Text Correction
The authors wish to make the following corrections to this paper [1]. We request correction of the MICs and gene possession ratios of the two strains TWCC 52027 and TWCC 53044. This correction does not affect any content in our paper at all, but only affects a p-value described in Section 2.2.4 (Page 8): “whereas the ratio was higher in the GH group than in G. morbillorum (p = 0.16).” This p-value (0.16) was replaced with 0.08. We need to correct the values in Abstract, Results, Figures 1 and 2, Tables 2–5, Supplementary Table S1, Sections 2.2.2–2.2.4, Section 2.2.6, Sections 2.3–2.5.
Abstract: Gemella is a catalase-negative, facultative anaerobic, Gram-positive coccus that is commensal in humans but can become opportunistic and cause severe infectious diseases, such as infective endocarditis. Few studies have tested the antimicrobial susceptibility of Gemella. We tested its antimicrobial susceptibility to 27 drugs and defined the resistant genes using PCR in 58 Gemella strains, including 52 clinical isolates and 6 type strains. The type strains and clinical isolates comprised 22 G. morbillorum, 18 G. haemolysans (GH) group (genetically indistinguishable from G. haemolysans and G. parahaemolysans), 13 G. taiwanensis, three G. sanguinis, and two G. bergeri. No strain was resistant to beta-lactams and vancomycin. In total, 6/22 (27.3%) G. morbillorum strains were erythromycin- and clindamycin-resistant ermB-positive, whereas 5/18 (27.8%) in the GH group, 6/13 (46.2%) G. taiwanensis, and 1/3 (33.3%) of the G. sanguinis strains were erythromycin-non-susceptible mefE- or mefA-positive and clindamycin-susceptible. The MIC90 of minocycline and the ratios of tetM-positive strains varied across the different species—G. morbillorum: 2 µg/mL and 27.3% (6/22); GH group: 8 µg/mL and 22.2% (4/18); G. taiwanensis: 8 µg/mL and 53.8% (7/13), respectively. Levofloxacin resistance was significantly higher in G. taiwanensis (8/13, 61.5%) than in G. morbillorum (2/22, 9.1%). Levofloxacin resistance was associated with a substitution at serine 83 for leucine, phenylalanine, or tyrosine in GyrA. The mechanisms of resistance to erythromycin and clindamycin differed across Gemella species. In addition, the rate of susceptibility to levofloxacin differed across Gemella spp., and the quinolone resistance mechanism was caused by mutations in GyrA alone.
