Abstract
Aminooxypentane (AOP)-RANTES efficiently and specifically blocks entry of non-syncytium-inducing (NSI), CCR5-tropic (R5) human immunodeficiency virus type 1 (HIV-1) into host cells. Inhibition appears to be mediated by increased intracellular retention of the CCR5 coreceptor- AOP-RANTES complex and/or competitive binding of AOP-RANTES with NSI R5 HIV-1 isolates for CCR5. Although AOP-RANTES and other β-chemokine analogs are potent inhibitors, the extreme heterogeneity of the HIV-1 envelope glycoproteins (gp120 and gp41) and variable coreceptor usage may affect the susceptibility of variant HIV-1 strains to these drugs. Using the same peripheral blood mononuclear cells (PBMC) with all isolates, we observed a significant variation in AOP-RANTES inhibition of 13 primary NSI R5 isolates; 50% inhibitory concentrations (IC50) ranged from 0.04 nM with HIV-1A-92RW009 to 1.3 nM with HIV-1B-BaL. Experiments performed on the same isolate (HIV-1B-BaL) with PBMC from different donors revealed no isolate-specific variation in AOP-RANTES IC50 values but did show a considerable difference in virus replication efficiency. Exclusive entry via the CCR5 coreceptor by these NSI R5 isolates suggests that variable inhibition by AOP-RANTES is not due to alternative coreceptor usage but rather differential CCR5 binding. Analysis of the envelope V3 loop sequence linked a threonine or arginine at position 319 (numbering based on the HXB2 genome) with AOP-RANTES resistance. With the exception of one isolate, A319 was associated with increased sensitivity to AOP-RANTES inhibition. Distribution of AOP-RANTES IC50 values with these isolates has promoted ongoing screens for new CCR5 agonists that show broad inhibition of HIV-1 variants.
Host cell entry of human immunodeficiency virus type 1 (HIV-1) and other primate lentiviruses is mediated by binding of viral envelope glycoproteins (gp120 and gp41) to the CD4 receptor and another host membrane protein (2, 9, 18, 21, 24, 29). The predominant coreceptors for HIV-1 are CCR5 and CXCR4, but these seven transmembrane G-coupled receptors are not utilized interchangeably by all HIV-1 isolates (5, 7, 54, 55). Identification of a major coreceptor was facilitated by the initial discovery that some β-chemokines (i.e., RANTES, macrophage inflammatory proteins 1α [MIP-1α], and MIP-1β), natural ligands of CCR5 but not MCP-1 (a ligand of CCR 2a and 2b), could block infection of macrophage-tropic, non-syncytium-inducing (NSI) R5 virus (13). In general, T-cell-line-tropic isolates that form cell syncytia during active replication (termed syncytium-inducing [SI] X4) utilize the CXCR4 coreceptor, whereas the CCR5 coreceptor is employed by NSI R5 isolates (2, 18, 21, 24). CCR5 versus CXCR4 coreceptor usage (abbreviated R5 and X4, respectively) can be mapped to neutral/acidic versus basic residues in the V3 loop of the HIV-1 envelope glycoprotein gp120 (10, 11, 14, 25, 47, 52). Other coreceptors have now been identified (e.g., CCR2b, CCR3, STRL33/BONZO, gpr15/BOB, gpr1, and APJ), but these support the entry of few HIV-1 isolates and do so less efficiently than CCR5 or CXCR4 (12, 20, 22, 23, 28, 34).
New HIV-1 infections are generally established by transmission of NSI R5 isolates even though both SI X4 and NSI R5 strains may coexist in the donor (56). Recently, it was reported that individuals who are at high risk for HIV-1 acquisition and homozygous for a 32-amino-acid deletion in the CCR5 allele (approximately 1% of the Caucasian population) remain uninfected (17). This deletion prevents proper surface expression of CCR5, rendering peripheral blood mononuclear cells (PBMC) resistant to infection by NSI R5 isolates but still sensitive to SI X4 HIV-1 strains (35, 41). Other polymorphisms in the promoter region of CCR5 or RANTES appear to have an effect on HIV-1 disease progression (37). Together, these data suggest that CCR5 agonists or chemokine analogs which block NSI R5 isolates may be effective in preventing HIV-1 transmission or reducing viral loads in early HIV-1 disease. One such analog, RANTES with an N-terminal Ser residue replaced by the n-pentane of glyoxylic acid (aminooxypentane [AOP]), is a potent inhibitor of macrophage-tropic HIV-1 laboratory isolates (48). In addition to increased antiviral potency potentially due to increased retention of the internalized CCR5 receptors (36), AOP-RANTES did not induce the chemotactic response characteristic of RANTES-receptor interactions (48). However, high AOP-RANTES concentrations (>100 nM) can increase replication of SI X4 isolates, suggesting possible drug-induced signaling and activation of some step in the virus life cycle (26; A. J. Marozsan and E. J. Arts, unpublished data).
Rapid emergence of drug-resistant HIV-1 variants during therapy remains an obstacle for all drugs targeting HIV replication (4, 19, 27). However, these antiretroviral drug-resistant variants appear less fit than the parental wild-type strain (19, 27). Reduced fitness is likely the dominant factor leading to rapid reversion of drug-resistant mutations upon removal of drug pressure (27). In contrast, resistance to β-chemokine analogs may be conferred by basic amino acid substitutions in the V3 loop and result in a common and stable switch from an NSI R5 to an SI X4 variant, a switch also associated with rapid disease progression (46). In hu-PBL-SCID mice models of infection with NSI R5 HIV-1, treatment with RANTES analogs rapidly selected for HIV-1 isolates utilizing the CXCR4 receptor (38). Combination therapies with CXCR4 antagonists or analogs to the native CXCR4 ligand SDF-1 (8, 31, 39, 51) could prevent this switch but may also select for variants using other coreceptors (e.g., CCR3 or CCR2b).
A switch in coreceptor usage may not be the favored mechanism of resistance to β-chemokines, considering the in vivo dominance of the NSI R5 isolates over the faster-replicating SI X4 virus in the absence of drug (11, 25, 46, 47). In vivo factors responsible for the switch in coreceptor usage and phenotype have not been resolved. An alternative resistance mechanism to β-chemokine analogs may result from altered binding to the CCR5 receptor. Recent findings show a lack of extensive overlap in the binding of HIV-1 gp120 and β-chemokines to CCR5. RANTES and MIP-1β bind predominantly to the second extracellular loop of CCR5, whereas gp120 interacts with the N terminus and first extracellular loop (1, 6, 33, 43). Although HIV-1 binding to CCR5 was less distinct (33), it is likely that subtle changes in the V3 loop sequence or other regions in gp120 and gp41 could lead to altered affinity for or binding to the CCR5 coreceptor and possible resistance to β-chemokine analogs.
