Abstract
The E7 oncoprotein encoded by human papillomavirus (HPV) type 16 repressed the transcription of fibronectin, a key component of the extracellular matrix. This repression, detected in several HPV-positive nontumorigenic and tumorigenic cell lines, was abolished when the Cys-X-X-Cys repeats in E7 were disrupted.
Fibronectins (FNs) are large glycoproteins (220 to 250 kDa) found in soluble form in plasma, as well as in insoluble form in the extracellular matrix. They have critical roles in cell adhesion, migration, differentiation, and proliferation (17). They interact with integrins and other cell surface receptors (17). Different regulatory molecules, e.g., serum, gamma interferon, cyclic AMP, and glucocorticoid hormones, affect FN expression at the transcriptional or posttranscriptional level (5–7, 10, 11). Loss of FN is well correlated with malignancy (17). In fact, loss of FN in hamster sarcoma virus-transformed cells was the original observation that led to the discovery and characterization of these glycoproteins (14, 16).
Among human papillomaviruses (HPVs), high-risk type 16 (HPV-16) and HPV-18 are usually detected in cervical cancers and derived cell lines (42, 43). Their major transforming proteins E6 and E7 (3, 24) bind and inactivate the tumor suppressor p53 and retinoblastoma (pRB) proteins, respectively, causing disruption of cell cycle control (43). The E7 oncoprotein can also interact with several other cellular proteins, e.g., AP-1, p130, and TATA box-binding protein (Los Alamos National Laboratory website; http://linker.lanl.gov/stdgen/virus/hpv/compendium/htdocs/HTML_FILES/), as well as act as a transactivator (22, 31, 41), properties that may be associated with the oncogenic potential of these viruses. Prior to this study, there was no information about the effect of the HPV-16 E7 oncoprotein on FN expression.
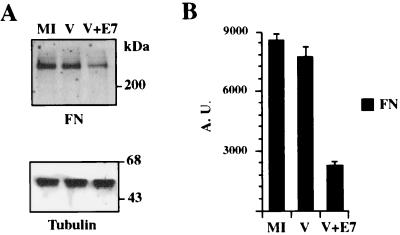
The FN protein level was analyzed in cells transiently expressing the HPV-16 E7 protein. CV-1 cells infected with a recombinant vaccinia virus (vTF7-3) encoding the T7 RNA polymerase protein (13) were transfected with the construct pGE7, which encodes the HPV-16 E7 oncoprotein under control of the T7 polymerase promoter. pGE7 was constructed by subcloning of a cDNA fragment containing the complete E7 open reading frame (nucleotides 505 to 1176) isolated from pE7Mo (12) into pGem4Z (Promega Corp., Madison, Wis.). A low multiplicity of infection (MOI) of 3 PFU/cell and a short time postinfection (6 h) were employed to minimize the cytopathic effect associated with vaccinia virus replication. As previously reported, the cytopatic effect associated with vaccinia virus replication occurs at early times postinfection only when a high MOI (>150 PFU/cell) is employed (1, 2). Six hours after transfection, the cells were lysed and the proteins were subjected to sodium dodecyl sulfate (SDS)–8% polyacrylamide gel electrophoresis (PAGE) and then transferred to a polyvinylidene difluoride membrane. The top and lower portions of the membrane were probed with murine monoclonal antibodies against FN (Transduction Laboratories, Lexington, Ky.) or β-tubulin (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.), respectively, and alkaline phosphatase-conjugated goat anti-mouse antibody. Signals were detected employing a chemifluorescent substrate (Amersham Pharmacia) and a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Signals were quantified with Image-Quant software, version 1.1 (Molecular Dynamics), and FN values were normalized to tubulin content. A reduction of approximately 60% in the amount of FN was detected in the cultures transfected with pGE7 compared with mock-infected cells or cells infected with vTF7-3 and transfected with the empty control vector pGem4Z (Fig. 1). This reduction was not related to vaccinia virus replication, because the same amount of FN was detected in mock-infected cells and vaccinia virus-infected cells transfected with the empty vector. A 60% reduction in FN expression was also detected in primary normal human oral keratinocytes (NHOK) as a result of transient E7 expression (data not shown). The steady state of tubulin (Fig. 1A), actin, or collagen type IV was not affected by E7 expression (data not shown).
FIG. 1.
Western blot analysis of FN in cells expressing WT E7. CV-1 cells infected with a recombinant vaccinia virus encoding T7 polymerase were transfected with the plasmid pGE7, encoding the HPV-16 E7 protein under control of the T7 promoter (V+E7) or with the empty control vector pGem4Z (V). Six hours after transfection, the cells were lysed and the proteins were subjected to SDS–8% PAGE and then transferred to a membrane. The top and lower portions of the membrane were decorated with murine monoclonal antibodies against FN or β-tubulin, respectively, and alkaline phosphatase-conjugated goat anti-mouse antibody (A). Signals were detected and quantified with a Storm 840 PhosphorImager. FN values were normalized to the tubulin content and represent the means of two different experiments (B). The bars represent standard deviations. The positions of molecular mass markers are indicated on the right of panel A. AU, arbitrary units; MI, mock-infected cells.
