | 1.6 × 105 |
| pHS1nZ | 8.5 × 103 | 8.4 × 103 |
| pHS1nZR | 8.1 × 103 | 8.8 × 104 |
| pHS3nZ | 1.2 × 105 | 1.4 × 105 |
| pHS3nZR | 2.5 × 103 | 4.8 × 105 |
| pHS7nZ | 7.3 × 103 | 7.0 × 103 |
| pHS7nZR | 3.4 × 103 | 5.8 × 103 |
The retroviral vector was generated as described in Materials and Methods. Five micrograms of vector genome, 5 μg of pSYNGP, and 2.5 μg of pHCMVG were used, and titers were determined in 293T cells. Experiments were performed at least twice, and the variation between experiments was less than 15%.
Rev was provided from pCMV-Rev (1 μg). Note that pH6nZ expresses Rev and contains the RRE. None of the pHS genomes expresses Rev, and only pHS1nZR, pHS3nZR, pHS7nZR, and pH6.1nZR contain the RRE. Gag-Pol expression from pSYNGP is Rev independent.
In addition, end point titers were determined for pHS3nZ and pH6nZ with pSYNGP in HeLa and HT1080 human cell lines. In both cases titers followed the pattern observed in 293T cells, with titers being two- to threefold lower for pHS3nZ than for pH6nZ (Fig. 10). Finally, the transduction efficiencies of the vector produced with pHS3nZ or pH6nZ and different amounts of pSYNGP or pGP-RRE3 at different multiplicities of infection (MOI) (as high as 1) were determined in HT1080 cells. This experiment was performed as the high-level Gag-Pol expression from pSYNGP may result in interference by genome-empty particles at high vector concentrations. As expected for VSV-G-pseudotyped retroviral particles (4), transduction efficiencies correlated with the MOI whether large or small amounts of pSYNGP were used with pH6nZ or pHS3nZ. For an MOI of 1 transduction efficiency was approximately 50 to 60% in all cases (Fig. 10). The above data indicate that no interference due to genome-empty particles is observed in this experimental system.
FIG. 10.
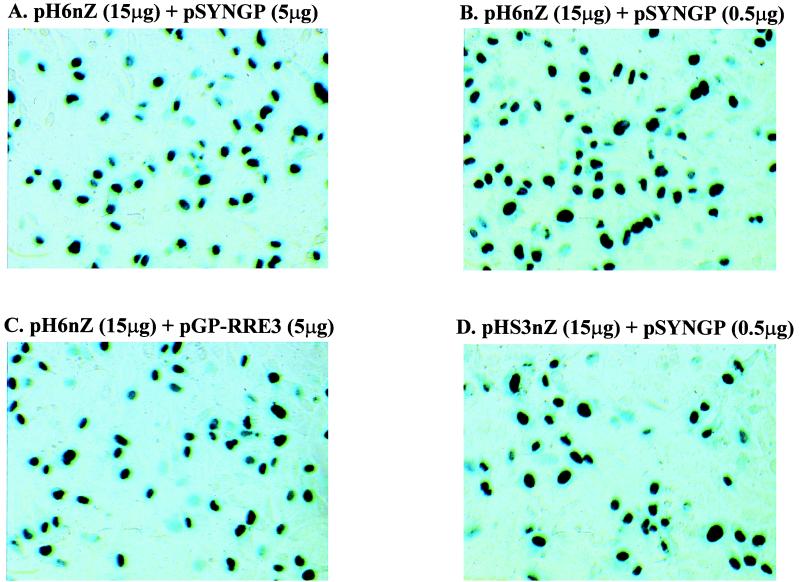
Transduction efficiency at an MOI of 1. Viral stocks were generated by cotransfection of each gag-pol expression plasmid (5 or 0.5 μg), 15 μg of pH6nZ or pHS3nZ (vector genome plasmid), and 5 μg of pHCMVG (VSV envelope expression plasmid) on 293T cells. Virus was concentrated as previously described (45), and transduction efficiency was determined at MOI from 0.01 to 1 on HT1080 cells. There was a linear correlation of transduction efficiency and MOI in all cases. An indicative picture at an MOI of 1 is shown here. Transduction efficiency was 50 to 60% with either genome, gag-pol, and either large or small amounts of pSYNGP. Titers before concentration (TU/ml) on 293T cells: 6.6 × 105 (A); 7.6 × 105 (B); 9.2 × 105 (C); 1.5 × 105 (D); titers before concentration (TU/ml) on HT1080 cells: 6.0 × 104 (A); 9.9 × 104 (B); 8.0 × 104 (C); 2.9 × 104 (D). Titers after concentration (TU/ml) on HT1080 cells: 6.0 × 105 (A); 2.0 × 106 (B); 1.4 × 106 (C); 2.0 × 105 (D).
