Abstract
The 96-amino-acid-long human immunodeficiency virus type 1 virion-encoded accessory protein Vpr is of particular interest, as this protein, which is found in association with viral particles, can exert a regulatory effect on both virus replication and host cell function. Evidently, Vpr, through interaction with several host regulatory proteins, can modulate transcription from the viral long terminal repeat promoter. Expression of Vpr in cells results in deregulation of cell proliferation during the cell cycle pathway at the G2 stage. Vpr has unique structural features consisting of multiple functional domains. In this study, we have focused on the leucine/isoleucine-rich domain near the carboxyl terminus of Vpr at residue 73 (arginine) and have demonstrated that alterations at this residue result in ablation of transcriptional activity of Vpr and its ability to block cell cycle events at the G2 stage. Interestingly, substitution mutations at R73 have resulted in a peptide with dominant negative activities on wild-type functions in transcription and host proliferation events. Results from in vitro and in vivo protein-protein interaction studies have revealed that functionally inactive mutant Vpr can be associated with wild-type protein, presumably through the N-terminal regions of the protein which have been shown to be important for Vpr oligomerization. Thus, it is likely that complexation of the mutant Vpr with wild-type protein functionally inactivates Vpr. The importance of these findings in light of the development of therapeutic strategies is discussed.
The human immunodeficiency virus type 1 (HIV-1) virion-associated accessory protein, Vpr, is a 96-amino-acid protein produced at the late phase of viral infection (6). Studies by several laboratories have revealed that Vpr is essential for optimum infection of primary target cells such as macrophages (1, 27) and positively affects virus replication in established T cells (4, 6). Vpr is a transcriptional activator which, through cooperative interaction with several cellular proteins including SP1 and p53, has the ability to stimulate transcription of the viral long terminal repeat (LTR) promoter (3, 10, 23). In addition, Vpr can cause proliferating cells to undergo an arrest or delay in the G2 phase of the cell cycle (6, 12, 13, 17). Though precise structural information for full-length Vpr is not available, studies with synthetic Vpr peptides utilizing circular dichroism spectroscopy and nuclear magnetic resonance revealed several features. These include helical domains I (residues 17 to 34) and II (residues 53 to 74), separated by a putative loop and an arginine-rich C-terminal (15, 21) domain. Mutational analysis of Vpr showed the importance of leucine-isoleucine-rich region (residues 60 to 81) which partly overlaps with the helical domain II (25). Results from several studies have implicated the N terminus of Vpr in virion incorporation and stability of the proteins. The C terminus of Vpr is linked to cell cycle arrest and transcriptional activation properties (7). Earlier and recent biochemical studies have shown that Vpr may exist in oligomeric form and that the N-terminal domain of the protein positioned between amino acid residues 1 and 52 is sufficient for Vpr oligomerization (26, 29). Recently, we evaluated the effect of wild-type Vpr and various Vpr mutants with altered amino acid residues within the three major domains of Vpr on regulation of the HIV-1 LTR (20). Our results have shown that while mutations within the N- and C-terminal domains of Vpr have little effect on HIV-1 transcriptional activity, residues within the leucine-rich domain, i.e., residue 73, may play an important role in transcriptional activation of the viral promoter. Here, we demonstrate that the Vpr variant with the mutation at position 73 (Vpr R73S) can function as a dominant negative mutant, blocking the ability of the wild-type protein to stimulate LTR transcription, and block cell proliferation at the G2 stage of the cell cycle. Of interest, the mutant protein which retains the critical N-terminal domain for oligomerization is found in association with wild-type Vpr, suggesting that complexation between wild-type and mutant Vpr abrogates the biological activity of the wild-type protein.
First we demonstrate that, unlike wild-type Vpr, mutant variants of this protein with changes in residue 73 that substitute serine for arginine (R73S) or alanine for arginine (R73A) are unable to significantly stimulate HIV-1 LTR transcription activity. In this study, human astroglial cells, T98G, were transfected with the LTR luciferase reporter plasmid alone or in combination with expression plasmids for Vpr or its mutant variants. As shown in Fig. 1B, ectopic expression of wild-type but not mutant Vpr elevated transcriptional activity of the LTR in the transfected cells. More interestingly, in the presence of mutant Vpr, wild-type protein was not able to augment transcription of the viral promoter, suggesting that these mutants may function as dominant negative proteins and abrogate transcriptional ability of the wild-type Vpr. Similar results were obtained in primary human astrocytic cells transfected with HIV-1 LTR reporter construct plus Vpr expression plasmid (B. Sawaya, unpublished observations).