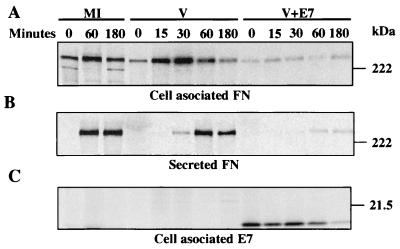
Since FN is a glycoprotein which, under normal conditions, is secreted into the external medium, a possible explanation for the observed decrease in its steady state was that E7 could affect the intracellular sorting of FN or accelerate its secretion. Alternatively, an increase in the rate of degradation of FN could also result in the same defect. Pulse-chase and immunoprecipitation of FN in cells expressing E7 were employed to investigate these possibilities. CV-1 cells infected with vTF7-3 with an MOI of 3 PFU/cell were transfected with pGE7 or pGem4Z. Three hours after transfection, the cells were incubated for 45 min in cysteine-free minimum essential medium with 10% dialyzed fetal bovine serum and then pulse-labeled with 250 μCi of [35S]cysteine (ICN Biomedicals, Inc., Irvine, Calif.) per ml for 15 min. Proteins were chased for different times with an excess (10×) of cold cysteine. At the indicated times, the culture supernatants were collected and the cells were lysed for 30 min on ice in 1 ml of 1× RIPA (50 mM Tris-HCl [pH 7.6], 150 mM NaCl: 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) and a protease inhibitor cocktail (Boehringer GmbH, Mannheim, Germany). The collected supernatants also were kept for 30 min on ice in 1× RIPA and the protease inhibitor cocktail. The cell lysates and culture supernatants were centrifuged at 10,000 × g for 10 min at 4°C. Pellets containing debris and unsolubilized proteins were discarded. Proteins were immunoprecipitated from the supernatant at 4°C first with a goat polyclonal antibody against FN (Santa Cruz Biotechnology, Santa Cruz, Calif.) and subsequently with a mixture of two murine monoclonal antibodies against E7 obtained from Zymed Laboratories Inc., San Francisco, Calif., and Santa Cruz Biotechnology. Alternatively, only E7 was immunoprecipitated from the cell lysates using the same mixture of murine monoclonal antibodies. The immune complexes, collected with a mixture of proteins A and G, were washed three times with ice-cold washing solution (50 mM Tris-HCl [pH 7.6], 500 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1% bovine serum albumin, protease inhibitor cocktail) and once with 1× RIPA. Proteins were extracted for 10 min at 95°C in 2× SDS sample buffer (1× SDS sample buffer contains 62.5 mM Tris-HCl [pH 6.8], 2% SDS, 2 mM EDTA, 5% 2-mercaptoethanol, 10% glycerol, and 0.001% bromophenol blue) and electrophoretically separated by SDS–8% PAGE (FN detection) or SDS–12% PAGE (E7 detection). Gels were fixed, dried, and exposed to PhosphorImager screens. Signals were detected with a Storm 840 PhosphorImager. There was a significant decrease in the amount of FN synthesized in the cells transfected with pGE7 compared to mock-infected cells or to cells infected with vTF-3 and transfected with pGem4Z lacking E7 (Fig. 2A). The chase pattern of the cell-associated FN in cells transfected with pGE7 was the same as that in the controls, indicating that the FN decrease was not due to a shortened half-life (Fig. 2A). Analysis of the secreted FN in the control cultures indicated that within 30 min FN appeared in the extracellular medium and reached a peak within 60 min after the pulse (Fig. 2B). In cells transfected with pGE7, a significant fraction of FN was detected in the extracellular medium, albeit approximately 30 min later that in control cells, indicating that there was no accelerated secretion (Fig. 2B). E7, which was immunoprecipitated after FN, was only found as a cell-associated protein with a half-life of approximately 90 min (Fig. 2C). No signals were detected when the immunoprecipitation was performed with preimmune goat or mouse serum (data not shown).
FIG. 2.
Pulse-chase and immunoprecipitation of FN in cells expressing WT E7. CV-1 cells infected with a recombinant vaccinia virus encoding T7 polymerase were transfected with plasmid pGE7, encoding the HPV-16 E7 protein under control of the T7 promoter (V+E7) or with empty control vector pGem4Z (V). Three hours after transfection, the cells were pulse-labeled with [35S]cysteine and chased for different times. At the indicated time, FN (A and B) and E7 (C) were sequentially immunoprecipitated from the culture supernatants (B) and cells lysates (A and C). Proteins were extracted from the immunocomplexes and subjected to SDS–8% PAGE (FN) or SDS–12% PAGE (E7). Gels were fixed, dried, and exposed to PhosphorImager screens. Signals were detected with a Storm 840 PhosphorImager. The positions of molecular mass markers are indicated on the right. MI, mock-infected cells.