2.2.2. Susceptibility to Erythromycin
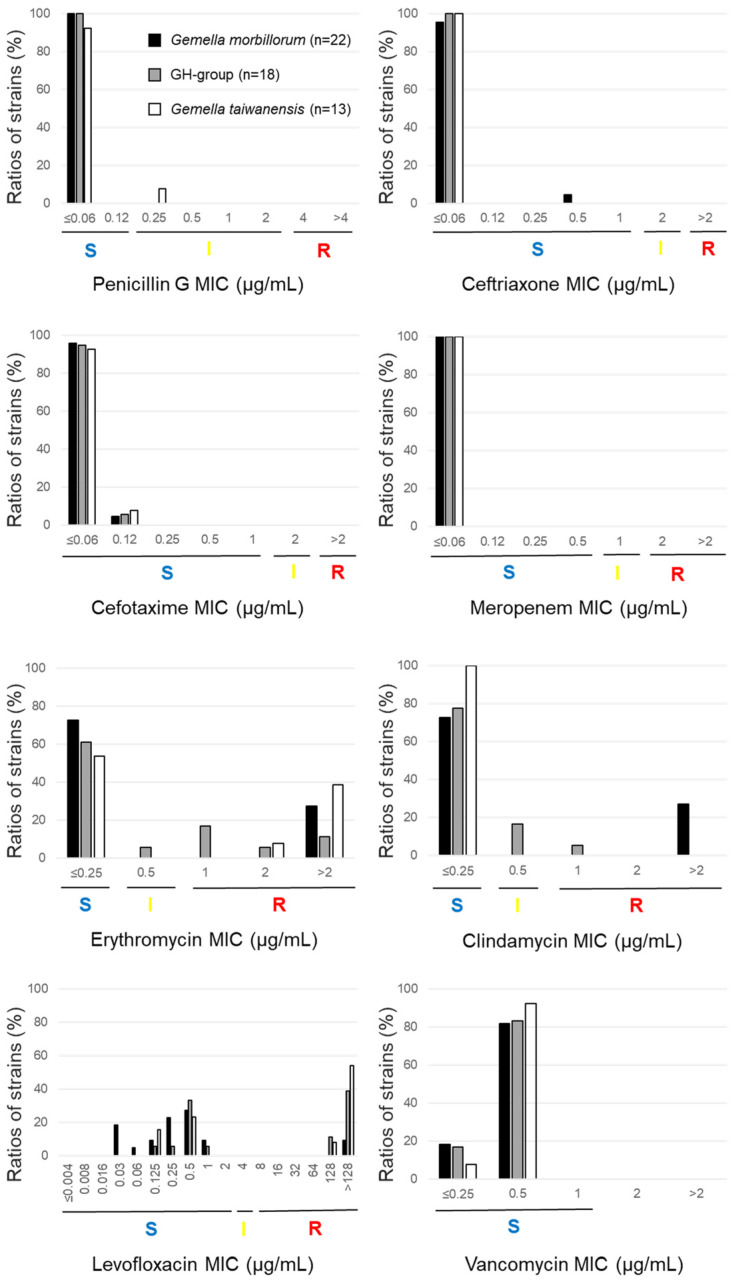
In total, 20/58 strains were erythromycin-non-susceptible (intermediate or resistant), with MIC90 > 2 µg/mL. Although the ratios of the erythromycin-non-susceptible isolates varied across species, there was no significant difference among G. morbillorum, the GH group, and G. taiwanensis—G. morbillorum: 27.3% (6/22), GH group: 38.9% (7/18), G. taiwanensis: 46.2% (6/13), G. sanguinis: 33.3% (1/3), and G. bergeri: 0.0% (0/2) (Table 2, Figures 1 and 2).
2.2.3. Susceptibility to Clindamycin
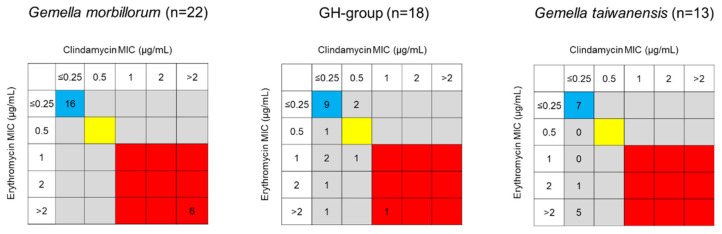
In total, 10/58 strains were clindamycin-non-susceptible, resulting in MIC90 > 2 µg/mL. Clindamycin-resistant G. taiwanensis, G. sanguinis, and G. bergeri isolates were not detected, and differences were not significant—G. morbillorum: 27.3% (6/22), GH group: 22.2% (4/18), G. taiwanensis: 0.0% (0/13), and G. sanguinis: 0.0% (0/3) (Table 2, Figures 1 and 2). Interestingly, all six erythromycin-resistant G. morbillorum strains were clindamycin-resistant. In contrast, 5/7 GH group strains, six strains of G. taiwanensis, and one G. sanguinis strain were erythromycin-non-susceptible and clindamycin-susceptible.
2.2.4. Susceptibility to Levofloxacin
In total, 21/58 strains were levofloxacin-resistant, resulting in MIC90 > 128 µg/mL. Ratios of the levofloxacin strains varied across species—G. morbillorum: 9.1% (2/22), GH group: 50.0% (9/18), G. taiwanensis: 61.5% (8/13), G. sanguinis: 66.7% (2/3), and G. bergeri: 0.0% (0/2). The ratio of the resistant strains was significantly higher in G. taiwanensis than in G. morbillorum (p < 0.05 using chi-squared test), whereas the ratio was higher in the GH group than in G. morbillorum (p = 0.08) (Table 2, Figure 1).