FIG. 1.
Transcriptional activity of Vpr and its mutant variants. (A) Structural organization of wild-type Vpr depicting the areas representing helical domains I and II and the leucine-rich domain. (B) Transcriptional activity of wild-type (wt) and mutant (R73S and R73A) Vpr upon the HIV-1 LTR promoter. The human astroglial cell line T98G was transfected with 1.0 μg of the reporter LTR-luciferase plasmid (LTR-Luc) alone or combined with 2.5 μg of the Vpr expression plasmids by calcium phosphate precipitation (11). All Vpr expression plasmids were created in pcDNA3 background plasmids, and empty pCDNA3 was added in all transfection mixtures in order to normalize the amounts of the DNA in each reaction. Luciferase activity was determined 48 h after transfection. The values shown on the top of each bar represent the fold activation over the basal HIV-1 LTR promoter arbitrarily set at 1. The data represent the mean value of at least three separate transfection experiments.
Next, we evaluated the effect of Vpr and its mutant variants on the cell cycle progression. In this respect, synchronized T98G cells were transfected with wild-type and mutant Vpr, singly or in combination, and after 20 to 24 and 36 to 40 h, cells were harvested and examined for their cell cycle states by fluorescence-activated cell sorter (FACS) analysis. A marker plasmid expressing enhanced green fluorescent protein (EGFP)-spectrin was included in the transfection mixture. The control cells were transfected only with EGFP-spectrin plasmid. After harvest, cells were fixed and stained with propidium iodide to determine the DNA content and were simultaneously examined for EGFP expression.
Cells that expressed EGFP were gated on the FACS, and the DNA profiles of both the EGFP-positive and EGFP-negative populations were determined and compared in transfected and nontransfected cells in the same culture.
In accord with previous observations, we found that expression of Vpr in human astroglial cells results in the accumulation of cells at the G2 phase (Table 1, compare GFP and Vpr wt at 36 h). However, mutant Vpr R73S failed to arrest cells at the G2 phase to the same extent as the wild-type (Table 1, compare Vpr wt and Vpr mut at 36 h). Interestingly, when wild-type Vpr was coexpressed with mutant R73S, progression of the cells through the G2 phase was not inhibited (Table 1, compare Vpr wt and Vpr wt + R73S), suggesting that the R73S mutant may function as a dominant negative mutant which interferes with the cell-cycle-inhibiting activity of wild-type Vpr in human astroglial cells. Similar results were obtained by using another astrocytic glial cell line, U87MG (data not shown).
TABLE 1.
Effect of Vpr and its mutants on cell cycle progressiona
| Plasmid transfected | % of cells in various stages at (h):
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0
|
20–24
|
36–40
|
|||||||
| G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | |
| Nontransfected (control) | 90.3 | 3.5 | 6.2 | 25.4 | 45.2 | 29.4 | 48.7 | 38.6 | 12.7 |
| GFP | 10.3 | 58.4 | 31.3 | 40.9 | 45.9 | 13.2 | |||
| GFP + Vpr wt | 19.1 | 45.0 | 35.9 | 12.5 | 8.9 | 78.6 | |||
| GFP + Vpr mut (R73S) | 8.8 | 76.0 | 15.2 | 57.1 | 19.6 | 23.3 | |||
| GFP + Vpr wt + R73S | 27.3 | 46.4 | 26.3 | 59.4 | 20.1 | 20.5 | |||
Human astroglial cells T98G were synchronized by serum starvation. After 60 h, complete Dulbecco modified Eagle medium containing fetal calf serum was added to the cells. The cells were transfected with 10 μg of plasmids expressing wild-type (wt) Vpr or mutant (R73S) proteins, along with 5 μg of plasmid expressing EGFP-spectrin. After 20 and 36 h, cells were harvested and prepared for the measurement of their DNA content and EGFP expression by FACS analysis. The percentages of cells in G1, S, and G2 are indicated in each panel. The data were analyzed using the SOBR program on a Becton Dickinson flow cytometer and represent the average of three independent experiments.