Few studies to date have performed quantitative assessments on the anti-HIV-1 activity of β-chemokine analogs. However, one study has shown differential RANTES inhibition of several NSI R5 isolates (50). We propose that the high degree of HIV-1 env heterogeneity could lead to variable inhibition by AOP-RANTES using different NSI R5 isolates. To test this hypothesis, we measured the 50% inhibitory concentration (IC50) values for AOP-RANTES with 13 different NSI R5 isolates in peripheral blood mononuclear cells (PBMC). Experiments with HOS or U87 cells expressing CD4 and different coreceptors confirmed exclusive use of the CCR5 coreceptor by these NSI R5 isolates. In addition, the sensitivity of NSI R5 isolates to AOP-RANTES in PBMC corresponded to that observed in CCR5+ CD4+ U87 cells. Sequence analysis of the V3 loop and other gp120 regions identified possible amino acid residues associated with decreased susceptibility to AOP-RANTES inhibition.
MATERIALS AND METHODS
Cell cultures.
PBMC were purified from blood from different HIV-negative human donors by Ficoll-Paque gradient centrifugation. Purified PBMC were resuspended in RPMI (Mediatech, Inc., Herndon, Pa.) medium supplemented with 10% fetal bovine serum (FBS; Life Technologies, Inc., Rockville, Md.), 100 U of penicillin and 100 μg of streptomycin (pen/strep; Mediatech, Inc.) per ml, 1 ng of recombinant human interleukin-2 (IL-2; Life Technologies, Inc.) per ml, and 1 U of phytohemagglutinin (PHA; Life Technologies, Inc.) per ml. Human osteosarcoma (HOS)-CD4 cell lines expressing CCR5 (HOS-CCR5), CXCR4 (HOS-CXCR4), CCR3 (HOS-CCR3), and CCR2b (HOS-CCR2b), obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health, from Nathaniel Landau) (18, 32) were grown in complete Dulbecco's modified Eagle's medium (10% FBS plus pen/strep) containing a hypoxanthine-xanthine-mycophenolic acid (HXM) supplement, consisting of 250 μg of xanthine (Aldrich, Milwaukee, Wis.), 13.5 μg of hypoxanthine (Aldrich), and 40 μg of mycophenolic acid (Life Technologies, Inc.) per ml to maintain CD4 expression and 0.5 μg of puromycin per ml to maintain coreceptor expression (CCR3, CCR2b, CXCR4 or CCR5). U87.CD4 (human glioma) cells expressing CCR5 or CXCR4 were obtained through D. Littman and the AIDS Reagent Project. U87.CD4-CCR5 and U87.CD4-CXCR4 cells were grown in complete DMEM (see above) containing 1 mg of G418 sulfate (Life Technologies, Inc.) per ml to maintain CD4 expression.
Viruses.
The following NSI R5 HIV-1 strains were obtained from the AIDS Research and Reagent Program for this study: A-92RW009, A-92RW008, A-93UG075, B-92BR021, B-92TH026, B-BaL, C-92BR025, C-93IN101, D-94UG108, E/A-92TH022, E-92TH001, B/F-93BR019, F-93BR029, G-92NG083-JV1083, and G-92NG003-G3. Two SI X4 strains (HXB2 and F-93BR020) were also obtained from the AIDS Reagent Program for use as controls. For most of the strains listed above, the letter before the dash indicates the subtype of the viral envelope and is followed by the year of isolation, country of origin, and strain number, e.g., A-92RW009 is a clade A HIV-1 strain isolated in Rwanda in 1992. All of these viruses were propagated in PBMC cultures until high virus titers (as determined by reverse transcriptase [RT] activity) were obtained in culture supernatants. The 50% tissue culture infective dose values were then calculated for each virus using the Reed-Muench technique (16).
Drugs.
This study focused on the β-chemokine analog AOP-RANTES (regulated upon activation normal T-cell expressed and secreted). Ten other RANTES derivatives with N-terminal modifications and N-nonanoyl (NNY)-RANTES were employed in this study. 3′-Azido-3′-deoxythymidine (AZT or zidovudine) was obtained from Sigma (St. Louis, Mo.).
Sensitivity to AOP-RANTES.
After PHA stimulation for 48 h, IL-2-treated PBMC were added to 96-well plates (2 × 105 cells/well) containing serially diluted (1:10) drugs: AOP-RANTES (125 to 0.003 nM), other RANTES derivatives (1,000 to 0.1 nM), or AZT (10 to 0.0001 nM). The appropriate HIV-1 isolate in complete RPMI medium (approximately 0.1 multiplicity of infection [MOI]) was then added to wells containing PBMC and the full range of drug dilutions. Triplicate experiments were performed with all NSI R5 HIV-1 isolates to test sensitivity to AOP-RANTES inhibition. On day 3 postinfection, each plate was centrifuged for 5 min at 1,200 × g in a swinging-bucket centrifuge. An aliquot (150 μl) of cell-free supernatant was then removed from each well and replaced with 150 μl of complete RPMI medium containing the appropriate concentrations of AOP-RANTES or other drugs. On days 5, 10, and 15 postinfection, each plate was centrifuged again for 5 min, and cell-free supernatant samples (25 μl) were removed and stored at −70°C for subsequent analysis. Cultures were discarded on day 15.
Virus production in the presence of AOP-RANTES and other inhibitors was measured in cell-free supernatants using RT assays as described before (15). Briefly, RT assays were performed on day 5 and day 10 supernatant samples. Supernatant samples (5 μl) clarified of cell debris by centrifugation at 2,500 × g for 5 min were added to 96-well plates along with 25 μl of RT master mix [50 mM Tris-HCl (pH 7.8), 75 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 5 μg of poly(rA) · poly(dT) per ml, 0.5% (vol/vol) NP-40, 1 μl of fresh 10-mCi/ml [α-32P]-dTTP per ml]. After incubation at 37°C overnight, 10 μl of the RT reaction mixtures were blotted onto a DEAE filtermat (Wallac Oy, Turku, Finland), washed five times with SSC (0.15 M NaCl, 0.015 M sodium citrate), rinsed in 80% ethanol, and dried. Radioactivity (counts per minute) from each well on the dried filters was measured with a Matrix 96 direct beta counter (Packard, Meriden, Conn.). Incorporation of [α-32P]dTTP by HIV-1 RT is a relative measure of RT activity and virus in the supernatant. A relative measure of RT activity (correcting for radioactive decay) was plotted against drug concentration to calculate the AOP-RANTES concentration required for 50% inhibition (IC50) of each HIV-1 isolate. All data were graphed and analyzed using SigmaPlot 4.0 (SPSS, Inc., Chicago, Ill.). Although similar MOIs were used for each isolate in these studies, all values for virus production were standardized due to the decay of the radiolabeled [α-32P]TTP utilized in the RT assays.