2.2.6. Susceptibility to Other Antimicrobial Agents
We tested the 18 antimicrobial agents whose breakpoints are not listed in CLSI M45-third edition. Gemella strains showed low MIC values for all beta-lactams: ampicillin, amoxicillin–clavulanic acid, sulbactam–ampicillin, cefazolin, cefdinir, cefepime, and imipenem (MIC90: ≤0.12, ≤0.25/0.12, ≤0.06/0.12, ≤0.25, ≤0.25, ≤0.06, and ≤0.06 μg/mL, respectively). The MIC90 values of clarithromycin and azithromycin were 8 and >4 µg/mL, respectively, consistent with those of erythromycin. The MIC90 values of clarithromycin varied among G. morbillorum (>16 μg/mL), the GH group (8 μg/mL), and G. taiwanensis (2 μg/mL), indicating the acquisition of high resistance to clarithromycin in G. morbillorum strains. The MIC90 value of moxifloxacin was high (>2 μg/mL) in Gemella strains. The MIC90 values of the aminoglycoside antibiotics gentamicin, gentamicin500 (to confirm tolerance to high concentrations of gentamicin), and arbekacin were 8, ≤500, and >8 μg/mL, respectively; sulfamethoxazole–trimethoprim, fosfomycin, and rifampicin were >38/2, ≤16, and ≤0.5 μg/mL, respectively; and the anti-MRSA agents teicoplanin, linezolid, and daptomycin were ≤0.5, 1, and 2 μg/mL, respectively (Table 2). Typically, streptococci are aminoglycoside-resistant. Therefore, we tested gentamicin500 to identify any Gemella strains that are highly resistant to aminoglycoside.
2.3. Phenotypes and Genotypes of Macrolide-Resistant Strains
The six erythromycin–clindamycin-resistant G. morbillorum strains exhibited constitutive resistance to macrolide, lincosamide, and streptogramin B (cMLSB). Their genotypes—mefA/E-negative, ermB-positive, and msrA-negative—were consistent with their phenotypes. Furthermore, five/seven strains of the GH group, six strains of G. taiwanensis, and one strain of G. sanguinis which were erythromycin-non-susceptible and clindamycin-susceptible, had macrolide-resistant (M) phenotypes and mefE- (four strains) or mefA-positive (one strain), erm-negative, and msrA-negative genotypes. In total, 2/7 GH group strains (TWCC 59567 and TWCC 59795) were erythromycin-resistant and clindamycin-non-susceptible and mefE-positive, but showed M phenotype. These results show that erythromycin-resistant G. morbillorum is associated with ermB, and erythromycin-non-susceptible GH-group, G. taiwanensis, and G. sanguinis are associated with mefE. The MIC values for clarithromycin were higher in the six ermB-positive G. morbillorum strains (8 or >16 µg/mL) (Table 3). All erythromycin-susceptible Gemella strains, except the G. sanguinis strain TWCC 70419, lacked mefA/E, erm, or msrA (Table S1).
2.4. Tetracycline Resistance
Next, we analyzed the possession rates of tet. Overall, 17/58 (29.3%) strains were tetM-positive; none of the other tet genes was detected. The ratios of tetM-positive strains in G. morbillorum, the GH group, G. taiwanensis, G. sanguinis, and G. bergeri were 27.3% (6/22), 33.3% (6/18), 38.5% (5/13), 0/3 (0.0%), and 0.0% (0/2), respectively. Among the 41 tetM-negative strains, one had minocycline (MIC = 2 µg/mL). The minocycline MIC values of the others were ≤1 µg/mL. The minocycline MIC of the 17 tetM-positive strains varied: ≤1 for five, 2 for five, and ≥8 µg/mL for seven strains, respectively (Table 4).
2.5. Mutations in gyrA and gyrB
We analyzed the gyrA and gyrB sequences. The 35 quinolone-susceptible strains possessed gyrA, encoding GyrA with a serine residue at 83 (S83). The serine residue was substituted with leucine (S83L), phenylalanine (S83F), or tyrosine (S83Y) in the 21 quinolone-resistant strains. Specifically, two G. morbillorum strains possessed GyrA/S83L, encoding gyrA. Seven of the GH group, seven G. taiwanensis, and two G. sanguinis strains contained S83F. Two in the GH group and one G. taiwanensis strains contained S84Y. GyrB mutations associated with levofloxacin resistance were not detected (Table 5).
Figure 1.
Ratios of resistant strains. S (blue), I (yellow), and R (red) indicate sensitive, intermediate, and resistant, respectively.