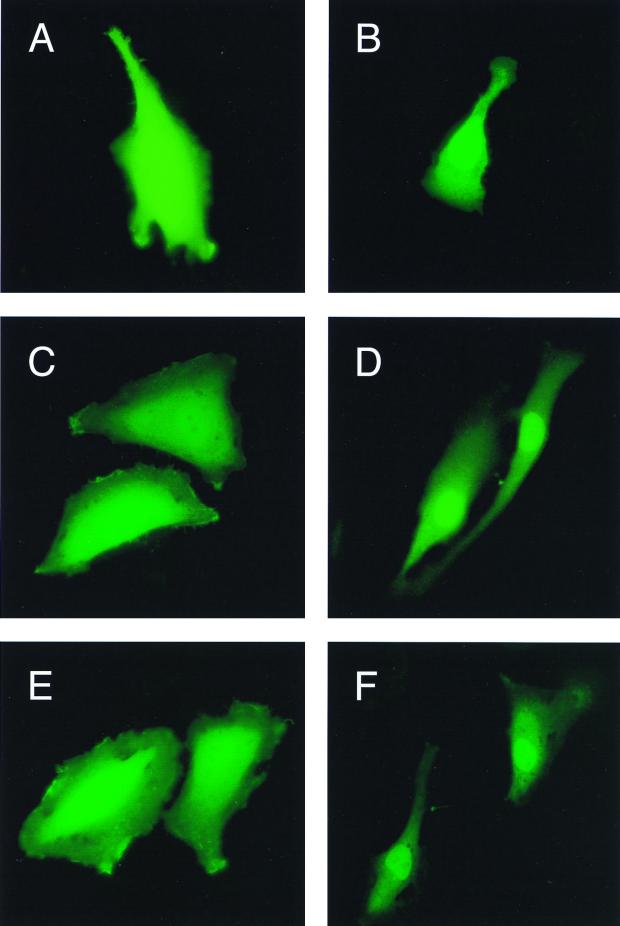
The lack of transcriptional activity and cell proliferation inhibition of mutant Vpr prompted us to examine cellular compartmentalization of this protein in the test cells. Toward that end, recombinant plasmids expressing Vpr fused to the green fluorescent protein (GFP) were constructed and introduced into T98G cells by transient transfection. Localization of the fluorescently labeled wild-type Vpr-GFP and mutant R73S-GFP and R73A fusion proteins was evaluated by fluorescence microscopy. The results shown in Fig. 2 revealed the presence of GFP-Vpr in the nuclear compartment as well as in the cytoplasmic compartment (Fig. 2B). A similar pattern was observed with GFP-Vpr mutants R73S and R73A (Fig. 2D and F, respectively). Cells transfected with control GFP plasmid or GFP with mutant Vpr in the antisense orientations (Fig. 2A, C, and E, respectively) did not demonstrate evidence of cellular compartmentalization. These data suggest that the inability of mutant Vpr to exert its biological activity on transcription and cell cycle progression may not be attributed to differential cellular distribution.
FIG. 2.
Subcellular localization of wild-type Vpr and its mutant variants. Approximately 2 μg of plasmids which express Vpr and its mutants R73S and R73A, in fusion with GFP (EGFP), were introduced into T98G cells by the calcium phosphate precipitation method. After 36 h, cells were examined by fluorescence microscopy for the presence of the EGFP-Vpr proteins in the transfected cells. Cells were transfected with EGFP alone (panel A) or with wild-type Vpr EGFP, EGFP-R73S, or EGFP-R73A (panels B, D, and F, respectively). Panels C and E show cells transfected with EGFP-R73S and EGFP-R73A in the antisense orientations, respectively.