HIV inhibition by AOP-RANTES in a CCR5-expressing cell line.
U87.CD4-CCR5 cells were removed from stock cultures by trypsin-EDTA treatment, then added to 24-well plates with 500 μl of complete DMEM and allowed to adhere and grow for approximately 60 h at 37°C with 5% CO2 prior to further handling. Following addition of no drug, AOP-RANTES (6.3 or 0.25 nM), or RANTES (6.3 nM) to the U87.CD4-CCR5 cells, 0.1 MOI of each NSI R5 HIV-1 isolate (A-93UG075, B-92BR021, B-BaL, E-92TH001, B/F-93BR019, and G-92NG003-G3) was added to wells containing each drug. On day 4 postinfection, each plate was centrifuged for 5 min at 1,200 × g. Supernatant (1 ml) was removed from each well and replaced with 1 ml of complete DMEM containing appropriate AOP-RANTES or RANTES concentrations. On days 5 and 11 postinfection, each plate was again centrifuged to harvest 25 μl of cell-free supernatant samples. The amount of HIV-1 in culture supernatants was then measured by RT activity as described above.
Coreceptor usage.
HOS cells expressing CD4 and different chemokine receptors (HOS-CCR5, HOS-CXCR4, HOS-CCR3, and HOS-CCR2b) and U87.CD4-CXCR4 cells were added to 24-well plates with 950 μl of complete DMEM (containing G418 for the U87 cell lines and HXM-puromycin for the HOS cell lines) and allowed to adhere and grow for approximately 2 days at 37°C in 5% CO2 prior to further handling. No additional drugs were added. Each of the following virus strains (0.1 MOI) was added to the appropriate wells: A-93UG075, B-92BR021, B-BaL, E-92TH001, B/F-93BR019, G-92NG003-G3, and B-HXB2 to U87.CD4-CXCR4 cells, and A-92RW009, B-92BR021, B-92TH026, B-BaL, C-92BR025, E/A-92TH022, B-HXB2, and F-93BR020 to HOS-CCR5, HOS-CXCR4, HOS-CCR3, and HOS-CCR2b cells. On day 4 postinfection, each plate was centrifuged for 5 min at 1,200 × g to replace all supernatant with 1 ml of complete DMEM. Supernatant samples (25 μl) were removed on days 5 and 11 postinfection. Virus production was measured using RT assays as described above.
Sequencing.
Proviral DNA was extracted directly from cocultured PBMC as previously described (42). A fragment of approximately 0.66 kb spanning the C2-V4 coding region of gp120 (env gene) was amplified by PCR with primers E80 and E105 (44). Nucleotide sequences corresponding to 70 amino acids in the gp120 env glycoprotein (i.e., the entire V3 loop and portions of the C2 and C3 regions) were sequenced in both directions using primers E110 and E125 (44) as previously described (42). These sequences were read using 1D Image Analysis Software (Eastman Kodak Company, New Haven, Conn.), edited and translated using DNASIS (Hitachi Genetic Systems, Alameda, Calif.), and then aligned using CLUSTAL X (49).
Fluorescence-activated cell sorting (FACS) analysis.
Unstimulated PBMC were left untreated for 12 h. Following this incubation, cells were centrifuged at 800 × g for 10 min and incubated on ice for 15 min with 5% bovine serum albumin (BSA; Sigma) in phosphate-buffered saline (PBS; BioWhittaker, Walkersville, Md.). Cells were again centrifuged at 800 × g for 10 min and resuspended in 50 μl of PBS. A 5-μl amount of Leu-3a conjugated anti-human CD4 antibody (Becton Dickinson Immunocytochemistry Systems, San Jose, Calif.), 20 μl of phycoerythrin (PE)-conjugated anti-human CCR5 antibody, or 5 μl of PE-conjugated mouse immunoglobulin G2a (κ isotype standard; PharmMingen, San Diego, Calif.) was added to the suspension and incubated in the dark on ice for 30 min. Cells were then washed with 5% BSA–PBS and 500 μl of PBS. After the final wash cells were fixed with 300 μl of 1% paraformaldehyde and analyzed with a FacsScan flow cytometer and Lysis II software (Becton Dickinson, Bedford, Ma.).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this study can be found in GenBank under the indicated accession numbers: V3 loop of A-92RW008 (AF231042), full genome of A-92RW009 (U88823), V3 loop of A-93UG075 (AF231043), V3 loop of B-92BR021 (AF231041), gp160 of B-92TH026 (U08802), full genome of B-BaL (M68893), full genome of B-HXB2 (K03455), full genome of C-92BR025 (U52953), V3 loop of C-93IN101 (AF231044), V3 loop of E-92TH001 (AF231045), gp160 of E-92TH022 (U09131), gp160 of B/F-93BR019 (U27404), full genome of F-93BR029 (AF005495), and full genome of G-92NG083-JV1083 (U88826).
RESULTS
Sensitivity of NSI R5 HIV-1 isolates to AOP-RANTES inhibition in PBMC.
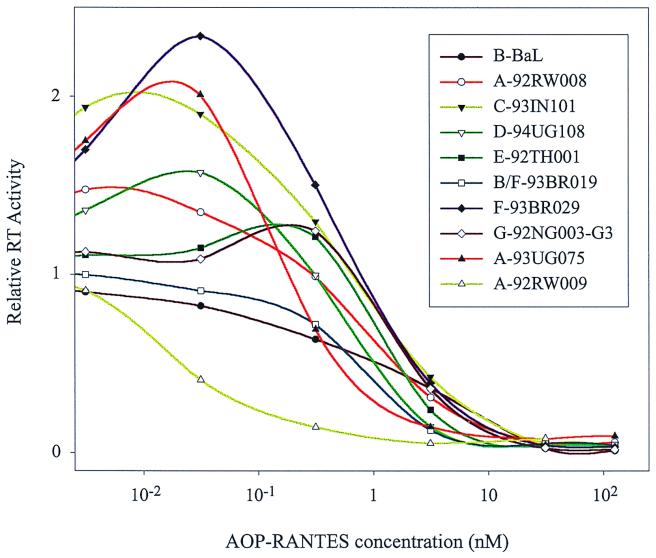
AOP-RANTES is a potent inhibitor of NSI R5 HIV-1 isolates (SF-162, M23, and E80) in PBMC and primary macrophage cultures (38, 48). This inhibition likely occurs at the level of HIV-1 entry, considering that AOP-RANTES could block fusion of cells expressing CCR5 and CD4 with cells expressing envelope glycoproteins derived from HIV-1B-Ada (O. Hartley and R. E. Offord, unpublished data). Although these studies have characterized an anti-HIV activity of AOP-RANTES, they have not measured IC50 values for AOP-RANTES, which may vary among different NSI R5 HIV-1 isolates. Unlike antiretroviral drugs, which inhibit more conserved HIV-1 enzymes, CCR5 agonists such as AOP- RANTES target an interaction between the highly variable HIV-1 envelope glycoproteins and CCR5. For these reasons, we have examined the inhibitory effects of AOP-RANTES on 13 NSI R5 HIV-1 isolates. These strains, isolated from different countries (see Materials and Methods), are subdivided into various clades based on env sequence. A significant range in sensitivity to AOP-RANTES inhibition was observed with these divergent NSI R5 HIV-1 isolates (Fig. 1). All curves were generated from experiments performed in triplicate with PBMC from one donor. Although day 5 and 10 samples resulted in similar curves for each virus, RT activity from day 10 samples provided the least deviation between replicates. The cytopathogenicity of HIV-1 and viral decay over 10 days resulted in a slight reduction in RT activity in untreated samples compared with infections treated with low AOP-RANTES concentrations (0.003 nM) (Fig. 1). It is important to note that pretreatment of PBMC with high AOP-RANTES concentrations (>125 nM) resulted in a slight breakthrough in virus replication (A. J. Marozsan and E. J. Arts, unpublished data). In this study, AOP-RANTES and virus were added simultaneously to cells.