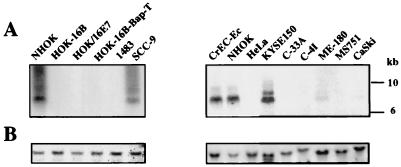
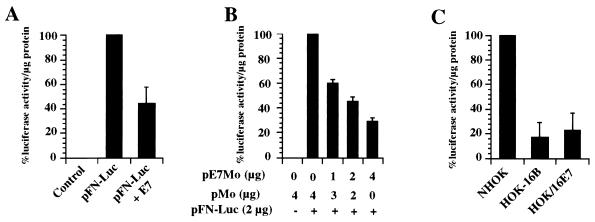
The above-described results suggested that FN downregulation by E7 occurred before its translation. Northern blot analysis showed a significant decrease in the amount of mRNA encoding FN in CV-1 cells transiently expressing E7 (data not shown). To confirm this result and rule out an artifact due to E7 overexpression, the content of endogenous FN mRNA was analyzed in a different system, in which E7 expression was more akin to natural conditions. We used a nontumorigenic immortal cell line (HOK-16B) which expressed a low level of the E7 oncoprotein (19). This cell line was established by transfection of primary NHOK with the cloned HPV-16 genome (28). The FN mRNA content was also determined in another nontumorigenic NHOK-derived cell line (HOK/16E7) which only expressed moderate levels of E7. HOK/16E7 cells were established by infecting NHOK with a retrovirus encoding the HPV-16 E7 protein under the control of the Moloney leukemia virus long terminal repeat (MLV LTR) (24). We also analyzed the FN mRNA content in seven HPV-positive and three HPV-negative tumorigenic cell lines. Total RNA was extracted from these cell lines, resolved in a 0.8% agarose-formaldehyde gel, and transferred to a nylon membrane as previously described (33). A cDNA for human FN (encompassing nucleotides 2323 to 2739) was used as a template for probe synthesis employing [α-32P]dCTP (ICN Biomedicals, Inc., Irvine, Calif.) and Prime-It RmT (Stratagene, La Jolla, Calif.). After prehybridization, hybridization, and washes, the signals were detected with a Storm 840 PhosphorImager. Strong signals corresponding to endogenous FN mRNA were only detected in NHOK, normal human cervical epithelial cells (CrEC-Ec), and the HPV-negative cell lines SCC-9 and KYSE150 (Fig. 3). Although we detected a very faint band corresponding to FN mRNA in the HPV-positives CaSki and ME-180 cells, none was detected in the other HPV-immortalized or HPV-positive tumor cells. The observed downregulation of FN mRNA in the immortalized cells expressing HPV-16 E7 or the tumor-derived HPV-positive cells could be due to an unstable FN mRNA and/or a decrease in FN promoter activity. Since E7 shares amino acid sequence similarity with portions of the AdE1A protein, which represses rat FN transcription (25, 26), we investigated whether E7 downregulates FN expression by repressing its transcription. A reporter vector encoding the luciferase protein under the control of the human FN (hFN) promoter was constructed by subcloning the hFN promoter isolated from pBShFN508 (39) into the BglII-HindIII sites of the pGL3-Control vector (Promega Corp.). The resulting construct, pFN-Luc, contained the hFN promoter, from nucleotide −508 to nucleotide +18, +1 being the transcription start site (39). The pFN-Luc construct was confirmed by DNA sequence analysis. The HPV-16 E7 protein was expressed using the construct pE7Mo, where E7 expression is under the control of the MLV LTR (12). The chloramphenicol acetyltransferase (CAT) reporter plasmid pOP13CAT (Stratagene Cloning Systems, La Jolla, Calif.) was used as a control for transfection efficiency. Subconfluent monolayers of CV-1 cells (5 × 105 cells) were cotransfected with pOP13CAT (0.5 μg), pFN-Luc (2 μg), and the empty control vector pMo (12) or pE7Mo (4 μg), and cell extracts were analyzed 48 h posttransfection (Fig. 4A). Luciferase activity was determined with the Luciferase Assay System (Promega Corp.) and a luminometer (Turner Designs, Sunnyvale, Calif.). CAT activity was assayed with [14C]chloramphenicol (ICN Biomedicals, Inc.) and the CAT Enzyme Assay System (Promega Corp.). An approximately 60% luciferase activity reduction was detected only in the cells cotransfected with pE7Mo. This reduction was dependent on the amount of pE7Mo used (Fig. 4B) and reached 95% 72 h posttransfection (see Fig. 5). E7 did not affect CAT activity (data not shown) or, as mentioned above, actin, tubulin, or collagen type IV expression, indicating that the inhibition of the hFN promoter was not due to a general effect of E7 on the transcription machinery. These results supported the conclusion that FN was repressed at the transcriptional level in cells expressing HPV-16 E7.
FIG. 3.
FN mRNA contents in cells expressing E7. Total RNA was extracted, resolved in an agarose-formaldehyde gel (15 μg of total RNA/lane), transferred to a membrane, and hybridized with a probe specific for hFN mRNA (A) or β-actin (B). Signals were detected using a PhosphorImager.
FIG. 4.
FN promoter activity in cells expressing WT E7. CV-1 cells were cotransfected with pOP13CAT as a control for transfection efficiency; pFN-Luc, encoding the luciferase protein under the control of the hFN promoter; and pE7Mo, encoding the HPV-16 E7 protein under control of the MLV LTR; or the empty control vector pMo (A and B). Forty-eight hours after transfection, the cells were lysed and assayed for luciferase and CAT activities. NHOK and HOK-16B and HOK/16E7 cells cotransfected with pOP13CAT and pFN-Luc were lysed 48 h later and assayed for luciferase and CAT activities (C). The luciferase activity expressed by pFN-Luc in the absence of pE7Mo or in NHOK was taken as 100% after normalization to CAT activity. The values shown represent the means of three different experiments, each done in duplicate. The bars represent standard deviations.