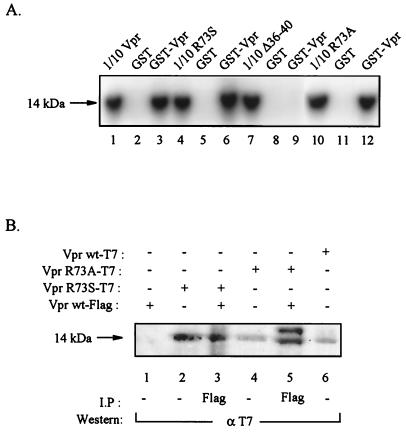
As mentioned above, Vpr can be oligomerized, and the N-terminal domain of the protein appears to be critical for this event (26, 29). Thus, it is likely that the association of wild-type Vpr, with its functionally inactive mutant variants including R73S, renders this protein nonfunctional. Thus, to examine the ability of mutant Vpr to form a complex with wild-type Vpr, we performed a glutathione S-transferase (GST) pull down assay utilizing bacterially produced wild-type or various Vpr mutants fused to GST. In this study, in-vitro-translated 35S-labeled Vpr was incubated with GST alone or GST-Vpr (wild-type) fusion proteins. After 1 h of incubation of the complexes at 4°C, the complexes bound to resin were pelleted and washed with binding buffer, as described previously (9). Bound proteins were diluted by boiling in Laemmli sample buffer, and after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were detected by autoradiography. As shown in Fig. 3A, the 14-kDa 35S-labeled Vpr was retained by the GST-Vpr fusion protein (Fig. 3, lane 3) but not by GST alone (lane 2). In lane 1, 1/10 of the in-vitro-synthesized Vpr which was used in GST assay was directly analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. These observations corroborate previous findings (26, 29) on the ability of Vpr to associate with itself and oligomerize. A similar set of experiments was performed with mutant R73S (Fig. 3A, lanes 4 to 6), mutant Δ36-40 (lanes 7 to 9), and mutant R73A (lanes 10 to 12). As shown in Fig. 3A, both R73S and R73A were able to form complexes with the wild-type Vpr. In contrast, mutant Δ36-40, which has small deletions in the N-terminal region of the protein (residues 36 to 40) was not able to associate with wild-type Vpr. This observation is in accord with the data pointing to the importance of the N terminus of Vpr in oligomerization (26, 29). Next, we evaluated the ability of wild-type and mutant Vpr to associate with each other in cells expressing these proteins. Toward this end, nuclear extracts from cells transfected with plasmids expressing wild-type Vpr or its mutant variants R73A and R73S were prepared and analyzed by a sequential immunoprecipitation-Western blot technique. In order to differentially detect wild-type and mutant proteins, we utilized two distinct expression vectors which permit detection of the wild-type protein by using anti-T7 antibody (Novagen, Madison, Wis.) and mutant protein via anti-FLAG antibody. First, nuclear extracts from the transfected cells were reacted with anti-FLAG antibody and immunocomplexes were analyzed by Western blotting by using anti-T7 antibody. In parallel, extracts from the transfected cells were also examined by direct Western blotting with appropriate antibodies. As shown in Fig. 3B, a 14-kDa band corresponding to wild-type Vpr was detected in extracts from Vpr wild type-T7-, Vpr R73A-T7-, and Vpr R73S-T7-transfected cells but not in extract from cells transfected with Vpr wt-FLAG (Fig. 3B, compare lanes 6, 4, and 2 with lane 1). Interestingly, anti-FLAG immunocomplexes derived from cells transfected with Vpr wt-FLAG plus Vpr R73S-T7 (lane 3) or Vpr wild type-FLAG plus Vpr R73A (lane 5) showed the 14-kDa band in Western blot analysis after treatment with anti-T7 antibody. These observations suggest that both mutants R73A and R73S can form a complex with wild-type Vpr in the transfected cells. The control complexes obtained upon treatment of cells with normal sera did not exhibit the 14-kDa band (data not shown).
FIG. 3.
