Abstract
Since the Marburg (MBG) and Ebola (EBO) viruses have sequence homology and cause similar diseases, we hypothesized that they associate with target cells by similar mechanisms. Pseudotype viruses prepared with a luciferase-containing human immunodeficiency virus type 1 backbone and packaged by the MBG virus or the Zaire subtype EBO virus glycoproteins (GP) mediated infection of a comparable wide range of mammalian cell types, and both were inhibited by ammonium chloride. In contrast, they exhibited differential sensitivities to treatment of target cells with tunicamycin, endoglycosidase H, or protease (pronase). Therefore, while they exhibit certain functional similarities, the MBG and EBO virus GP interact with target cells by distinct processes.
The Marburg virus (MBG) and the Ebola virus (EBO) are filoviruses that have caused lethal outbreaks of hemorrhagic fever (8). Both are RNA viruses that carry host-derived envelopes and unique but related transmembrane glycoproteins (GP) that likely mediate cellular binding and fusion. The highly glycosylated GP exhibit conservation in the N- and C-terminal regions but more variability in the middle region (9, 17). Similarities in virus organization, GP structure (15), and pathogenesis suggest that MBG and EBO use similar mechanisms to enter target cells. However, since these viruses share only 31% identity in GP amino acid sequence (11) and exhibit differences in GP transcriptional processing (10), it is also conceivable that different filoviruses use alternate mechanisms to infect cells and incite disease.
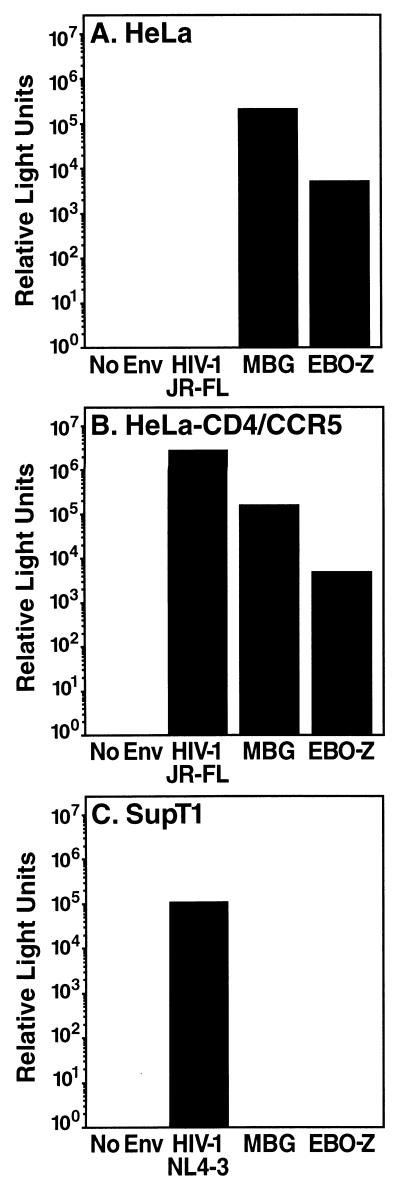
To construct a system for comparing these mechanisms of infection, genes encoding the MBG GP and the Zaire subtype EBO (EBO-Z) GP (provided by A. Sanchez, Centers for Disease Control and Prevention), cloned into the mammalian expression vector pCMV4neo (3), were incorporated into replication-incompetent pseudotype viruses carrying a human immunodeficiency virus type 1 (HIV-1) provirus NL4-3 lacking env but carrying a luciferase reporter gene (2) as previously described (1). Both MBG and EBO-Z pseudotypes infected HeLa cells to a significant degree, whereas no infection was observed by pseudovirions expressing the CCR5- and CD4-dependent envelope of HIV-1 JR-FL (provided by N. Landau, Salk Institute) (Fig. 1A). Expression of CD4 and CCR5 restored JR-FL infection with no change in the infection patterns of MBG or EBO-Z (Fig. 1B). Furthermore, no infection of the T-cell line SupT1 was observed for the MBG and EBO-Z pseudotypes, while robust infection was mediated by the envelope of CXCR4-dependent HIV-1 NL4-3 (Fig. 1C). Thus, MBG and EBO-Z GP can package HIV-1 virions and dictate distinct specificities of cellular infection.
FIG. 1.
HIV-1 pseudotypes packaged by HIV-1 (JR-FL or NL4-3), MBG, or EBO-Z GP display distinct specificities for virus entry. To determine range of infection by pseudotype viruses, HeLa cells (A), HeLa-CD4/CCR5 cells (B), or SupT1 T cells (C) were challenged with constant inocula of HIV-1 Luc+ pseudotypes. After 48 h, luciferase expression was assessed as previously described (1). Displayed values are typical of three separate infections.