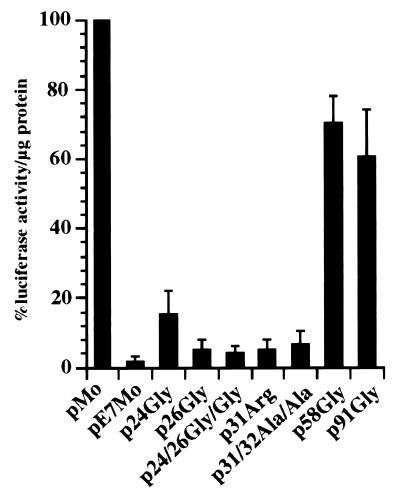
FIG. 5.
FN promoter activity in cells expressing mutant E7 proteins. CV-1 cells were cotransfected with pOP13CAT and pFN-Luc plus pMo or with pFN-Luc plus pE7Mo or pFN-Luc plus several constructs encoding mutant E7 proteins (p24Gly, p26Gly, p24/26Gly/Gly, p31Arg, p31/32Ala/Ala, p58Gly, and p91Gly). Seventy-two hours after transfection, the cells were lysed and assayed for luciferase and CAT activities. The luciferase activity expressed by pFN-Luc in the absence of pE7Mo and normalized to CAT activity was taken as 100%. The values shown represent the means of three different experiments, each done in duplicate. The bars represent standard deviations.
The activity of the hFN promoter was also analyzed in NHOK, HOK-16B, and HOK/16E7 cells cotransfected with pOP13CAT and pFN-Luc. Cell extracts were obtained 48 h later and analyzed for luciferase and CAT activities (Fig. 4C). A luciferase activity reduction of approximately 80% was detected in HOK-16B cells; as well as in HOK/16E7 cells, indicating that the repression of the hFN promoter detected with the transient-expression system was not due to E7 overexpression. Moreover, when the hFN promoter activity was assayed in two immortal nontumorigenic HPV-18-positive cell lines (36), similar results were obtained (Table 1). These two cell lines, HOK-18A and HOK-18C, which also expressed the HPV E7 gene (36), showed 60 and 90% reductions, respectively, in luciferase activity compared to NHOK.
TABLE 1.
Luciferase activity expressed by pFN-Luc in HPV-positive and -negative cells
| Cells | Origin | HPV status | % Luciferase activitya |
|---|---|---|---|
| NHOK | Normal human gingival tissue | Negative | 100 |
| HOK-16Bb | NHOK | Positive (HPV-16) | 26 ± 12 |
| HOK-18Ab | NHOK | Positive (HPV-18) | 40 ± 3 |
| HOK-18Cb | NHOK | Positive (HPV-18) | 10 ± 2 |
| HOK/16E7b | NHOK | Positive (HPV-16E7) | 30 ± 5 |
| HOK-16B-Bap-T | HOK-16B | Positive (HPV-16) | 36 ± 7 |
| CaSkic | Human epidermoid carcinoma, cervical | Positive (HPV-18) | 27 ± 1 |
| 1483d | Human squamous carcinoma, oral | Positive (HPV-16) | 2 ± 0.2 |
| SiHac | Human squamous carcinoma, cervical | Positive (HPV-16) | 39 ± 4 |
| KBc | Human epidermoid carcinoma, oral | Positive (HPV-18) | 42 ± 10 |
| HeLac | Human epithelioid carcinoma, cervical | Positive (HPV-18) | 52 ± 1 |
| KYSE150e | Human epidermoid carcinoma, esophageal | Negative | 1,080 ± 15 |
| SCC-9c | Human squamous cell carcinoma, lingual | Negative | 240 ± 15 |
| FaDuc | Human squamous cell carcinoma; pharynxgeal | Negative | 152 ± 42 |
| U-20sc | Human osteogenic sarcoma, osteal | Negative | 2,176 ± 154 |
| Saos2c | Human osteogenic sarcoma, osteal | Negative | 422 ± 54 |
| RKOf | Human carcinoma, colorectal | Negative | 290 ± 12 |
The amount of luciferase activity per microgram of protein expressed by pFN-Luc in NHOK was taken as 100% after normalization to CAT activity. The luciferase activities in the other cell lines are relative to that 100% value. The values shown represent means of two different experiments, each done in duplicate, ± the standard deviations. Luciferase and CAT activities were determined 48 h after transfection.
Human immortal nontumorigenic cells.
Cell line obtained from the American Type Culture Collection.
Cell line obtained from P. Sacks (34).
Cell line obtained from Y. Shimada (35).
Cell line obtained from M. Kastan (21).