Figure 2.
Distribution of erythromycin/clindamycin resistance in Gemella strains. Blue, yellow, and red boxes indicate sensitive, intermediate, and resistant, respectively.
Table 2.
Susceptibility to antimicrobial agents with breakpoints listed in CLSI M45-third edition.
| Antimicrobial Agents/ Gemella spp. |
MIC (μg/mL) | Interpretive Breakpoint (μg/mL) a
or % of Isolates |
||||
|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | Susceptible | Intermediate | Resistant | |
| Penicillin G c | ≤0.06–>4 | ≤0.12 | 0.25–2 | ≥4 | ||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06–0.25 | ≤0.06 | ≤0.06 | 92.3 | 7.7 | 0.0 |
| Gemella sanguinis | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06–0.25 | ≤0.06 | ≤0.06 | 98.3 | 1.7 | 0.0 |
| Ampicillin | ≤0.12–>4 | |||||
| Gemella morbillorum | ≤0.12–0.25 | ≤0.12 | ≤0.12 | NA b | NA | NA |
| GH group | ≤0.12 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–0.5 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.12 | – | – | NA | NA | NA |
| Total | ≤0.12–0.5 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Amoxicillin–clavulanic acid | ≤0.25/0.12–>4/2 | |||||
| Gemella morbillorum | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| GH group | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.25/0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25/0.12 | – | – | NA | NA | NA |
| Total | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Sulbactam–ampicillin | ≤0.06/0.12–>2/4 | |||||
| Gemella morbillorum | ≤0.06/0.12 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| GH group | ≤0.06/0.12 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06/0.12–0.25/0.5 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.06/0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06/0.12 | – | – | NA | NA | NA |
| Total | ≤0.06/0.12–0.25/0.5 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Cefazolin | ≤0.25–>2 | |||||
| Gemella morbillorum | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| GH group | ≤0.25–0.5 | ≤0.25 | 0.5 | NA | NA | NA |
| G emellataiwanensis | ≤0.25–0.5 | ≤0.25 | 0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.25 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25 | – | – | NA | NA | NA |
| Total | ≤0.25–0.5 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Cefdinir | ≤0.25–>1 | |||||
| G emella morbillorum | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| GH group | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Gememlla sanguinis | ≤0.25–0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25 | – | – | NA | NA | NA |
| Total | ≤0.25–0.5 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Ceftriaxone c | ≤0.06–>2 | ≤1 | 2 | ≥4 | ||
| Gemella morbillorum | ≤0.06–0.5 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | 0.25–1 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | 100.0 | 0.0 | 0.0 | ||
| Total | ≤0.06–1 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Cefotaxime c | ≤0.06–>2 | ≤1 | 2 | ≥4 | ||
| Gemella morbillorum | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | 0.25–1 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06–1 | ≤0.06 | 0.12 | 100.0 | 0.0 | 0.0 |
| Cefepime | ≤0.06–>2 | |||||
| G emella morbillorum | ≤0.06–0.5 | ≤0.06 | ≤0.06 | NA | NA | NA |
| GH group | ≤0.06–0.12 | ≤0.06 | 0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06–0.12 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella sanguinis | 0.25–1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06 | – | – | NA | NA | NA |