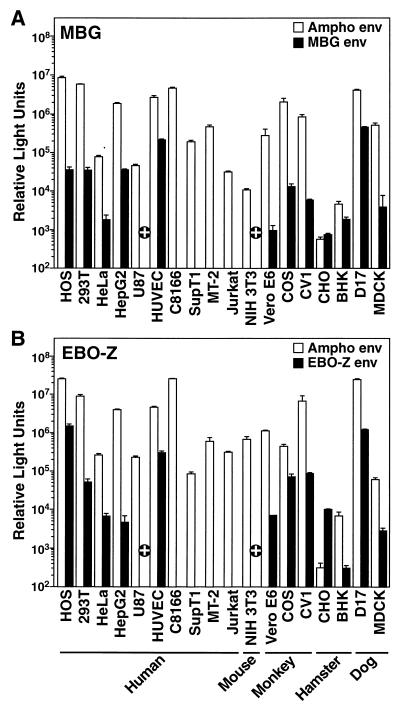
To define the cellular range of infection controlled by MBG and EBO-Z GP, a panel of mammalian cells was tested as targets for entry. The MBG pseudotype yielded variable but significant signals (up to 105 relative light units) in diverse target cells, including human osteosarcoma (HOS), 293T, HeLa, HepG2, and primary HUVEC cells (provided by A. van Zante and S. Rosen, University of California, San Francisco), as well as all adherent monkey, hamster, and dog cell lines (Fig. 2A). In contrast, human U87, murine NIH 3T3, and all suspension T-cell lines (C8166, SupT1, MT-2, and Jurkat) were nonpermissive for MBG. The hamster cell lines CHO and BHK were exceptional in supporting low levels of infection by both MBG and amphotropic Moloney leukemia virus (Ampho) (5) pseudotypes. Because the activity of the luciferase gene promoter, the HIV-1 long terminal repeat, in these pseudotype viruses is compromised in human astrocytes and murine cells, the lack of detectable infection of U87 and NIH-3T3 cells may have resulted from weak promoter function rather than failure of entry. Therefore, infections of U87, NIH-3T3, and Jurkat control cells were repeated by using MBG pseudotypes carrying an HIV-1 vector (HIV-puro) containing a puromycin resistance gene driven by the simian virus 40 promoter (provided by R. Sutton, Baylor University). After 10 days of selection of infected U87 and NIH-3T3 cells with puromycin (1 μg/ml), a significant number of antibiotic-resistant colonies survived in the MBG-infected samples, while none survived in the mock-infected cultures (data not shown). In contrast, while a significant number of Jurkat cells were viable after infection with pseudotypes carrying the vesicular stomatitis virus G protein (VSV-G) (provided by J. Burns, University of California, San Diego) and subsequent puromycin selection, no MBG-infected Jurkat cells survived. Therefore, both U87 and NIH 3T3 cells are, in fact, susceptible to entry mediated by MBG GP (Fig. 2A), and human T cells were the only cells identified as nonpermissive for MBG.
FIG. 2.
MBG and EBO-Z GP dictate similar ranges of infection in a panel of mammalian cell types. Infections with constant inocula of MBG (A) and EBO-Z (B) luciferase pseudotypes were performed for 48 h in parallel with infections by Ampho pseudotype virus to estimate cell type variability in luciferase reporter expression after virus entry. Data are the means derived from three separate infections (± standard error of the mean). +, cell line permissive to infection by pseudotype virions carrying the puromycin resistance gene driven by a simian virus 40 promoter but nonpermissive as assessed through infection by pseudovirions carrying the luciferase reporter gene driven by the HIV-1 long terminal repeat promoter.
Similarly, EBO-Z pseudotypes selectively infected various human cell lines, including HOS, 293T, HeLa, HepG2, and HUVEC, and all tested monkey, hamster, and dog cells (Fig. 2B). Like the MBG pattern, U87 and NIH 3T3 cells and all T-cell lines were nonpermissive for EBO-Z entry, as assessed by luciferase expression. However, puromycin selection studies with HIV-puro virions carrying EBO-Z GP as described above indicated that both U87 and NIH 3T3 cells were susceptible to EBO-Z pseudotypes while Jurkat controls were not (Fig. 2B). Therefore, both MBG and EBO-Z pseudotype virions display similarly broad, yet selective, ranges of infectivity.