The inhibitory effect of HPV upon the hFN promoter was further confirmed when the luciferase activity of pFN-Luc was assayed in several HPV-positive or -negative human tumor cell lines (Tables 1 and 2). A reduction in luciferase activity was detected in all nine of the HPV-positive tumor cell lines analyzed compared to NHOK or CrEC-Ec cells. This reduction varied among different cell lines, possibly reflecting dissimilar expression of the E7 protein (4, 19). On the contrary and surprisingly, some of the HPV-negative tumor cell lines analyzed showed much higher luciferase activity than normal cells. Nevertheless, FN was only detected in SCC-9 and U-20s cells (data not shown). This observation suggested that in these HPV-negative cell lines, FN downregulation may result from mechanisms other than repression of its transcription, such as alterations in the FN biosynthetic rate or in FN mRNA splicing, posttranslational modifications, or transformation-induced proteolysis (17). We cannot rule out the possibility that cellular alteration, other than that caused by E7 expression, associated with tumor development also plays a role in the observed FN downregulation in HPV-positive tumor cells.
TABLE 2.
Luciferase activity expressed by pFN-Luc in HPV-positive and -negative cells
| Cells | Origin | HPV status | % Luciferase activitya |
|---|---|---|---|
| CrEC-Ecb | Normal human cervical epithelial cells | Negative | 100 |
| C-4Ic | Human carcinoma, cervix | Positive (HPV-18) | 15 ± 2.8 |
| ME-180c | Human carcinoma, cervix | Positive (HPV-18/39) | 10.5 ± 2.1 |
| CaSkic | Human epidermoid carcinoma, cervix | Positive (HPV-18) | 50 ± 14.8 |
| MS751c | Human carcinoma, cervix | Positive (HPV-18) | 13.5 ± 2.1 |
| C-33 Ac | Human carcinoma cervix | Negative | 143.5 ± 14.8 |
| HT-3c | Human carcinoma, cervix | Negative | 16.5 ± 3.5 |
The amount of luciferase activity per microgram of protein expressed by pFN-Luc in CrEC-Ec cells was taken as 100% after normalization to CAT activity. The luciferase activities in the other cell lines are relative to that 100% value. The values shown represent the means of two different experiments, each done in duplicate, ± the standard deviations. Luciferase and CAT activities were determined 48 h after transfection.
Cells obtained from Clonetics Corp., San Diego, Calif.).
Cells obtained from the American Type Culture Collection.
E7 proteins, with mutations that partially disrupt their oncogenic potential, were tested for the ability to repress the hFN promoter. Plasmids p24Gly, p26Gly, p31Arg, p58Gly, and p91Gly encode mutant HPV-16 E7 proteins under the control of the MLV LTR (12). The plasmids encoding the mutant E7 proteins, p24/26Gly/Gly and p31/32Ala/Ala, were constructed by PCR site-directed mutagenesis of pE7Mo using mutagenic primers 5′ ACAACTGATCTCTACGGTTATGGGCAATTAAATGACAGC 3′ and 5′ GAGCAATTAATGACGCCGCAGAGGAGGAGGATGAA 3′, respectively. Plasmid p24/26Gly/Gly encodes a protein in which the 24Cys and 26Glu amino acid residues of wild-type (WT) E7 were mutated to 24Gly and 26Gly. Plasmid p31/32Ala/Ala encodes a protein in which Ser amino acid residues 31 and 32 of WT E7 were mutated to Ala. All of the constructs generated were confirmed by DNA sequence analysis. Subconfluent monolayers of CV-1 cells were cotransfected with pOP13CAT, pFN-Luc, and the constructs encoding the WT or mutant E7 proteins. Cell extracts were analyzed 72 h posttransfection for luciferase and CAT activities as described above (Fig. 5). Cells transfected with plasmid pE7Mo, which encoded the WT E7 protein, showed a 95% reduction in luciferase activity compared to cells transfected with empty control vector pMo. Cells transfected with plasmids encoding mutant E7 proteins in which the pRB-binding site was disrupted, i.e., 24Gly, 26Gly, and 24/26Gly/Gly, showed luciferase activity similar to that of cells transfected with the pE7Mo. Cells transfected with plasmids encoding mutant E7 protein 31Arg or 31/32Ala/Ala, in which the E7 proteins cannot be phosphorylated by casein kinase II, also showed less than 10% luciferase activity. On the contrary, cells transfected with the plasmids encoding E7 proteins with mutations in the Cys-X-X-Cys repeats, i.e., 58Gly and 91Gly, failed to repress the hFN promoter to the same extent as cells transfected with pE7Mo. The luciferase activity in cells transfected with p58Gly and p91Gly was, on average, approximately 70 and 60% higher, respectively, than that detected in cells transfected with a vector encoding WT E7. This suggests that both Cys-X-X-Cys repeats are involved in the repression of the hFN promoter. We cannot rule out the possibility that other domains of E7 also play a role in this phenomenon, but the Cys-X-X-Cys domains appear to be the most significant.