| Total | ≤0.06–1 | ≤0.06 | 0.12 | NA | NA | NA |
| Imipenem | ≤0.06–>4 | |||||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella sanguinis | ≤0.06 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06 | – | – | NA | NA | NA |
| Total | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Meropenem c | ≤0.06–>2 | ≤0.5 | 1 | ≥2 | ||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Erythromycin c | ≤0.25–>2 | ≤0.25 | 0.5 | ≥1 | ||
| Gemella morbillorum | ≤0.25–>2 | ≤0.25 | >2 | 72.7 | 0.0 | 27.3 |
| GH group | ≤0.25–>2 | ≤0.25 | >2 | 61.1 | 5.6 | 33.3 |
| Gemella taiwanensis | ≤0.25–>2 | ≤0.25 | >2 | 53.8 | 0.0 | 46.2 |
| Gemella sanguinis | ≤0.25–1 | – | – | 66.7 | 0.0 | 33.3 |
| Gemella bergeri | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–>2 | ≤0.25 | >2 | 65.5 | 1.7 | 32.8 |
| Clarithromycin | ≤0.12–>16 | |||||
| Gemella morbillorum | ≤0.12–>16 | ≤0.12 | >16 | NA | NA | NA |
| GH group | ≤0.12–16 | ≤0.12 | 8 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–8 | ≤0.12 | 2 | NA | NA | NA |
| Gemella sanguinis | ≤0.12–0.25 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.12 | – | – | NA | NA | NA |
| Total | ≤0.12–>16 | ≤0.12 | 8 | NA | NA | NA |
| Azithromycin | ≤0.12–>4 | |||||
| Gemella morbillorum | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| GH group | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–>4 | 0.25 | >4 | NA | NA | NA |
| Gemella sanguinis | 0.25–4 | – | – | NA | NA | NA |
| Gemella bergeri | 0.25 | – | – | NA | NA | NA |
| Total | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| Clindamycin c | ≤0.25–>2 | ≤0.25 | 0.5 | ≥1 | ||
| Gemella morbillorum | ≤0.25–>2 | ≤0.25 | >2 | 72.7 | 0.0 | 27.3 |
| GH group | ≤0.25–1 | ≤0.25 | 0.5 | 77.8 | 16.7 | 5.6 |
| Gemella taiwanensis | ≤0.25 | ≤0.25 | ≤0.25 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–>2 | ≤0.25 | >2 | 82.8 | 5.2 | 12.1 |
| Erythromycin/clindamycin | ≤1/0.5–>1/0.5 | |||||
| G. morbillorum | ≤1/0.5–>1/0.5 | ≤1/0.5 | >1/0.5 | NA | NA | NA |
| GH group | ≤1/0.5–>1/0.5 | ≤1/0.5 | ≤1/0.5 | NA | NA | NA |
| G. taiwanensis | ≤1/0.5 | ≤1/0.5 | ≤1/0.5 | NA | NA | NA |
| G. sanguinis | ≤1/0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤1/0.5 | – | – | NA | NA | NA |
| Total | ≤1/0.5–>1/0.5 | ≤1/0.5 | >1/0.5 | NA | NA | NA |
| Levofloxacin c | ≤0.004–>128 | ≤2 | 4 | ≥8 | ||
| Gemella morbillorum | 0.03–>128 | 0.25 | 1 | 90.9 | 0.0 | 9.1 |
| GH group | 0.125–>128 | 1 | >128 | 50.0 | 0.0 | 50.0 |
| Gemella taiwanensis | 0.125–>128 | >128 | >128 | 38.5 | 0.0 | 61.5 |
| Gemella sanguinis | 0.5–>128 | – | – | 33.3 | 0.0 | 66.7 |
| Gemella bergeri | 0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Total | 0.03–>128 | 0.5 | >128 | 63.8 | 0.0 | 36.2 |
| Moxifloxacin | ≤0.5–>2 | |||||
| G emella morbillorum | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| GH group | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5–>2 | >2 | >2 | NA | NA | NA |
| Gemella sanguinis | ≤0.5–>2 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | – | – | NA | NA | NA |
| Total | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| Minocycline | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–>8 | ≤1 | 2 | NA | NA | NA |
| GH group | ≤1–8 | ≤1 | 8 | NA | NA | NA |
| Gemella taiwanensis | ≤1–8 | ≤1 | 8 | NA | NA | NA |
| Gemella sanguinis | ≤1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤1 | – | – | NA | NA | NA |
| Total | ≤1 | ≤1 | 8 | NA | NA | NA |
| Sulfamethoxazole– trimethoprim |
≤9.5/0.5–>38/2 | |||||