The extensive range of infection by these viruses is consistent with other studies that have reported the in vitro tropism dictated by EBO-Z GP (18, 19), but there have been no previous reports of a similar comprehensive review for MBG GP. The broad target range correlates well with the widespread tissue necrosis after MBG and EBO infections (8). These similarities suggest that the cellular receptor(s) that mediates infection by these viruses not only is expressed in a variety of different tissues but also is highly conserved among mammalian species. Interestingly, all four suspension cell lines tested were not infectable by either virus, in agreement with previous reports regarding both EBO-Z and the Reston (EBO-R) subtypes of EBO (18, 19). Therefore, we postulated that the cellular receptor(s) mediating filovirus infection may play a role in cellular attachment and perhaps is a member of the highly conserved integrin family. However, antibody neutralization across a range of integrin complexes did not reproducibly inhibit entry by either pseudotype virus (data not shown). Nonetheless, the comparable infection profiles support the hypothesis that both filoviruses cause disease in part by infection and cytopathicity in a broad range of body tissues. Furthermore, the identification of both infectable and noninfectable cells should prove useful in combination with these pseudotype viruses to identify the cellular receptor(s) for these filoviruses.
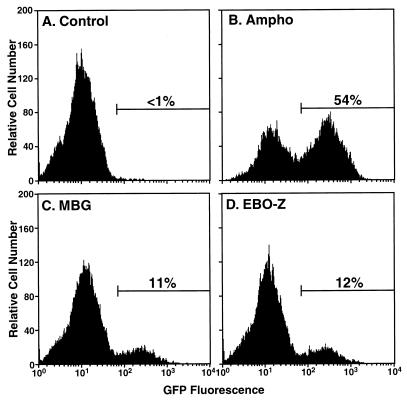
To determine the efficiency of single-round infection by MBG and EBO-Z pseudotypes, we utilized HOS cells that express a green fluorescent protein (GFP) reporter only in the presence of HIV-1 Tat protein (14) (GHOST cells), and thus after successful infection by HIV-1 pseudotype virions. While negligible basal GFP expression was observed in these cells as assessed by flow cytometry (Fig. 3A), Ampho pseudotype virus infected 54% of cells (Fig. 3B) and MBG and EBO-Z pseudotypes infected 11 and 12% of target cells, respectively (Fig. 3C and D). Since different viral stocks were used in the studies to determine range (Fig. 2) and efficiency (Fig. 3) of infection, percentages of infected cells cannot be correlated directly with luciferase activities in these two experiments. Nonetheless, both MBG and EBO-Z GP package HIV-1 genomes relatively efficiently, yielding significant and comparable titers of infectious viruses.
FIG. 3.
Efficient single-round infection of GHOST cells by pseudotypes of HIV-1 carrying Ampho, MBG, or EBO-Z GP. To determine the percentage of infected cells, GHOST cells were infected for 48 h with no virus (A) or Ampho (B), MBG (C), or EBO-Z (D) pseudotypes and analyzed for GFP expression by flow cytometry.
Previous studies have reported analogous strategies to pseudotype a murine leukemia virus vector with EBO-Z GP (18, 19) or a VSV vector with EBO-R GP (13), but no such result has been described with MBG GP. We also attempted to use murine leukemia virus for MBG or EBO-Z pseudotyping but were unable to obtain titers that supported >1% infection. Nonetheless, modest efficiency may not have precluded their use for entry studies due to the high sensitivity of the luciferase assay system. In the present study, MBG and EBO-Z pseudotypes based on the HIV-1 backbone infected >10% of target cells when the highest achievable titers were utilized. The discovery of an efficient viral packaging system for filoviruses will likely prove useful in future studies of virus infection that can be performed without a Biosafety Level 4 facility.
To identify processes critical to viral entry by MBG and EBO-Z, HeLa-CD4/CCR5 cells were subjected to chemical treatment to alter functions (e.g., receptor presentation) that may influence infection. First, cells were preincubated (2 h) and incubated during infection with ammonium chloride (3 h), a lysosomotropic reagent that prevents acidification of endosomes and vesicles. After 48 h, infection indicated by luciferase expression would be expected to be inhibited for viruses internalized by endosomes (e.g., VSV) but not for viruses gaining access through plasma membrane fusion at the cell surface (e.g., HIV-1). Infection by the HIV-1 JR-FL pseudotype did not exhibit a dose-dependent decrease compared to the untreated condition (Fig. 4A). We did note that treatment by ammonium chloride decreased infection by 30 to 40% of the untreated signal, but this dose-independent phenomenon is most likely attributable to nonspecific effects. In contrast, the VSV pseudotype demonstrated a dose-dependent and complete abrogation of infection. Likewise, infections by both the MBG and EBO-Z pseudotypes exhibited marked, dose-dependent decreases following treatment. Therefore, both MBG and EBO-Z GP mediate viral entry into target cells by a pH-dependent process. Similar findings were reported for pseudotype viruses carrying EBO-R (13) and EBO-Z (18) GP, and acidification has been implicated in an undefined aspect of the MBG replication cycle (7). Furthermore, the recently solved crystal structure of the transmembrane portion of EBO GP2 reveals structural similarities to the low-pH-induced HA2 protein that regulates influenza virus fusion (6, 16). Together, these data suggest that virus fusion by both MBG and EBO-Z likely depends on postendocytic acid-dependent conformational changes in the virus GP. Thus, our studies extend the set of diverse virus GP that appears to rely on a common final pathway for mediating entry.