FIG. 1.
Sensitivity of 10 NSI R5 HIV-1 isolates to AOP-RANTES inhibition. PBMC were exposed to each virus in the presence of various concentrations of AOP-RANTES. Supernatant samples were harvested on day 10 postinfection and analyzed for RT activity. To facilitate comparison, each curve was standardized to the 3 × 10−3 nM concentration of AOP-RANTES, which was assigned a relative RT activity of 1 for all viruses. Error bars representing 1 standard deviation were not placed on points to reduce complexity.
IC50 values of AOP-RANTES for each isolate were calculated from three separate experiments using the same donor PBMC. With the 13 isolates tested, the IC50 of AOP-RANTES ranged from 0.04 ± 0.02 nM (A-92RW009) to 1.25 ± 0.22 nM (B-BaL), representing a 31-fold difference (Table 1). There were no distinct groups of more or less sensitive NSI R5 HIV-1 strains but rather a distribution of the IC50 of AOP-RANTES for different isolates. Subtype A isolates did show an increased sensitivity to AOP-RANTES inhibition compared with non-subtype A isolates. However, specific resistance or sensitivity was not observed with any other subtype.
TABLE 1.
Amino acid sequence alignment of V3 region; summary of AOP-RANTES and AZT IC50 values
| Virus | V3 net charge | AOP-RANTES
|
AZT
|
V3 sequenceb | ||
|---|---|---|---|---|---|---|
| IC50 (nM) ± SD | Fold differencea | IC50 (nM) | Fold difference | |||
| NSI | ∗▿ ∗ | |||||
| A-92RW009 | +3 | 0.04 ± 0.02 | 1 | 3.3 ± 0.6 | 1 | CSRPNNNTRKSVHI––GPGQAFYATGDVIGDIRQAYC |
| A-93UG075 | +3 | 0.14 ± 0.02 | 3.5 | NAc | NA | CTRPNNNTRKSVRI––GPGQAFYATGDIIGDTRQAYC |
| C-92BR025 | +4 | 0.16 ± 0.05 | 4.0 | NA | NA | CTRPNNNTRKSIRI––GPGQAFYATGEIIGDIRQAHC |
| B-92BR021 | +6 | 0.20 ± 0.07 | 5.0 | NA | NA | CTRPNNNTRKSIHM––GWGRAFYATGEIIGNIRQAHC |
| G-92NG083-JV1083 | +3 | 0.22 ± 0.10 | 5.5 | NA | NA | CIRPNNNTRKSIPI––GPGQAFYATGDIIGDIRQAHC |
| A-92RW008 | +4 | 0.56 ± 0.17 | 14 | NA | NA | CTRPNNNTRTSIRI––GPGQSFHATGDIIGDIRQAHC |
| B/F-93BR019 | +5 | 0.73 ± 0.16 | 18 | NA | NA | CARPNNNTRKSIHI––GPGQAFYTTGEIIGDIRKAHC |
| E-92TH022 | +3 | 0.81 ± 0.09 | 20 | NA | NA | CTRPSNNTRTSITI––GPGQVFYRTGDIIGDIRRAYC |
| B-92TH026 | +4 | 0.85 ± 0.07 | 21 | NA | NA | CTRPNNNTRKSIPL––GPGQAWYTTGQIIGNIRQAHC |
| C-93IN101 | +3 | 0.96 ± 0.14 | 24 | 1.3 ± 0.6 | 0.39 | CTRPNNNTRKSIRV––GPGQTFYATGDIIGDIRQAHC |
| E-92TH001 | +2 | 1.21 ± 0.10 | 30 | NA | NA | CTRPSNNTRTSINI––GPGQVFYRTGDIIGDIRQAYC |
| F-93BR029 | +5 | 1.25 ± 0.20 | 31 | NA | NA | CTRPNNNTRKSIQI––GPGRAFYTTGEIIGDIRKAHC |
| B-BaL | +5 | 1.25 ± 0.22 | 31 | NA | NA | CTRPNNNTRKSIHI––GPGRAFYTTGEIIGDIRQAHC |
| SI | ||||||
| B-HXB2 | +10 | NA | NA | 2.5 ± 0.4 | 0.76 | CTRPNNNTRKRIRIQRGPGRAFVTIGKIGN–MRQAHC |
Fold difference is calculated from the IC50 value for each virus and is based on the IC50 value for virus A-92RW009.
Multiple alignment of amino acid sequences of the V3 loop (gp120 coding region). Numbering positions are based on the HIV-1 B-HXB2 isolate. Positions associated with viral phenotype (306 and 322) are indicated by asterisks. Position 319, which may be associated with variable sensitivity to AOP-RANTES, is indicated by ▵.
NA, not available.
Although sensitivity to AOP-RANTES inhibition varies considerably with different NSI R5 HIV-1 isolates, two NSI R5 HIV-1 isolates (C-93IN101 and A-92RW009) and one SI X4 isolate (B-HXB2) showed no significant difference in sensitivity to AZT inhibition (Table 1). In contrast, the IC50 of AOP-RANTES for C-93IN101 was 25-fold greater (P < 0.00005) than that for A-92RW009 (Table 1). To date, we have observed less than a fivefold difference in the IC50 of AZT for various wild-type HIV-1 strains (data not shown). A greater than fivefold increase in the IC50 of AZT is commonly associated with AZT-resistant HIV-1, generally isolated from AZT-treated patients (3). In contrast to AZT-resistant isolates selected under direct drug pressure and containing specific amino acid substitutions in the RT coding region, these NSI R5 HIV-1 isolates were isolated from infected individuals never exposed to AOP-RANTES. However, it is possible that native RANTES may impose a selective force on HIV evolution in infected patients (see Discussion).