| Gemella morbillorum | ≤9.5/0.5–>38/2 | 19/1 | >38/2 | NA | NA | NA |
| GH group | ≤9.5/0.5–>38/2 | 38/2 | >38/2 | NA | NA | NA |
| Gemella taiwanensis | ≤9.5/0.5–>38/2 | 19/1 | 19/1 | NA | NA | NA |
| Gemella sanguinis | 19/1–>38/2 | – | – | NA | NA | NA |
| Gemella bergeri | ≤9.5/0.5 | – | – | NA | NA | NA |
| Total | ≤9.5/0.5–>38/2 | 19/1 | >38/2 | NA | NA | NA |
| Gentamicin | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–8 | 2 | 8 | NA | NA | NA |
| GH group | ≤1–2 | ≤1 | 2 | NA | NA | NA |
| Gemella taiwanensis | ≤1–4 | 2 | 4 | NA | NA | NA |
| Gemella sanguinis | ≤1–8 | – | – | NA | NA | NA |
| Gemella bergeri | 2, 4 | – | – | NA | NA | NA |
| Total | ≤1–8 | 2 | 8 | NA | NA | NA |
| Gentamicin 500 | ≤500–>500 | |||||
| Gemella morbillorum | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| GH group | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Gemella taiwanensis | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Gemella sanguinis | ≤500 | – | – | NA | NA | NA |
| Gemella bergeri | ≤500 | – | – | NA | NA | NA |
| Total | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Arbekacin | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–8 | 8 | >8 | NA | NA | NA |
| GH group | ≤1–8 | 4 | 8 | NA | NA | NA |
| Gemella taiwanensis | 2–>8 | 4 | 8 | NA | NA | NA |
| Gemella sanguinis | 4–>8 | – | – | NA | NA | NA |
| Gemella bergeri | 4, >8 | – | – | NA | NA | NA |
| Total | ≤1–8 | 4 | >8 | NA | NA | NA |
| Fosfomycin | ≤16–>128 | |||||
| G emella morbillorum | ≤16–32 | ≤16 | ≤16 | NA | NA | NA |
| GH group | ≤16 | ≤16 | ≤16 | NA | NA | NA |
| Gemella taiwanensis | ≤16 | ≤16 | ≤16 | NA | NA | NA |
| Gemalla sanguinis | ≤16 | – | – | NA | NA | NA |
| Gemella bergeri | ≤16 | – | – | NA | NA | NA |
| Total | ≤16–32 | ≤16 | ≤16 | NA | NA | NA |
| Rifampicin | ≤0.5–>2 | |||||
| Gemella morbillorum | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| GH group | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | NA | NA | NA | ||
| Total | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Vancomycin c | ≤0.25–>2 | ≤1 | ||||
| Gemella morbillorum | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.25–0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | 0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Teicoplanin | ≤0.5–>16 | |||||
| Gemella morbillorum | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| GH group | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | – | – | NA | NA | NA |
| Total | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Linezolid | ≤0.5–>4 | |||||
| Gemella morbillorum | ≤0.5–1 | ≤0.5 | 1 | NA | NA | NA |
| GH group | ≤0.5–1 | ≤0.5 | 1 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5–1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5, 2 | – | – | NA | NA | NA |
| Total | ≤0.5 | ≤0.5 | 1 | NA | NA | NA |
| Daptomycin | ≤0.25–>4 | |||||
| Gemella morbillorum | ≤0.25–4 | 2 | 2 | NA | NA | NA |
| GH group | 0.5–2 | 1 | 2 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25–2 | 1 | 2 | NA | NA | NA |
| Gemella sanguinis | 1–4 | – | – | NA | NA | NA |
| Gemella bergeri | 2, 4 | – | – | NA | NA | NA |
| Total | ≤0.25–4 | 1 | 2 | NA | NA | NA |
a Interpretive breakpoints are shown in bold for each antibiotic. b NA, not applicable (breakpoints not established). c Antimicrobial agents with breakpoints listed in CLSI M45-third edition.
Table 3.
Distribution of macrolides and clindamycin MICs and possession of the mef, erm, and msrA genes in erythromycin-non-susceptible Gemella isolates.