In vitro and in vivo association of wild-type Vpr with mutant R73A and R73S. (A) In-vitro-translated 35S-labeled wild-type Vpr (lanes 1 to 3) or its mutant variants R73S (lanes 4 to 6), Δ36-40 (lanes 7 to 9), and R73A (lanes 10 to 12) were incubated with either GST or GST wild-type Vpr as indicated above the lanes. The position of the 14-kDa Vpr bands bound to the GST fusion proteins is shown. (B) Cell lysates were prepared from T98G cells transfected with plasmids expressing wild-type Vpr fused to T7 or FLAG or mutant Vpr fused to pEBV-His-Vpr or pCDNA-Vpr-FLAG (Invitrogen). Approximately 300 μg of extract was utilized in immunoprecipitations followed by Western blotting utilizing anti-FLAG and anti-T7 antibodies, respectively (lanes 3 and 5). In parallel, 100 μg of extracts were utilized by direct Western blot assay (lanes 1, 2, 4, and 6). The position of the 14-kDa Vpr is depicted by an arrow. The immunoprecipitation and Western blot analyses were carried out according to the procedure described previously (19).
The results presented in this report provide evidence supporting the importance of the residue 73 of Vpr for its transcriptional activity and its role in the control of the cell cycle at the G2 phase. As reported previously, mutation at this residue of Vpr, however, has no effect on the incorporation of this protein into virions or on the stability of Vpr (5, 28). Of note, the ability of HIV-1 carrying the mutant R73S to replicate is significantly decreased in comparison to that of the wild-type virus (data not shown).
In our effort to develop agents specifically targeting Vpr, we undertook the analysis of Vpr mutants for their effect on functions mediated by wild-type Vpr. The underlying rationale is that mutant Vpr, depending on the location of the mutation on the molecule, may exert a dominant negative effect on the function carried out by wild-type Vpr. This is based on the notion that a mixed (wild-type and mutant Vpr) oligomeric protein may not have the same property as the wild-type oligomeric protein. Such a scenario was reported with respect to HIV-1-encoded Rev, Gag, and Env proteins (8, 16, 24). The results presented here show that the residue R73 in the Vpr molecule is essential for Vpr-mediated functions, as substitutions at this residue led to the elimination of Vpr-induced transcriptional activation and cell cycle arrest. Further, it was also noted that the mutant Vpr exerted a transdominant effect on wild-type Vpr activity. Earlier studies suggested that a desirable feature of a candidate protein with a transdominant phenotype would be distinct functional domains. This allows the protein to have a defect in the effector domain without abrogating the biochemical feature such as oligomerization (2). Interestingly, Vpr R73S mutant fits this criterion. The mechanism by which Vpr R73S mutant brings about the dominant negative effect on wild-type Vpr is not clear. Based on our data, a hypothetical model is proposed. This model predicts three forms of oligomeric Vpr which may be present when wild-type and mutant Vpr are coexpressed: a homomeric wild-type protein, which carries out the various functions of Vpr, and a heteromeric (wild-type and mutant Vpr) and a homomeric mutant Vpr, which lack the functions of wild-type Vpr due to a deficit at the effector domain.
As the functions of Vpr are directly linked to HIV-1 replication which is crucial for the induction of HIV-1-associated diseases, it has been suggested that Vpr should also be considered as one of the targets for the development of antiviral strategies. In this regard, it was reported that RU486, an antagonist of glucocorticoid, inhibited Vpr-induced HIV-1 replication, implicating the involvement of GR or a closely related molecule in the Vpr-induced effect (14, 20). Shimura et al. (22) identified Quercetin, a flavonoid extracted from Houttuyniae herve, to effectively interfere with Vpr functions by utilizing a conditional Vpr-expressing cell line. The identification of Vpr mutant with transdominant properties may provide an additional avenue to explore genetic approaches to inhibit HIV-1 replication.
Acknowledgments
We thank Andrew Beavis for kindly providing EGFP-spectrin and for helpful comments, Nathaniel Landau for kindly providing reagents used in these studies, Dave Decker and Reiner Class for carrying out FACS analysis, and Maggie Kasten for helpful discussions. We also thank past and present members of the Center for Neurovirology and Cancer Biology for insightful discussions and sharing of ideas and reagents, particularly Irina Feldman for technical assistance and Cynthia Schriver for preparation of the manuscript.
This work was made possible by grants awarded by NIH to K.K. and S.A.