Donor effect on AOP-RANTES inhibition in PBMC.
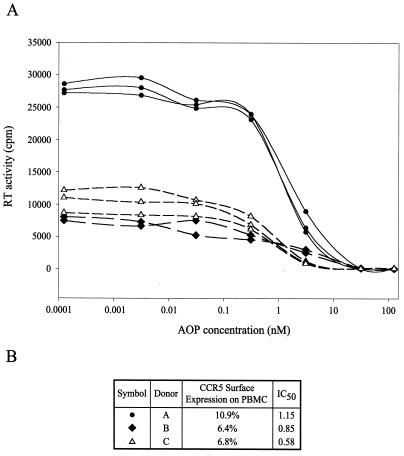
Lower levels of CCR5 surface expression on cells from different human donors could reduce the number of CCR5 receptors available for AOP-RANTES binding. Using PBMC from three separate donors, the same MOI did lead to different amounts of HIV-1B-BaL production in the presence (Fig. 2A) and absence (data not shown) of drug. As determined by FACS analysis, this difference in HIV-1 replication did not correlate with the level of CCR5 surface expression on PBMC from these three donors (data not shown). In addition, the extent of inhibition or AOP-RANTES IC50 values for HIV-1B-BaL did not vary significantly or in relation to CCR5 expression. With HIV-1B-BaL, the IC50 of AOP-RANTES was 0.58 nM to 1.15 nM with PBMC from these three donors (Fig. 2B). Lack of donor effects (e.g., CCR5 surface expression) on AOP-RANTES inhibition cannot be inferred from this limited data set. It is important to note that the data shown in Table 1 and Fig. 1 were generated from experiments employing several HIV-1 strains and different AOP-RANTES concentrations but only one batch of PBMC from a single donor. Thus, variations in AOP-RANTES sensitivity were virus specific and not host specific.
FIG. 2.
Comparison of NSI R5 HIV-1B-BaL inhibition by AOP-RANTES using PBMC from three different donors. (A) PBMC from three different HIV-1-negative donors were exposed to HIV-1B-BaL in the presence of various concentrations of AOP-RANTES. Supernatant samples were harvested on day 10 postinfection and analyzed for RT activity. For each donor, either two or three curves were plotted, each corresponding to a separate replication of the assay. (B) Summary of percentage of PBMC from each donor expressing CCR5 (determined by FACS analysis) and average AOP-RANTES IC50 values (nanomolar) calculated for each set of donor PBMC.
Inhibition by other RANTES analogs.
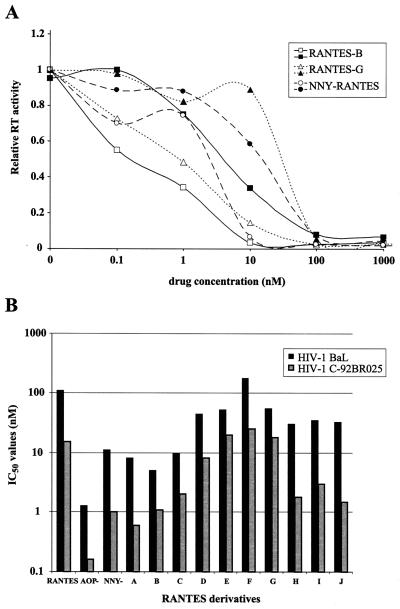
We have tested the anti-HIV activity of several RANTES analogs, including NNY-RANTES (Fig. 3). All of these RANTES derivatives had variable inhibitory effects on two NSI R5 HIV-1 isolates, B-BaL and C-92BR025. IC50 values ranged from 5 to 180 nM for HIV-1B-BaL, with RANTES derivative F being the least inhibitory and RANTES derivative B being the most inhibitory (Fig. 3B). The range in IC50 values for HIV-1C-92BR025 was 0.6 to 25 nM, with a similar distribution in sensitivity to the different drugs. When comparing IC50 values, the majority of compounds (9 of 12) inhibited replication of HIV-1C-92BR025 approximately 5- to 10-fold more (lower IC50 values) than HIV-1B-BaL (Fig. 3B). This is consistent with the lower sensitivity of HIV-1B-BaL to AOP-RANTES inhibition (Table 1). As indicated by the drug sensitivity curves of Fig. 3A, HIV-1C-92BR025 was consistently more sensitive than HIV-1B-BaL to inhibition by all RANTES analogs.
FIG. 3.
Sensitivity of two NSI R5 HIV-1 isolates to inhibition by various RANTES analogs. The sensitivity of HIV-1B-BaL and HIV-1C-92BR025 to inhibition by RANTES analogs with various N-terminal modifications was tested in PBMC. (A) Sensitivity of these HIV-1 isolates to three RANTES derivatives. To facilitate comparison, each curve was standardized to the highest data point (assigned a relative RT activity of 1). (B) Log10 plot of the IC50 values of several RANTES derivatives for HIV-1B-BaL and HIV-1C-92BR025.
Coreceptor usage and sensitivity to AOP-RANTES.
All NSI R5 HIV-1 isolates appear to preferentially utilize CD4 and the CCR5 coreceptor for host cell entry. However, several isolates are capable of utilizing (i) both major coreceptors (CXCR4 and CCR5) for entry (e.g., HIV-189.6) (20) or (ii) another seven-transmembrane G-coupled protein as a coreceptor (e.g., CCR2b and CCR3), albeit less efficiently than CCR5 (12, 20, 22, 23, 28, 34). Most of these coreceptors would be expressed on some subset of CD4+ cells in the PBMC population but may not interact with RANTES analogs. Thus, AOP-RANTES or other RANTES analogs would not block HIV infection of these cells. Given the variable AOP-RANTES inhibition of NSI R5 HIV-1 isolates, it was necessary to determine if alternative coreceptor usage plays a role in decreased sensitivity to RANTES analogs. For these studies, we employed two cell lines (HOS and U87 human glioma cells) expressing CD4 and a chemokine receptor (CCR5, CXCR4, CCR2b, or CCR3).