| Strain No. | Identification | MIC (μg/mL) | Macrolide Phenotype a,b |
mefA/E | erm | msrA | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Erythromycin | Clindamycin | Erythromycin/Clindamycin | Clarithromycin | Azithromycin | ||||||
| TWCC 57201 | Gemella morbillorum | >2 | >2 | >1/0.5 | 8 | >4 | cMLSB | - | ermB | - |
| TWCC 57818 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 57944 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 59111 | Gemella morbillorum | >2 | >2 | >1/0.5 | 8 | >4 | cMLSB | - | ermB | - |
| TWCC 71703 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 72266 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 52027 | GH group | 0.5 | ≤0.25 | ≤1/0.5 | 8 | 2 | M | mefE | - | - |
| TWCC 59566 | GH group | 2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 59567 | GH group | >2 | 1 | >1/0.5 | 16 | >4 | M | mefE | - | - |
| TWCC 59795 | GH group | 1 | 0.5 | ≤1/0.5 | 0.5 | 2 | M | mefE | - | - |
| TWCC 70939 | GH group | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 71200 | GH group | 1 | ≤0.25 | ≤1/0.5 | 2 | 2 | M | mefA | - | - |
| TWCC 71814 | GH group | 1 | ≤0.25 | ≤1/0.5 | 0.5 | 1 | M | mefE | - | - |
| TWCC 55344 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 8 | >4 | M | mefE | - | - |
| TWCC 58522 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | 4 | M | mefE | - | - |
| TWCC 70386 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | 4 | M | mefE | - | - |
| TWCC 72085 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 70387L | Gemella taiwanensis | 2 | ≤0.25 | ≤1/0.5 | 0.5 | >4 | M | mefE | - | - |
| TWCC 70387S | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 54965 | Gemella sanguinis | 1 | ≤0.25 | ≤1/0.5 | 0.25 | 4 | M | mefE | - | - |
| TWCC 70419 | Gemella sanguinis | ≤0.25 c | ≤0.25 | ≤1/0.5 | ≤0.12 | 0.25 | not M | mefE | - | - |
a cMLSB: macrolide–lincosamide–streptogramin B-resistant phenotype. b M: macrolide-resistant phenotype. c Erythromycin-susceptible.
Table 4.
Distribution of minocycline MIC and ermB in Gemella isolates harboring the tetM gene.
| Strain No. | Identification | tetM | Minocycline MIC (μg/mL) |
ermB |
|---|---|---|---|---|
| TWCC 57944 | Gemella morbillorum | + | 2 | + |
| TWCC 57987 | Gemella morbillorum | + | ≤1 | − |
| TWCC 59111 | Gemella morbillorum | + | 2 | + |
| TWCC 70937 | Gemella morbillorum | + | >8 | − |
| TWCC 71703 | Gemella morbillorum | + | 2 | + |
| TWCC 72266 | Gemella morbillorum | + | ≤1 | + |
| TWCC 51800 | GH group | + | 8 | − |
| TWCC 59795 | GH group | + | ≤1 | − |
| TWCC 70939 | GH group | + | 8 | − |
| TWCC 71814 | GH group | + | 2 | − |
| TWCC 53044 | Gemella taiwanensis | + | 8 | − |
| TWCC 56546 | Gemella taiwanensis | + | 2 | − |
| TWCC 58522 | Gemella taiwanensis | + | 8 | − |
| TWCC 70386 | Gemella taiwanensis | + | 8 | − |
| TWCC 72085 | Gemella taiwanensis | + | 8 | − |
| TWCC 70387L | Gemella taiwanensis | + | ≤1 | − |
| TWCC 70387S | Gemella taiwanensis | + | ≤1 | − |
Table 5.
Distribution of MIC of tested quinolones and amino acid substitutions in gyrA gene in quinolone-resistant Gemella isolates.
| Strain | n | MIC (μg/mL) | GyrA Amino Acid Substitutions a | |
|---|---|---|---|---|
| Levofloxacin | Moxifloxacin | |||
| Gemella morbillorum | 2 | >128 | >2 | Ser83 > Leu83 (n = 2) |
| GH group | 9 | 128–>128 | >2 | Ser83 > Phe83 (n = 7), Ser83 > Tyr83 (n =2) |
| Gemella taiwanensis | 8 | 128–>128 | >2 | Ser83 > Phe83 (n = 7), Ser83 > Tyr83 (n = 1) |
| Gemella sanguinis | 2 | 128–>128 | >2 | Ser83 > Phe83 (n = 2) |
a gyrA-Ser83 Leu: serine to leucine at codon 83; Ser83 Phe: serine to phenylalanine at codon 83; Ser83 Tyr; serine to tyrosine at codon 83.
The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Reference
- 1.Furugaito M., Arai Y., Uzawa Y., Kamisako T., Ogura K., Okamoto S., Kikuchi K. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics. 2023;12:1538. doi: 10.3390/antibiotics12101538. [DOI] [PMC free article] [PubMed] [Google Scholar]