FIG. 4.
Comparison of alterations in infectability of MBG and EBO-Z pseudotypes after chemical modification of target cells. Shown are treatments of HeLa-CD4/CCR5 cells with ammonium chloride (A), with tunicamycin (B), with endoglycosidase H (D), with endoglycosidase H at 4°C and in the presence of protease inhibitors (E), and with pronase protease (F) and a treatment of HOS cells with tunicamycin (C). Displayed values are means (± standard error of the mean) of luciferase activity from three separate infections (A to C, F) or are typical of two separate infections (D and E).
Second, to investigate the role of N-glycosylation of surface proteins on target cells, HeLa cells were preincubated with tunicamycin (24 h) or endoglycosidase H (2 h) in serum-free media before inoculation with the pseudotype viruses. Tunicamycin inhibits intracellular N-glycosylation of proteins, while endoglycosidase H cleaves high-mannose type N-glycosylated carbohydrate moieties at the cell surface. After inoculation (48 h), infection by MBG was not altered in cells treated with a range of tunicamycin concentrations (Fig. 4B). However, infection by EBO-Z decreased by >90% at a concentration of either 3 or 15 μg/ml. To rule out the possibility that variability in the virus titers allowed for this distinction, cells were challenged with equivalent inocula of MBG and EBO-Z pseudotypes and normalized by luciferase expression, and essentially the same pattern of inhibition was observed (data not shown). Identical profiles of inhibition were also observed in HOS cells (Fig. 4C). Similarly, infection by neither the MBG nor the VSV pseudotype was altered after pretreatment of cells (37°C for 2 h) with endoglycosidase H. In contrast, infection by the EBO-Z pseudotype decreased by >90% (Fig. 4D). Separate experiments with endoglycosidase H were performed at 4°C to prevent internalization of the enzyme and thus to ensure carbohydrate cleavage only at the cell surface. In addition, a protease inhibitor cocktail (leupeptin [10 μg/ml], pepstatin A [1 μg/ml], aprotinin [10 μg/ml], and phenylmethylsulfonyl fluoride [1 mM]) was used to ensure neutralization of undetected proteases; specific inhibition of EBO-Z was observed (Fig. 4E). These complementary experiments utilizing tunicamycin and endoglycosidase H revealed that alterations in N-glycosylation in target cells selectively impact virus entry mediated by EBO-Z GP, but not by MBG GP, and therefore suggest that MBG and EBO-Z are dependent on different cell surface moieties for cellular entry.
Third, target cells were preincubated with pronase protease to cleave surface proteins nonspecifically prior to infection (13). Although VSV infection may not utilize a protein receptor (12), infection by the VSV pseudotype decreased by 40% compared to that of the untreated culture (Fig. 4F), a previously reported nonspecific effect of pronase treatment on cells (13). Similarly, infection by MBG decreased by 40%. In contrast, entry mediated by EBO-Z GP was more significantly inhibited, decreasing by 73%. These effects of pronase were quantitatively variable across multiple experiments, but the pattern of specific inhibition of EBO-Z GP above the background effects was reproducible. Therefore, since alteration of proper protein presentation on target cells via treatment with both pronase and inhibitors of N-glycosylation suppressed EBO-Z but not MBG entry, their infection processes must not be fully identical.
Treatment of target cells with tunicamycin, endoglycosidase H, and pronase delineated potentially important distinctions between MBG and EBO-Z GP. While inhibition of infection by EBO-Z after loss of either surface proteins or N-glycosylated moieties on target cells was consistent with earlier findings regarding the EBO-R subtype (13), these treatments had little effect upon infection by MBG. It is also possible that variability in GP incorporation into MBG and EBO-Z pseudotype virions may contribute to these distinctions. However, the same pattern of infection was observed when using equivalent MBG and EBO-Z inocula normalized by luciferase expression in target cells. Furthermore, since MBG GP and EBO-Z GP were expressed at significant levels in 293T cells for virus preparations (data not shown), and both pseudotype viruses infected with similar efficiencies when used at high titers (Fig. 3), it is likely that they carry similar amounts of GP in their envelopes and that variability in pseudotype virus titers was not the cause of this distinction.