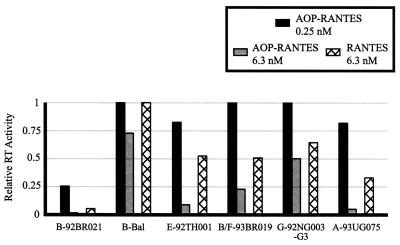
Using the U87.CD4-CCR5 and -CXCR4 cell lines, we compared CCR5 usage of six NSI R5 HIV-1 isolates in the presence of AOP-RANTES at two concentrations (0.25 and 6.3 nM) and of RANTES at 6.3 nM (Fig. 4). Treatment with 6.3 nM AOP-RANTES resulted in variable inhibition (25 to 95%) of all six isolates, whereas RANTES and the lower concentration of AOP-RANTES (0.25 nM) had little or no inhibitory effect (Fig. 4). Although each of these NSI R5 HIV-1 isolates utilized CCR5 for entry, none were able to infect U87.CD4-CXCR4 cells. In contrast, SI X4 HIV-1 isolate B-HXB2 replicated efficiently in U87.CD4-CXCR4 cells but not in U87.CD4-CCR5 cells (data not shown). Thus, variable inhibition by AOP-RANTES in PBMC (Table 1) was not due to CXCR4 usage (i.e., dual tropism) by these NSI R5 isolates.
FIG. 4.
Sensitivity of NSI R5 HIV-1 isolates to inhibition by AOP-RANTES and RANTES in U87.CD4 cells expressing CCR5. Six NSI R5 isolates were cultured in the presence of 6.3 nM AOP-RANTES, 0.25 nM AOP-RANTES, 6.3 nM RANTES, or no drug. Supernatant samples harvested on day 11 postinfection were analyzed for RT activity. Values obtained for each virus were standardized to the RT activity observed in the no-drug controls. Relative RT values above 1 are not shown.
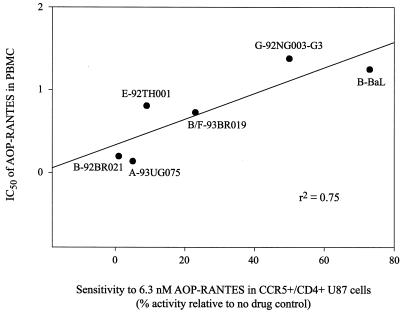
If CCR5 usage by HIV-1 strains were associated with variable sensitivity to AOP-RANTES, a similar level of AOP-RANTES inhibition should be observed in PBMC as is observed in U87.CD4-CCR5 cells. For each of the same six NSI R5 HIV-1 isolates, the IC50 of AOP-RANTES obtained from PBMC experiments was plotted against the sensitivity to 6.3 nM AOP-RANTES in U87.CD4-CCR5 cells (Fig. 5). This analysis revealed a linear correlation between the inhibitory effects of AOP-RANTES in both primary cells (PBMC) and CCR5-expressing lines (U87.CD4-CCR5) (r2 = 0.75) (Fig. 5).
FIG. 5.
Comparison of inhibitory effect of AOP-RANTES on NSI R5 HIV-1 isolates in both PBMC and U87.CD4-CCR5 cells. The y axis values represent the IC50 of AOP-RANTES for each virus. The x axis values represent the sensitivity of each virus to 6.3 nM AOP-RANTES relative to the no-drug control, as measured by RT activity (obtained from Fig. 4).
To further verify that this variable sensitivity to AOP-RANTES inhibition was not due to alternative coreceptor usage, six NSI R5 HIV-1 isolates and two SI X4 HIV-1 isolates were added to HOS-CD4 cell cultures expressing either CXCR4, CCR2b, or CCR3. None of the NSI R5 HIV-1 isolates tested could utilize any coreceptor other than CCR5 (Table 2). The low RT activity in the supernatants of HOS-CCR5 cell cultures exposed to HIV-1B-HXB2 or HIV- 1F-93BR020 was likely due to the low constitutive expression of CXCR4 on these cells (45). HIV-1 entry through other putative coreceptors (STRL33, gpr15, gpr1, or APJ) was not examined. Considering the comparable levels of HIV-1 inhibition by AOP-RANTES in PBMC and in U87.CD4-CCR5 cells, variable sensitivity to RANTES analogs among the different NSI R5 HIV-1 isolates was likely due to altered binding of CCR5 and not to alternative coreceptor usage.
TABLE 2.
NSI and SI HIV-1 isolates specifically utilize coreceptors CCR5 and CXCR4, respectively
| Virus | RT activitya (102 cpm/ml) with coreceptor:
|
|||
|---|---|---|---|---|
| CCR5 | CXCR4 | CCR2b | CCR3 | |
| NSI | ||||
| A-92RW009 | 54 | 0 | 0 | 0 |
| B-92BR021 | 69 | 2 | 0 | 0 |
| B-92TH026 | 51 | 0 | 0 | 0 |
| C-92BR025 | 58 | 5 | 0 | 0 |
| E-92TH022 | 50 | 0 | 0 | 0.2 |
| B-BaL | 49 | 6 | 0 | 0 |
| SI | ||||
| B-HXB2 | 16 | 157 | 0 | 0 |
| F-93BR020 | 12 | 86 | 0 | 0 |
RT activity minus background.
Comparison of env V3 sequence and sensitivity to AOP-RANTES.
Considering the high degree of variability in both HIV-1 sequence and sensitivity to AOP-RANTES, we examined whether specific HIV-1 env sequences may correspond to the sensitivity of the isolates to AOP-RANTES. Although other env sequences were compared (data not shown), these analyses focused on the hypervariable V3 loop sequences (i.e., the major determinant for coreceptor usage and biological phenotype) (14, 25, 47). The V3 region was sequenced for 13 NSI R5 isolates and is compared in Table 1 with the V3 loop sequences of HIV-1B-HXB2 (numbering on the V3 loop corresponds to that of the HXB2 sequence). Variations in AOP-RANTES IC50 values were not related to changes in the net charge of the V3 loop or the number of positively charged amino acids in this region (Table 1). In addition, no positively charged amino acids were observed in the NSI R5 HIV-1 isolates at positions 306 or 322 (i.e., the basic amino acid residues associated with an SI X4 T-cell-line-tropic phenotype). This analysis predicts that these NSI R5 isolates would preferentially or exclusively use CCR5 as a coreceptor for cell entry. We did observe one sequence modification in the V3 loop that may be related to increased or decreased sensitivity to AOP-RANTES (Table 1). Six of seven NSI R5 HIV-1 isolates with a threonine or arginine substitution at position 319 (highlighted in bold on Table 1) had an AOP-RANTES IC50 of >0.73 nM. In contrast, HIV-1 isolates with an A319 in the V3 loop, with the exception of C-93IN101, were at least 1.3- to 18-fold more susceptible to AOP-RANTES inhibition (Table 1).