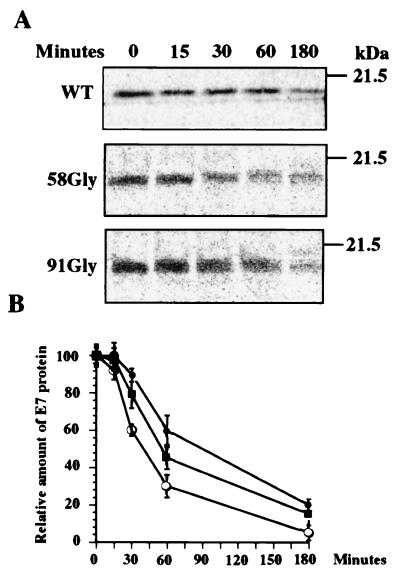
Previous reports have indicated that mutations affecting the Cys-X-X-Cys repeats caused a decrease in the half-life of E7 (23, 30, 40). This phenomenon raises the possibility that the lack of repression of the hFN promoter detected with the 58Gly and 91Gly mutants is due to a significant decrease in the steady state of these E7 proteins. To investigate this possibility, we carried out pulse-chase and immunoprecipitation experiments to determine the half-lives of the two Cys mutant proteins in CV-1 cells. CV-1 cells transfected with pMo, pE7Mo, p58Gly, or p91Gly were pulse-labeled 72 h later for 15 min and chased for different time periods, and the E7 proteins were detected by immunoprecipitation and SDS-PAGE as indicated above. Representative electrophoretic profiles of the WT and 58Gly and 91Gly mutant E7 proteins are shown in Fig. 6A. While the half-life of WT E7 in CV-1 cells was approximately 90 min, those of 58Gly E7 and 91Gly E7 were close to 40 and 60 min, respectively (Fig. 6B). Signals were not detected in the immunocomplexes obtained from CV-1 cells or CV-1 cells transfected with pMo (data not shown). The 58Gly mutant E7 protein showed a 65% decrease in its half-life compared to WT E7, while the 91Gly mutant E7 protein showed a 35% reduction in its stability. However, our results suggested that this difference in stability between WT E7 and both of the E7 Cys mutant proteins was insufficient to account for the loss of FN repression, since the two mutant proteins showed similar levels of pFN-Luc repression despite the difference in their half-lives. Although a defect in cellular localization was not detected by immunofluorescence assay when these two mutant proteins were transiently expressed using the vaccinia virus-T7 expression system (data not shown), we cannot rule out the possibility that other defects resulting from these mutations, e.g., dimerization and zinc binding, could also contribute to their failure to repress the hFN promoter.
FIG. 6.
Stability of WT and mutant E7 proteins. CV-1 cells transfected with pMo, pE7Mo, p58Gly, or p91Gly were cultured for 72 h and then incubated for 45 min in cysteine-free minimum essential medium. Cultures were pulse-labeled for 15 min with [35S]cysteine and chased for different times. At the indicated times, the E7 protein was immunoprecipitated from the cell lysates. Proteins were extracted from the immunocomplexes and subjected to SDS–12% PAGE (A). Gels were fixed, dried, and exposed to PhosphorImager screens. Signals were detected and quantified with a Storm 840 PhosphorImager. (B) E7 values determined after different times (minutes) relative to the amount of WT or mutant E7 protein detected at chase time zero. Each value represents the mean of two different experiments. Symbols: ●, WT E7; ○, 58Gly; ■, 91Gly. The bars represent standard deviations. The positions of molecular mass markers are indicated on the right of panel A.
Loss of FN on the surface of transformed cells appears to be involved in oncogenic progression (17). Transforming agents responsible for this effect include viral oncogenes, e.g., src (Rous sarcoma virus), T antigens (Simian virus 40), E1A, and E1B (adenovirus), as well as chemical carcinogens (17). Our results showed that high-risk HPV E7 was also able to downregulate FN expression by repressing its transcription. The mechanism responsible for this phenomenon may involve the Sp1 transcription factor. The adenovirus E1A protein, which shares amino acid sequence similarity with portions of E7, represses rat FN transcription. This repression is mediated by G10BP, a negative regulator of Sp1, and is abrogated by disruption of the pRB-binding domain in E1A (25–27, 38). Although the human and rat FN promoters seem to be activated by Sp1, the two promoters differ in terms of the arrangement and C content of their G-rich sequences. These differences are such that G10BP was unable to bind any of the GC boxes found in the hFN promoter (39), an indication that different factors or mechanisms may be involved in its regulation. Furthermore, as shown in this study, mutations of the pRB-binding domain in the HPV-16 E7 protein did not abrogate the hFN promoter transcription repression induced by E7 despite the fact that pRB stimulated Sp1-mediated transcription (9, 20). However, mutations affecting the E7 Cys-X-X-Cys repeats resulted in a protein unable to repress the hFN promoter. The Cys-X-X-Cys repeats play different roles in the biological properties of E7, such as transactivation (37, 40), immortalization (18), protein stability, dimerization, and zinc binding (23, 30, 32, 40). In addition, the Cys-X-X-Cys repeats can interact with pRB in the absence of high pRB affinity conserved region 2 of E7, which encompasses amino acids 20 to 30 (29). These repeats seem to be responsible for disruption of the pRB-E2F complex mediated by HPV E7, while the region encompassing only the pRB-binding domain in E1A appears to be sufficient to disrupt the same complex (15, 29). The mechanism by which pRB stimulates Sp1-mediated transactivation may involve its physical associations or, more likely, the interaction of pRB with regulators of Sp1 function (9). Since the binding of E7 to pRB affects its biological properties, it is tempting to speculate that the normal interaction between pRB and Sp-1 regulators is also affected in the presence of E7, which can lead to FN downregulation. The Cys-X-X-Cys repeats are also involved in the association of E7 with Mi2 and histone deacetylase activity (8), a phenomenon that could be involved in FN repression. For example, a defective histone deacetylase complex, resulting from its interaction with E7, could cause inappropriate derepression of the gene(s) encoding regulators of FN transcription.