In conclusion, this is the first report of an HIV-1-based vector system, composed of pSYNGP, pHS3nZ, and pHCMVG, where significant vector production can be achieved in the absence of all accessory proteins. These data indicate that, in order to achieve maximum titers, the HIV vector genome must be configured to retain efficient packaging and that this requires the retention of gag sequences and SD. By reducing the gag sequence to 360 nt in pHS3nZ and combining this with pSYNGP, it is possible to achieve a titer of at least 105 TU/ml, which is only fivefold lower than the maximum levels achieved in the presence of Rev.
DISCUSSION
We have shown that codon optimization of the HIV-1 gag-pol gene results in a 10-fold increase in steady-state levels of cognate RNA with an equivalent 10-fold increase in protein production. Furthermore, unlike that for the wild-type gag-pol gene, expression occurs independently of the Rev/RRE system. Rev-independent expression of HIV-1 genes has been previously shown (24, 56). We have further demonstrated that, consistent with Rev independence, LMB has no effect on expression, implying that the exportin-1 (CRM1) nuclear export pathway is not involved.
In previous studies increased protein production upon codon optimization has been attributed to more-efficient translation (24). The molecular basis for the increased RNA levels of our codon-optimized gene is not yet known. Elimination of INS residing within the coding sequence (36, 57, 58) is one possible explanation. Another possibility is that inefficient translation can lead to increased probability of mRNA degradation and that this has been obviated by the change in codon bias. For example, it has been shown that mRNAs that contain premature termination codons are easily degraded (6; reviewed in reference 27) and that sequences involved in rapid mRNA turnover can be active only when they are part of the translation open reading frame (e.g., in c-myc) (69). However, the increase in levels of protein paralleled rather than exceeded the increase in RNA levels. It is possible that Gag is autoregulating its expression to allow viral RNA packaging as has been proposed for Rous sarcoma virus (61). This is further supported by the fact that HIV-1 Gag interacts with translation factors and inhibits translation in vitro (13, 67), and it could also explain why codon optimization for HIV-1 env, but not for gag-pol, results in a significant augmentation in protein levels and equal RNA levels (24).
In this study we have also demonstrated that HIV-1 gag INS can act in a position-dependent manner irrespective of translation and that their effects are INS size dependent. However, the molecular mechanism underlying the action of these INS elements was not determined. One hypothesis for the action of INS elements in general is that they may induce mRNA deadenylation and rapid degradation in the cytoplasm. This is the favored scenario for AU-rich elements found in the 3′ UTR of cellular labile mRNAs (71). Alternatively, they may exert their effects prior to the exit of the mRNA from the nucleus (42). The results obtained here are in agreement with the second hypothesis, as the observed effect seemed to be translation independent, and indicate that HIV-1 gag INS elements may also act by retaining the mRNA in the nucleus (47) and possibly directing it towards a nuclear degradation pathway (37, 42). This is further supported by previous reports showing that HIV-1 pol INS elements interact with several cellular factors, one of which has been identified as hnRNP C, a component of the splicing machinery (47). However, a translational effect cannot be ruled out at this stage.
With our codon-optimized gene we achieved higher vector particle production than with the wild-type gene, but this did not convert into higher vector titers, indicating that Gag-Pol is not limiting. As expected, however, in plasmid ratios where the Gag-Pol is limiting, i.e., one gag-pol gene per 50 genomes, a 10-fold increase in titers was observed with the codon-optimized gene. These data suggest that further improvements in vector yield could be made by increasing genome levels in combination with pSYNGP.
The codon-optimized gene may be of particular use for the production of simpler and safer HIV vectors. To date these still require the provision of Rev to facilitate gag-pol expression (17, 31), and this requirement is now removed by using the codon-optimized gene. There are alternative export sequences, such as constitutive transport elements (CTEs) (23, 74) and parts of histone RNAs (28), that are functional in Gag-Pol production, but these have not yet been proven to be as efficient as Rev/RRE, especially in the context of a vector system (23, 31). We have now demonstrated the feasibility of making a fully Rev-independent vector system without a significant compromise in titer. Our results indicate that the vector genome is also Rev dependent when large portions of gag or gag and env are present, but also that retention of more than 260 bases of gag is required for efficient infectious vector production. Further manipulations, such as inclusion of a CTE in the vector genome, may be necessary to overcome these limitations and fully restore titers. Another safety feature of vectors produced using the codon-optimized gag-pol gene is that there is no homology with the residual gag elements in the vector genome that are of necessity retained for packaging. More significantly, there is also no homology with the circulating natural strains of HIV-1 or HIV-2, thus eliminating the possibility of homologous recombination. The ability to render the genome Rev independent also removes the RRE homology between the vector and human retroviruses. These features should render the vector safer.