DISCUSSION
HIV-1 entry into human cells is mediated by CD4 and a seven-transmembrane G-coupled protein receptor. Several HIV-1 coreceptors have now been identified, but only two, CCR5 and CXCR4, are preferentially utilized by NSI R5 and SI X4 isolates, respectively (18, 21, 24). The initial observation that natural ligands of CCR5 (e.g., RANTES, MIP-1α, and MIP-1β) could block infection by macrophage-tropic NSI R5 HIV-1 strains (13) has led to rapid development of β-chemokine and SDF-1α analogs as potential anti-HIV compounds (8, 31, 38, 39, 51, 56). In this study, we have examined the inhibitory effect of AOP-RANTES and other RANTES analogs on primary HIV-1 isolates. Although the chemokine analog AOP-RANTES is a potent inhibitor of NSI R5 HIV-1 replication, the level of inhibition varied considerably with different HIV-1 strains. IC50 values ranged from 0.04 to 1.25 nM, or greater than a log10 difference. However, the donor PBMC used for these experiments did not affect the IC50 value derived from AOP-RANTES inhibition of an NSI R5 HIV-1 isolate. This variability to AOP-RANTES inhibition among distinct NSI R5 HIV-1 is related to differential CCR5 binding rather than alternative or dual coreceptor usage.
Several NSI R5 isolates (E-92TH001, B-BaL, and F-93BR029) had at least a >8.6-fold increase in the IC50 of AOP-RANTES compared with others (A-92RW009 and A-93UG075). A greater than fivefold increase in the IC50 values of several antiretroviral drugs (e.g., 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine) is often interpreted as resistance and attributable to specific drug-resistant substitutions (L74V and K65R, respectively, both found in the RT coding region) (4). In most instances, resistant HIV-1 strains arise from antiretroviral selection in HIV-infected patients or in HIV-infected cultures. This drug selection appears to be necessary for a resistant isolate, less fit than the wild type, to compete with and predominate over a heterogeneous, intrapatient HIV-1 population (19, 27). In contrast to HIV-1 resistance to other antiretrovirals, AOP-RANTES resistance appears to be intrinsically acquired in some HIV-infected individuals. The native RANTES may act as a selective force, considering that levels of this β-chemokine can vary considerably between infected individuals. Although the RANTES concentration required for effective inhibition of NSI R5 HIV-1 isolates (IC50 > 15 nM) often exceeds even the highest circulating levels of this β-chemokine (13, 40), a weak selective pressure by RANTES may lead to resistance mechanisms other than a switch in coreceptor usage. As previously described (38), treatment with the potent analogs NNY-RANTES and AOP-RANTES results in rapid selection of X4 isolates in hu-PBL-SCID mice originally exposed to an NSI R5 HIV-1 isolate. It is also important to note that variable sensitivity of HIV-1 to AOP-RANTES inhibition may or may not have an impact on drug activity in vivo. A high therapeutic index, sufficient drug tolerance, and a long half-life may ensure a high circulating concentration of a RANTES analog and negate the effects of even a 30-fold variation in sensitivity.
Increased affinity for CCR5 or a change in the gp120 or gp41 binding site on CCR5 may be responsible for the “intrinsic” AOP-RANTES resistance observed with some R5 isolates. Evidence for this hypothesis was derived from several experiments. First, the level of HIV-1 inhibition by AOP-RANTES in PBMC correlated with that observed in U87.CD4-CCR5 cells. Unlike the U87.CD4-CCR5 cell clones, PHA- and IL-2-treated PBMC are a mixture of cells (mainly lymphocytes) expressing several of the putative HIV-1 coreceptors. Alternative coreceptor usage by one of the isolates may have lead to replication in non-CCR5-expressing cells and escape from AOP-RANTES inhibition. However, similar AOP-RANTES inhibition of several HIV-1 isolates in PBMC and U87.CD4-CCR5 cells suggests that only CCR5 was mediating entry of these viruses into PBMC. A second set of experiments also showed that none of the tested isolates could utilize CXCR4, CCR3, or CCR2b as coreceptors. It is important to note that, among this set of coreceptors, RANTES and possibly AOP-RANTES can only bind to CCR3. Considering that none of the known coreceptors are utilized more efficiently than CCR5 and CXCR4, it is also unlikely that usage of any other putative coreceptor (e.g., gpr14, gpr1, or APJ) by these HIV-1 isolates could contribute to variable sensitivity to AOP-RANTES inhibition.
The V3 loop in the gp120 envelope glycoprotein is characterized as the major determinant for biological phenotype (NSI versus SI) and coreceptor usage (CCR5 versus CXCR4) (14, 25, 47). Basic amino acids at positions 306 and 322 and/or a positively charged V3 loop is generally associated with an SI X4, CXCR4-tropic isolate (14, 25, 47). Therefore, V3 sequence analysis is often used to predict biological phenotype. In this study, the predicted biological phenotype based on the average V3 loop sequence corresponded to the actual tropism for the CCR5 or CXCR4 coreceptor. Sensitivity to AOP-RANTES among the various NSI R5 isolates was not related to a basic amino acid residue at 306 and 322 or the net charge of the V3 loop. A scan of the entire C2-V3 region of gp120 revealed only one position (residue 319) which may be associated with variable sensitivity to AOP-RANTES. An alanine at position 319 was present in all NSI R5 isolates with increased sensitivity to AOP-RANTES (IC50, 0.04 to 0.56 nM), whereas a threonine or arginine at this position was associated with decreased sensitivity to AOP-RANTES (IC50, 0.73 to 1.3 nM). This residue is found in the proposed crown of the V3 loop, four residues downstream of the semiconserved GPGQ sequence (30). Interestingly, an NSI R5 HIV-1 isolate with a single V3 mutation (i.e., one residue upstream [H318R] of 319) was recovered from an HIV-infected hu-PBL-SCID mouse treated with NNY-RANTES (38). Although there was no switch in coreceptor usage, the appearance of this H318R substitution resulted in failure of NNY-RANTES to contain HIV replication in the mouse. In future studies, we will examine the impact of A319T/R, H318R, and other gp120 and gp41 substitutions in NSI R5 HIV-1 isolates on sensitivity to AOP-RANTES inhibition and CCR5 usage. Clones of five primary isolates are being used for this extensive mutagenesis and subsequent analysis of drug sensitivity. We suspect that several regions or residues in the gp120 and gp41 coding region, aside from or in addition to positions 318 and 319, may be required or synergistic for resistance.
Several studies now indicate that the binding site for β-chemokines on CCR5 does not overlap significantly with the binding site for either recombinant gp120 or the entire virus. RANTES and MIP-1α bind predominantly to the second extracellular loop of CCR5, whereas gp120JRFL interacts with the N terminus and first extracellular loop of CCR5 (1, 6, 33, 43). Variable gp120-CCR5 binding can only be confirmed in similar studies employing a recombinant gp120 derived from AOP-RANTES-resistant NSI R5 isolates. The role of these coreceptors in HIV-1 entry is unclear. However, recent findings suggest that (i) a CD4-CCR5 association at the cell surface and (ii) the small extracellular loops of these coreceptors (i.e., relative to the CD4 extracellular domain) may augment virus-cell membrane fusion via the env gp41 (53). Chemokine analogs may prevent or disrupt this process by blocking the HIV-1 binding site on CCR5 and/or removing this coreceptor from the cell surface.