This study suggested that FN downregulation precedes oncogenic transformation in HPV-positive tumors, since we found that FN was downregulated in three immortal nontumorigenic cell lines which expressed the high-risk HPV E7 protein. These cells should provide a critical tool with which to study the changes in cell adhesion, migration, differentiation, and proliferation that may occur as a result of FN repression before tumor development.
Acknowledgments
O. Rey and S. Lee contributed equally to this work.
We are very grateful to K. Vousden for the constructs pMo, pE7Mo, p24Gly, p26Gly, p31Arg, p58Gly, and p91Gly; to K. Oda for pBShFN508; and to D. Chang for the hFN cDNA. We are also very grateful to M. Kastan for the RKO cells, to P. Sacks for the 1483 cells, and to Y. Shimada for the KYSE150 cells. We thank M. A. Baluda, R. Chiu, and A. Dasgupta for helpful suggestions and critical reading of the manuscript.
This work was, in part, supported by grants DE10598, DE11229, and DE07296 from the National Institute for Dental and Craniofacial Research, National Institutes of Health, Bethesda, Md.
REFERENCES
- 1.Bablanian R. Structural and functional alterations in cultured cells infected with cytocidal viruses. In: Melnick J, editor. Progress in medical virology. 19. S. Basel, Switzerland: Karger; 1975. pp. 41–75. [PubMed] [Google Scholar]
- 2.Bablanian R, Baxt B, Sonnabend J A, Esteban M. Studies on the mechanisms of vaccinia virus cytopathic effects. II. Early cell rounding is associated with virus polypeptide synthesis. J Gen Virol. 1978;39:403–413. doi: 10.1099/0022-1317-39-3-403. [DOI] [PubMed] [Google Scholar]
- 3.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard H U, Oltersdorf T, Seedorf K. Expression of the human papillomavirus type 18 E7 gene by a cassette-vector system for the transcription and translation of open reading frames in eukaryotic cells. EMBO J. 1987;6:133–138. doi: 10.1002/j.1460-2075.1987.tb04730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernath V A, Muro A F, Vitullo A D, Bley M A, Baranao J L, Kornblihtt A R. Cyclic AMP inhibits fibronectin gene expression in a newly developed granulosa cell line by a mechanism that suppresses cAMP-responsive element-dependent transcriptional activation. J Biol Chem. 1990;265:18219–18226. [PubMed] [Google Scholar]
- 6.Blumberg P M, Driedger P E, Rossow P W. Effect of a phorbol ester on a transformation-sensitive surface protein of chick fibroblasts. Nature. 1976;264:446–447. doi: 10.1038/264446a0. [DOI] [PubMed] [Google Scholar]
- 7.Bowlus C L, McQuillan J J, Dean D C. Characterization of three different elements in the 5′-flanking region of the fibronectin gene which mediate a transcriptional response to cAMP. J Biol Chem. 1991;266:1122–1127. [PubMed] [Google Scholar]
- 8.Brehm A, Nielsen S J, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y-H, Grünwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czaja M J, Weiner F R, Eghbali M, Giambrone M A, Zern M A. Differential effects of gamma-interferon on collagen and fibronectin gene expression. J Biol Chem. 1987;262:13348–13351. [PubMed] [Google Scholar]
- 11.Dean D C, McQuillan J J, Weintraub S. Serum stimulation of fibronectin gene expression appears to result from rapid serum-induced binding of nuclear proteins to a cAMP response element. J Biol Chem. 1990;265:3522–3527. . (Erratum, 265:8346, 1990.) [PubMed] [Google Scholar]
- 12.Edmonds C, Vousden K H. A point mutational analysis of human papillomavirus type 16 E7 protein. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gahmberg C G, Hakomori S I. Altered growth behavior of malignant cells associated with changes in externally labeled glycoprotein and glycolipid. Proc Natl Acad Sci USA. 1973;70:3329–3333. doi: 10.1073/pnas.70.12.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang P S, Patrick D R, Edwards G, Goodhart P J, Huber H E, Miles L, Garsky V M, Oliff A, Heimbrook D C. Protein domains governing interactions between E2F, the retinoblastoma gene product, and human papillomavirus type 16 E7 protein. Mol Cell Biol. 1993;13:953–960. doi: 10.1128/mcb.13.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes R O. Alteration of cell-surface proteins by viral transformation and by proteolysis. Proc Natl Acad Sci USA. 1973;70:3170–3174. doi: 10.1073/pnas.70.11.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hynes R O. Fibronectins. New York, N.Y: Springer-Verlag; 1990. [Google Scholar]
- 18.Jewers R J, Hildebrandt P, Ludlow J W, Kell B, McCance D J. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J Virol. 1992;66:1329–1335. doi: 10.1128/jvi.66.3.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke L D, Adler-Storthz K, Mitchelll M F, Clayman G L, Chen Z. Expression of human papillomavirus E7 mRNA in human oral and cervical neoplasia and cell lines. Oral Oncol. 1999;35:415–420. doi: 10.1016/s1368-8375(99)00015-9. [DOI] [PubMed] [Google Scholar]