These results indicate that EBO-Z GP either interacts with a cell surface protein receptor to initiate viral entry or relies on the function of a surface protein to increase infection efficiency, as exemplified by disruption of HIV-1 infection by inhibition of LFA-1 (4). On the other hand, like MBG, infection by HIV-1 JR-FL was not inhibited by pronase (data not shown) despite the fact that entry by this virus is dependent on the binding of two protein receptors (CD4 and CCR5). Therefore, the unaffected MBG infection profiles do not exclude the possibility that MBG GP utilizes a surface protein as a receptor. Rather, these data highlight the fact that MBG and EBO-Z infections depend differentially on the presentation of target cell surface proteins, which may uniquely influence the viral life cycle and/or pathogenesis. Because only partial identity in amino acid sequence exists between MBG GP and EBO-Z GP, it is not unreasonable to expect functional differences in entry requirements to have evolved. Future investigations aimed at identifying the cellular receptor(s) for these filoviruses are necessary to characterize these mechanisms definitively.
ACKNOWLEDGMENTS
We thank J. Burns, D. Kabat, N. Landau, D. Littman, K. Page, S. Rosen, A. Sanchez, R. Sutton, and A. van Zante for kindly providing reagents, J. Carroll and N. Shea for preparation of the figures, and H. Livesay for administrative assistance. We also thank O. Keppler and D. Sheppard for scientific advice.
This work was supported by the J. David Gladstone Institutes (M.A.G.). S.Y.C. was supported by the NIH Medical Scientist Training Program and the UCSF Biomedical Sciences Graduate Program, and R.F.S. was supported by the UCSF-Gladstone Center for AIDS Research.
REFERENCES
- 1.Chan S, Speck R, Power C, Gaffen S, Chesebro B, Goldsmith M. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connor R, Chen B, Choe S, Landau N. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 3.Goldsmith M, Xu W, Amaral M, Kuczek E, Greene W. The cytoplasmic domain of the interleukin-2 receptor beta chain contains both unique and functionally redundant signal transduction elements. J Biol Chem. 1994;269:14698–14704. [PubMed] [Google Scholar]
- 4.Hildreth J, Orentas R. Involvement of a leukocyte adhesion receptor (LFA-1) in HIV-induced syncytium formation. Science. 1989;244:1075–1078. doi: 10.1126/science.2543075. [DOI] [PubMed] [Google Scholar]
- 5.Landau N, Page K, Littman D. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malashkevich V, Schneider B, McNally M, Milhollen M, Pang J, Kim P. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mar'iankova R, Glushakova S, Pyzhik E, Lukashevich I. The penetration of the Marburg virus into eukaryotic cells. Vopr Virusol. 1993;38:74–76. [PubMed] [Google Scholar]
- 8.Peters C, Sanchez A, Rollin P, Ksiazek T, Murphy F. Filoviridae: Marburg and Ebola viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1161–1176. [Google Scholar]
- 9.Sanchez A, Kiley M, Holloway B, Auperin D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez A, Trappier S, Mahy B, Peters C, Nichol S. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez A, Trappier S, Stroher U, Nichol S, Bowen M, Feldmann H. Variation in the glycoprotein and VP35 genes of Marburg virus strains. Virology. 1998;240:138–146. doi: 10.1006/viro.1997.8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlegel R, Tralka T, Willingham M, Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell. 1983;32:639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- 13.Takada A, Robison C, Goto H, Sanchez A, Murti K, Whitt M, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trkola A, Ketas T, Kewalramani V, Endorf F, Binley J, Katinger H, Robinson J, Littman D, Moore J. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volchkov V, Feldmann H, Volchkova V, Klenk H. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissenhorn W, Carfi A, Lee K, Skehel J, Wiley D. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 17.Will C, Muhlberger E, Linder D, Slenczka W, Klenk H, Feldmann H. Marburg virus gene 4 encodes the virion membrane protein, a type I transmembrane glycoprotein. J Virol. 1993;67:1203–1210. doi: 10.1128/jvi.67.3.1203-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wool-Lewis R, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Delgado R, Xu L, Todd R, Nabel E, Sanchez A, Nabel G. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]