There may be other advantages of employing a codon-optimized gag-pol; for example, therapeutic ribozymes can now be targeted against gag and pol and yet can still be delivered by HIV vectors that will be resistant to their effects during production.
The codon-optimized gene could also be used for DNA vaccination approaches. The codon-optimized HIV env and gp120 (3) and the Rev-independent gag where the INS have been eliminated (52) have already been shown to significantly increase both cellular cytotoxic T-lymphocyte responses and humoral antibody titers when they were used for DNA immunization of BALB/c mice. The use of codon-optimized HIV genes for DNA vaccines has the advantage of higher expression levels that enhance the stimulation of immune responses and also eliminates risks of homologous recombination with viral genomes.
In conclusion, we have shown that codon optimization of the HIV-1 gag-pol gene leads to higher expression without the requirement for Rev/RRE. Furthermore, based on this codon-optimized gene, we have constructed an HIV-1-based vector system that now lacks all the accessory proteins, retains only minimal HIV-1 sequences (required for packaging and reverse transcription) in the vector genome construct, and exhibits no homology regions between its components. This vector system should now be safer than murine leukemia virus-based vectors and should facilitate the clinical evaluation of this new class of gene therapy vectors.
ACKNOWLEDGMENTS
We thank Barbara Wolff for the kind provision of LMB. We also thank Mark Uden, Melvyn Yap, and Enca Martin-Rendon for useful discussions and suggestions and Said Ismail and Enca Martin-Rendon for the construction of pH6nZ.
E. Kotsopoulou is a Marie Curie Fellow and has been supported by the Onassis foundation.
REFERENCES
- 1.Adachi A, Gendelman H, Koenig S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonina E, Neumann M, Pavlakis G. Preferential binding of poly(A)-binding protein I to an inhibitory RNA element in the HIV-1 gag mRNA. J Biol Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 3.Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai T, Takada M, Ui M, Iba H. Dose-dependent transduction of vesicular stomatitis virus G protein-pseudotyped retrovirus vector into human solid tumor cell lines and murine fibroblasts. Virology. 1999;260:109–115. doi: 10.1006/viro.1999.9773. [DOI] [PubMed] [Google Scholar]
- 5.Arrigo S J, Chen I S. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 6.Barker G F, Beemon K. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11:2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg O, Kurland C. Growth-rate optimised tRNA abundance and codon usage. J Mol Biol. 1997;270:1705–1711. doi: 10.1006/jmbi.1997.1142. [DOI] [PubMed] [Google Scholar]
- 8.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brighty D, Rosenberg M. A cis-acting repressive sequence that overlaps the Rev responsive element of HIV-1 regulates nuclear retention of env mRNAs independently of known splice signals. Proc Natl Acad Sci USA. 1994;91:8314–8318. doi: 10.1073/pnas.91.18.8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassan M, Delaunay N, Vaquero C, Rousset J P. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J Virol. 1994;68:1501–1508. doi: 10.1128/jvi.68.3.1501-1508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 12.Chang L-J, Urlacher V, Iwakama T, Cui Y, Zucali J. Efficacy and safety analysis of a recombinant HIV-1 derived vector system. Gene Ther. 1999;6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- 13.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y, Iwakama T, Chang L-J. Contribution of viral splice sites and cis regulatory elements to lentivirus vector function. J Virol. 1999;73:6171–6176. doi: 10.1128/jvi.73.7.6171-6176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Agostino D M, Felber B K, Harrison J E, Pavlakis G N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuBridge R B, Tang P, Hsia H C, Leong P-M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dull T, Zufferey R, Kelly M, Mandel R, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Favaro J, Maldarelli F, Arrigo S, Schmidt M. Effect of Rev on the cytoplasmic localization of intron-containing HIV-1 RNA. Virology. 1999;255:237–249. doi: 10.1006/viro.1998.9584. [DOI] [PubMed] [Google Scholar]
- 19.Felber B K, Hadzopoulou Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral messenger RNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher A, Collalti E, Ratner L, Gallo R, Wong-Staal F. A molecular clone of HTLV-III with biological activity. Nature. 1985;316:262–265. doi: 10.1038/316262a0. [DOI] [PubMed] [Google Scholar]