- 20.Kim S-J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam E W, Morris J D, Davies R, Crook T, Watson R J, Vousden K H. HPV16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J. 1994;13:871–878. doi: 10.1002/j.1460-2075.1994.tb06330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntyre M C, Frattini M G, Grossman S R, Laimins L A. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J Virol. 1993;67:3142–3150. doi: 10.1128/jvi.67.6.3142-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munger K, Phelps W C. The human papillomavirus E7 protein as a transforming and transactivating factor. Biochim Biophys Acta. 1993;1155:111–123. doi: 10.1016/0304-419x(93)90025-8. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima T, Nakamura T, Tsunoda S, Nakada S, Oda K. E1A-responsive elements for repression of rat fibronectin gene transcription. Mol Cell Biol. 1992;12:2837–2846. doi: 10.1128/mcb.12.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T, Nakajima T, Tsunoda S, Nakada S, Oda K, Tsurui H, Wada A. Induction of E1A-responsive negative factors for transcription of the fibronectin gene in adenovirus E1-transformed rat cells. J Virol. 1992;66:6436–6450. doi: 10.1128/jvi.66.11.6436-6450.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda E, Shirasuna K, Suzuki M, Nakano K, Nakajima T, Oda K. Cloning and characterization of a GC-box binding protein, G10BP-1, responsible for repression of the rat fibronectin gene. Mol Cell Biol. 1998;18:4772–4782. doi: 10.1128/mcb.18.8.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park N H, Min B M, Li S L, Huang M Z, Cherick H M, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–1631. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- 29.Patrick D R, Oliff A, Heimbrook D C. Identification of a novel retinoblastoma gene product binding site on human papillomavirus type 16 E7 protein. J Biol Chem. 1994;269:6842–6850. [PubMed] [Google Scholar]
- 30.Phelps W C, Münger K, Yee C L, Barnes J A, Howley P M. Structure-function analysis of the human papillomavirus type 16 E7 oncoprotein. J Virol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phelps W C, Yee C L, Munger K, Howley P M. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 32.Rawls J A, Pusztai R, Green M. Chemical synthesis of human papillomavirus type 16 E7 oncoprotein: autonomous protein domains for induction of cellular DNA synthesis and for trans activation. J Virol. 1990;64:6121–6129. doi: 10.1128/jvi.64.12.6121-6129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey O, Baluda M A, Park N-H. Differential gene expression in neoplastic and human papillomavirus-immortalized oral keratinocytes. Oncogene. 1999;18:827–831. doi: 10.1038/sj.onc.1202328. [DOI] [PubMed] [Google Scholar]
- 34.Sacks P G, Parnes S M, Gallick G E, Mansouri Z, Lichtner R, Satya-Prakash K L, Pathak S, Parsons D F. Establishment and characterization of two new squamous cell carcinoma cell lines derived from tumors of the head and neck. Cancer Res. 1988;48:2858–2866. [PubMed] [Google Scholar]
- 35.Shibata K, Tanaka S, Shiraishi T, Kitano S, Mori M. G-protein gamma 7 is downregulated in cancers and associated with p27kip1-induced growth arrest. Cancer Res. 1999;59:1096–1101. [PubMed] [Google Scholar]
- 36.Shin K H, Min B M, Cherrick H M, Park N H. Combined effects of human papillomavirus-18 and N-methyl-N′-nitro-N- nitrosoguanidine on the transformation of normal human oral keratinocytes. Mol Carcinog. 1994;9:76–86. doi: 10.1002/mc.2940090205. [DOI] [PubMed] [Google Scholar]
- 37.Storey A, Almond N, Osborn K, Crawford L. Mutations of the human papillomavirus type 16 E7 gene that affect transformation, transactivation and phosphorylation by the E7 protein. J Gen Virol. 1990;71:965–970. doi: 10.1099/0022-1317-71-4-965. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki M, Kuroda C, Oda E, Tsunoda S, Nakamura T, Nakajima T, Oda K. G10BP, and E1A-inducible negative regulator of Sp1, represses transcription of the rat fibronectin gene. Mol Cell Biol. 1995;15:5423–5433. doi: 10.1128/mcb.15.10.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki M, Oda E, Nakajima T, Sekiya S, Oda K. Induction of Sp1 in differentiating human embryonal carcinoma cells triggers transcription of the fibronectin gene. Mol Cell Biol. 1998;18:3010–3020. doi: 10.1128/mcb.18.5.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe S, Kanda T, Sato H, Furuno A, Yoshiike K. Mutational analysis of human papillomavirus type 16 E7 functions. J Virol. 1990;64:207–214. doi: 10.1128/jvi.64.1.207-214.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerfass K, Schulze A, Spitkovsky D, Friedman V, Henglein B, Jansen-Dürr P. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16 E7 through sequences necessary for transformation. J Virol. 1995;69:6389–6399. doi: 10.1128/jvi.69.10.6389-6399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.zur Hausen H. Molecular pathogenesis of cancer of the cervix and its causation by specific human papillomavirus types. Curr Top Microbiol Immunol. 1994;186:131–156. doi: 10.1007/978-3-642-78487-3_8. [DOI] [PubMed] [Google Scholar]
- 43.zur Hausen H, de Villiers E M. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]