REFERENCES
- 1.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of human immunodeficiency virus type 1 accessory genes vpr, vpu and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 2.Bogerd H P, Fridell R A, Blair W S, Cullen B R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouhamdan M, Benichou S, Rey F, Navarro J M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracin DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 5.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N. Mutational analysis of cell cycle arrest, nuclear localization and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 7.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallia G L, Safak M, Khalili K. Interaction of single-stranded DNA binding protein, Purα, with human polyomavirus, JCV, early protein, T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 10.Gragerov A, Kino T, Ilyina-Gragerova G, Chrousos G P, Pavlakis G N. HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology. 1998;245:323–330. doi: 10.1006/viro.1998.9138. [DOI] [PubMed] [Google Scholar]
- 11.Graham F L, van der Eb A J. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1973;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He J, Choe S, Walker R, DiMarzio P, Morgan D O, Landau N. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jowett J B, Planelles V, Poon B, Shah N P, Chen M, Chen I S. The human immunodeficiency virus type 1 Vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kino T, Gragerov A, Kopp J B, Stauber R H, Pavlakis G N, Chrousos G P. The HIV-1 virion-associated protein vpr is a coactivator of the human glucocorticoid receptor. J Exp Med. 1999;189:51–62. doi: 10.1084/jem.189.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Z, Butcher D J, Murali R, Srinivasan A, Huang Z. Structural studies of synthetic peptide fragments derived from the HIV-1 Vpr protein. Biochem Biophys Res Commun. 1998;244:732–736. doi: 10.1006/bbrc.1998.8330. [DOI] [PubMed] [Google Scholar]
- 16.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. Stable expression of transdominant Rev proteins in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Re R, Braten D, Frank E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Refaeli Y, Levy D N, Weiner D B. The glucocorticoid receptor type II complex is a target of the HIV-1 Vpr gene product. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawaya B E, Khalili K, Mercer W E, Denisova L, Amini S. Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J Biol Chem. 1998;273:20052–20057. doi: 10.1074/jbc.273.32.20052. [DOI] [PubMed] [Google Scholar]
- 20.Sawaya B E, Khalili K, Rappaport J, Serio D, Chen W, Srinivasan A, Amini S. Suppression of HIV-1 transcription and replication by a Vpr mutant. Gene Ther. 1999;6:947–950. doi: 10.1038/sj.gt.3300907. [DOI] [PubMed] [Google Scholar]
- 21.Schuler W, Wecker K, deRocquigny H, Baudat Y, Sire J, Roques B P. NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions. J Mol Biol. 1999;285:2105–2117. doi: 10.1006/jmbi.1998.2381. [DOI] [PubMed] [Google Scholar]
- 22.Shimura M, Zhou Y, Asada Y, Yoshikawa T, Helake K, Takaku F, Ishizaka Y. Inhibition of Vpr-induced cell cycle abnormality by quercetin: a novel strategy for searching compounds targeting Vpr. Biochem Biophys Res Commun. 1999;261:308–316. doi: 10.1006/bbrc.1999.0994. [DOI] [PubMed] [Google Scholar]
- 23.Stark L A, Hay R T. Human immunodeficiency virus type 1 (HIV-1) viral protein R (Vpr) interacts with Lys-tRNA synthetase: implications for priming of HIV-1 reverse transcription. J Virol. 1998;72:3037–3044. doi: 10.1128/jvi.72.4.3037-3044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trono D, Feinberg M B, Baltimore D. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 26.Wecker K, Rogues B. NMR structure of the (1-51) N-terminal domain of the HIV-1 regulatory protein Vpr. Eur J Biochem. 1999;266:359–369. doi: 10.1046/j.1432-1327.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- 27.Westervelt P, Henkel T, Trowbidge D B, Orenstein J, Heuser J, Gendelman H E, Ratner L. Dual regulation of silent and productive infection in monocytes by distinct human immunodeficiency virus type 1 determinants. J Virol. 1992;66:3925–3931. doi: 10.1128/jvi.66.6.3925-3931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao X J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L J, Wang L, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Oligomerization mediated by the N-terminal domain. J Biol Chem. 1994;269:32131–32137. [PubMed] [Google Scholar]