- 21.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 22.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasmi M, Glynn J, Jin M-J, Jolly D, Yee J-K, Chen S-T. Requirements for efficient production and transduction of human immunodeficiency virus type 1-based vectors. J Virol. 1999;73:1828–1834. doi: 10.1128/jvi.73.3.1828-1834.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas J, Park E-C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 25.Hadzopoulou Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison G, Miele G, Hunter E, Lever A. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentze M, Kulozik A. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Carmichael G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1 related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 30.Kim S Y, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 33.Kypr J, Mrazek J. Unusual codon usage of HIV. Nature. 1987;327:20. doi: 10.1038/327020a0. [DOI] [PubMed] [Google Scholar]
- 34.Kypr J, Mrazek J, Reich J. Nucleotide composition bias and CpG dinucleotide content in the genomes of HIV and HTLV 1 and 2. Biochim Biophys Acta. 1989;1009:280–282. doi: 10.1016/0167-4781(89)90114-0. [DOI] [PubMed] [Google Scholar]
- 35.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 39.Maurer F, Tierney M, Medcalff R. An AU-rich sequence in the 3′ UTR of PAI-2 mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res. 1999;27:1664–1673. doi: 10.1093/nar/27.7.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medappa K C, McLean C, Rueckert R R. On the structure of rhinovirus 1A. Virology. 1971;44:259–270. doi: 10.1016/0042-6822(71)90258-3. [DOI] [PubMed] [Google Scholar]
- 42.Mikaelian I, Krieg M, Gait M, Karn J. Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNAs. J Mol Biol. 1996;257:246–264. doi: 10.1006/jmbi.1996.0160. [DOI] [PubMed] [Google Scholar]
- 43.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr Opin Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 44.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 46.Nasioulas G, Zolotukhin A S, Tabernero C, Solomin L, Cunningham C P, Pavlakis G N, Felber B K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen H, Cochrane A, Rosen C. Interaction of cellular factors with intragenic cis-acting repressive sequences within the HIV genome. Virology. 1992;191:709–715. doi: 10.1016/0042-6822(92)90246-l. [DOI] [PubMed] [Google Scholar]
- 48.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkin N T, Chamorro M, Varmus H E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollard V, Malim M. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 52.Qiu J-T, Song R, Dettenhofer M, Tian C, August T, Felber B, Pavlakis G, Yu X-F. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J Virol. 1999;73:9145–9152. doi: 10.1128/jvi.73.11.9145-9152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabbits P, Forster A, Stinson M, Rabbits T. Truncation of exon 1 from the c-myc gene results in prolongued c-myc mRNA stability. EMBO J. 1985;4:3727–3733. doi: 10.1002/j.1460-2075.1985.tb04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rouwendal G J, Mendes O, Wolbert E J, Douwe de Boer A. Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage. Plant Mol Biol. 1997;33:989–999. doi: 10.1023/a:1005740823703. [DOI] [PubMed] [Google Scholar]
- 55.Sagerstrom C, Sive H. RNA blot analysis. In: Krieg P, editor. A laboratory guide to RNA: isolation, analysis and synthesis. Vol. 1. New York, N.Y: Wiley-Liss Inc.; 1996. pp. 83–104. [Google Scholar]
- 56.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharp P M. What can AIDS virus codon usage tell us? Nature. 1986;324:114. doi: 10.1038/324114a0. [DOI] [PubMed] [Google Scholar]
- 60.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sostengard T, Hackett P. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76Gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 62.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 63.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 64.van Hemert F J, Berkhout B. The tendency of lentiviral open reading frames to become A-rich: constraints imposed by viral genome organization and cellular tRNA availability. J Mol Evol. 1995;41:132–140. doi: 10.1007/BF00170664. [DOI] [PubMed] [Google Scholar]
- 65.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a nonuniform process: effect of transfer RNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 66.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 67.Wilson S, Sieiro-Vasquez C, Edwards N, Iourin O, Byles E, Kotsopoulou E, Adamson C, Kingsman S, Kingsman A, Martin-Rendon E. Cloning and characterisation of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem J. 1999;342:97–103. [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 69.Wisdom R, Lee W. The protein coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 70.Wolff B, Sanglier J-J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleocytoplasmic translocation of the HIV-1 Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 71.Xu N, Chen C-Y, Shyu A-B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang T T, Cheng L, Kain S R. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yee J K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]