In conclusion, the variable sensitivity of NSI R5 isolates to inhibition by β-chemokine analogs may affect the potential therapeutic benefit of these drugs. Aside from a switch from an NSI R5 to an SI X4 phenotype, requiring multiple substitutions in the V3 loop, resistance to AOP-RANTES or other CCR5 agonists may be conferred by single substitutions in the V3 loop (e.g., A319T). In HIV-infected individuals, the latter may be the preferred resistance mechanism selected by circulating RANTES. The predominance of CCR5-tropic virus prior to severe immunodeficiency and AIDS suggests in vivo preference in maintaining this phenotype over SI X4.
ACKNOWLEDGMENTS
This work was supported by Projects I (R.E.O.) and II (E.J.A.) of the NIH program project (AI-43645) entitled Development of HIV Co-receptor Inhibitors.
We thank M. M. Lederman at Case Western Reserve University, D. E. Mosier at the Scripps Research Institute, and Charles Flexner at the Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases, for their assistance and critical comments.
REFERENCES
- 1.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structural determinants. Critical residues in the third extracellular loop support HIV-1 fusion. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Arts E J, Quiñones-Mateu M E, Albright J L. Mechanisms of clinical resistance by HIV-1 variants to zidovudine and the paradox of reverse transcriptase sensitivity. Drug Resistance Updates. 1998;1:21–28. doi: 10.1016/s1368-7646(98)80211-2. [DOI] [PubMed] [Google Scholar]
- 4.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 15.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, Coico R. Current protocols in immunology, CD-ROM version. New York, N.Y: John Wiley & Sons, Inc.; 1999. Detection and analysis of HIV; detection assays for HIV proteins, basic protocol: assay for HIV reverse transcriptase activity; p. 12.5.8. [Google Scholar]
- 16.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, Coico R. Current protocols in immunology, CD-ROM version. New York, N.Y: John Wiley & Sons, Inc.; 1999. Detection and analysis of HIV; isolation and quantitation of HIV in peripheral blood, alternate protocol: assessment of HIV titer using the Reed-Muench accumulative method; p. 12.2.5. [Google Scholar]
- 17.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. . (Erratum, 274:1069.) [DOI] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Domingo E, Menendez-Arias L, Quiñones-Mateu M E, Holguin A, Gutierrez-Rivas M, Martinez M A, Quer J, Novella I S, Holland J J. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog Drug Res. 1997;48:99–128. doi: 10.1007/978-3-0348-8861-5_4. [DOI] [PubMed] [Google Scholar]
- 20.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 21.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 22.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 25.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon C J, Muesing M A, Proudfoot A E, Power C A, Moore J P, Trkola A. Enhancement of human immunodeficiency virus type 1 infection by the CC-chemokine RANTES is independent of the mechanism of virus-cell fusion. J Virol. 1999;73:684–694. doi: 10.1128/jvi.73.1.684-694.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrigan P R, Bloor S, Larder B A. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J Virol. 1998;72:3773–3778. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 29.Hill C M, Deng H, Unutmaz D, Kewal-Ramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J, Sodroski J. Human retroviruses and AIDS 1998. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1998. [Google Scholar]
- 31.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Durell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 34.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Chao D, Nakayama E E, Taguchi H, Goto M, Xin X, Takamatsu J K, Saito H, Ishikawa Y, Akaza T, Juji T, Takebe Y, Ohishi T, Fukutake K, Maruyama Y, Yashiki S, Sonoda S, Nakamura T, Nagai Y, Iwamoto A, Shioda T. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci USA. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mack M, Luckow B, Nelson P J, Cihak J, Simmons G, Clapham P R, Signoret N, Marsh M, Stangassinger M, Borlat F, Wells T N, Schlondorff D, Proudfoot A E. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 38.Mosier D E, Picchio G R, Gulizia R J, Sabbe R, Poignard P, Picard L, Offord R E, Thompson D A, Wilken J. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J Virol. 1999;73:3544–3550. doi: 10.1128/jvi.73.5.3544-3550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. . (Erratum, 384:288.) [DOI] [PubMed] [Google Scholar]
- 40.Polo S, Veglia F, Malnati M S, Gobbi C, Farci P, Raiteri R, Sinicco A, Lusso P. Longitudinal analysis of serum chemokine levels in the course of HIV-1 infection. AIDS. 1999;13:447–454. doi: 10.1097/00002030-199903110-00002. [DOI] [PubMed] [Google Scholar]
- 41.Quillent C, Oberlin E, Braun J, Rousset D, Gonzalez-Canali G, Metais P, Montagnier L, Virelizier J L, Arenzana-Seisdedos F, Beretta A. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet. 1998;351:14–18. doi: 10.1016/S0140-6736(97)09185-X. [DOI] [PubMed] [Google Scholar]
- 42.Quiñones-Mateu M E, Dopazo J, Este J A, Rota T R, Domingo E. Molecular characterization of human immunodeficiency virus type 1 isolates from Venezuela. AIDS Res Hum Retroviruses. 1995;11:605–616. doi: 10.1089/aid.1995.11.605. [DOI] [PubMed] [Google Scholar]
- 43.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 44.Sanders-Buell E, Salminen M O, McCutchan F. Sequencing primers for HIV-1, p. III-15–III-21. In Human retroviruses and AIDS 1995. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. [Google Scholar]
- 45.Schols D, Este J A, Cabrera C, De Clercq E. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1 alpha contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E, van Steenwijk R P, Lange J M, Schattenkerk J K, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 48.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola A, Paxton W A, Monard S P, Hoxie J A, Siani M A, Thompson D A, Wu L, Mackay C R, Horuk R, Moore J P. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda H, Siani M A, Gong W, Thompson D A, Brown G G, Wang J M. Chemically synthesized SDF-1alpha analogue, N33A, is a potent chemotactic agent for CXCR4/fusin/LESTR-expressing human leukocytes. J Biol Chem. 1997;272:24966–24970. doi: 10.1074/jbc.272.40.24966. [DOI] [PubMed] [Google Scholar]
- 52.Wang W K, Dudek T, Essex M, Lee T H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci USA. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao X, Wu L, Stantchev T S, Feng Y R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, Dimitrov D S. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, He T, Huang Y, Chen Z, Guo Y, Wu S, Kunstman K J, Brown R C, Phair J P, Neumann A U, Ho D D, Wolinsky S M. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:9307–9312. doi: 10.1128/jvi.72.11.9307-9312.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, Kewal-Ramani V N, Moore J P. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]