Abstract
Background
Prior research exploring the correlation between the XRCC3 Thr241Met polymorphism and the susceptibility to pancreatic cancer has yielded conflicting outcomes. To date, there has been a notable absence of studies examining this polymorphism. The primary aim of the current investigation is to elucidate the potential role of the XRCC3 Thr241Met polymorphism as a risk factor in the development of pancreatic cancer.
Methods
The comprehensive literature search was meticulously conducted across primary databases, including PubMed, Embase, and CNKI (China National Knowledge Infrastructure), spanning from the inception of each database through January 2024. To synthesize the data, a meta-analysis was performed using either a fixed or random-effects model, as appropriate, to calculate the odds ratios (ORs) and their corresponding 95% confidence intervals (CIs).
Results
The analysis revealed significant associations between the XRCC3 Thr241Met polymorphism and an increased risk of pancreatic cancer. This was evidenced through various genetic model comparisons: allele contrast (T vs. C: OR = 0.77, 95% CI = 0.70–0.86, P < 0.001), homozygote comparison (TT vs. CC: OR = 0.71, 95% CI = 0.58–0.88, P = 0.001), heterozygote comparison (TC vs. CC: OR = 0.67, 95% CI = 0.52–0.87, P = 0.003), and a dominant genetic model (TT/TC vs. CC: OR = 0.68, 95% CI = 0.57–0.81, P < 0.001). Additionally, subgroup analyses based on ethnicity disclosed that these associations were particularly pronounced in the Caucasian population, with all genetic models showing significance (P < 0.05).
Conclusions
The XRCC3 Thr241Met polymorphism has been identified as contributing to a reduced risk of pancreatic cancer in the Caucasian population. This finding underscores the need for further research to validate and expand upon our conclusions, emphasizing the urgency for continued investigations in this domain.
Keywords: Pancreatic cancer, XRCC3, Gene polymorphism, Meta-analysis
Introduction
Pancreatic cancer ranks as one of the most lethal malignancies globally, currently holding the fourth position in cancer-related fatalities [1]. In 2012, there were approximately 178,000 new cases reported [2]. Forecasts suggest that by 2030, pancreatic cancer may become the second leading cause of death from malignant tumors [3, 4]. The incidence and mortality rates of pancreatic cancer exhibit considerable variation across different regions. A study encompassing 54 countries and regions from 1980 to 2007 revealed the highest mortality rates in Northern Europe and the Baltic Sea region, averaging 9.5 per 100,000, with Lithuania reporting the highest at 11.1 per 100,000. Conversely, lower mortality rates were observed in Hong Kong, Japan, Latin America, the United States, and Russia, with Venezuela having the lowest rate in Latin America at 2.9 per 100,000.
The insidious onset of pancreatic cancer, coupled with its nonspecific early symptoms, poses significant diagnostic challenges. It is often misdiagnosed as gastroduodenal ulcers, diabetes, or other conditions. Consequently, most patients are diagnosed at an advanced stage. The low rate of surgical resection, coupled with limited treatment options and negligible efficacy, results in a dismal five-year survival rate of less than 6% [5], posing a grave threat to public health. Understanding the factors related to the development of pancreatic cancer is therefore crucial for reducing its incidence and mortality.
The exact pathogenesis of pancreatic cancer remains elusive, and there is a lack of effective screening and early diagnosis techniques. Pancreatic cancer arises from a complex interplay of genetic and environmental factors. During the development of pancreatic cancer, mutations in repair genes alter their function and expression levels, triggering mutations in oncogenes and tumor suppressor genes that regulate the disease. This accumulation of changes eventually leads to pancreatic cancer.
XRCC3 is a DNA repair gene primarily involved in the repair of double-strand DNA breaks. Its deletion or mutation significantly increases susceptibility to DNA damage factors, contributing to the onset of malignant tumors like lung cancer. The C to T mutation at nucleoside acid 18,067 in exon of XRCC3 results in a codon change at position 241 from threonine (Thr) to methionine (Met), disrupting the protein’s normal conformation and potentially enhancing susceptibility to certain tumors. The XRCC3 Thr241Met gene polymorphism has been linked to pancreatic cancer development. However, due to variations in study populations, sample sizes, genetic backgrounds, and environmental exposures, the results of independent case-control studies have been inconsistent. Therefore, this study aims to apply evidence-based medicine principles and methods to conduct a comprehensive analysis of all published studies on the relationship between XRCC3 Thr241Met polymorphism and genetic susceptibility to pancreatic cancer.
Materials and methods
Data Collection
Using a variety of well-known databases, including PubMed, EMBASE, the Cochrane Library, Google Scholar, and CNKI (China National Knowledge Infrastructure), two separate writers carried out extensive searches. The search period ran from each database’s creation until January 2024. The terms “pancreatic cancer,” “polymorphism” or “polymorphisms,” and “XRCC3” were used to deliberately narrow the search.
Inclusion and exclusion criteria
The following exacting standards were established for inclusion in this analysis: Studies that explicitly evaluate the relationship between the XRCC3 Thr241Met polymorphism and pancreatic cancer fall into two categories: (a) case-control studies; and (b) studies with enough information to compute odds ratios (ORs) and their 95% confidence intervals (CIs). On the other hand, the exclusion criteria were similarly strict and included: (a) studies that did not follow the case-control design; (b) studies that did not have enough data to calculate the OR and 95% CI; and (c) studies that used animals as experimental subjects.
Data extraction and methodological quality assessment
Following strict adherence to the predetermined inclusion and exclusion criteria, the first and second authors carefully went through and assessed all available data and information. When the first and second writers couldn’t agree on anything, they discussed it and came to a mutually agreed-upon decision by consulting the corresponding author. Three primary components comprised the main assessment criteria: the evaluation of exposure outcomes and variables (0–3 points); comparability between groups (0–2 points); and the selection of cases and controls (0–4 points). References 6–18 provide transparent methods and procedures used in this review that are cross-referenceable with previously published material [6–18].
Statistical analysis
With the aid of odds ratios (ORs) and 95% confidence intervals (CIs), the degree of correlation between the XRCC3 Thr241Met polymorphism and the risk of pancreatic cancer was statistically determined. Both the Q-statistic and I2 statistics were used to assess the level of heterogeneity among the studies. Four genetic models were used to investigate this relationship, in line with other research. The degree of observed heterogeneity dictated which model to use: a fixed-effects model or a random-effects model [19, 20]. Sensitivity analyses and publication bias evaluations were carried out according to the protocols set forth in earlier meta-analyses [13–18]. The software Stata 15.0 was used for all statistical analyses. According to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2009 standards, this meta-analysis was carried out meticulously and reported.
Results
General information
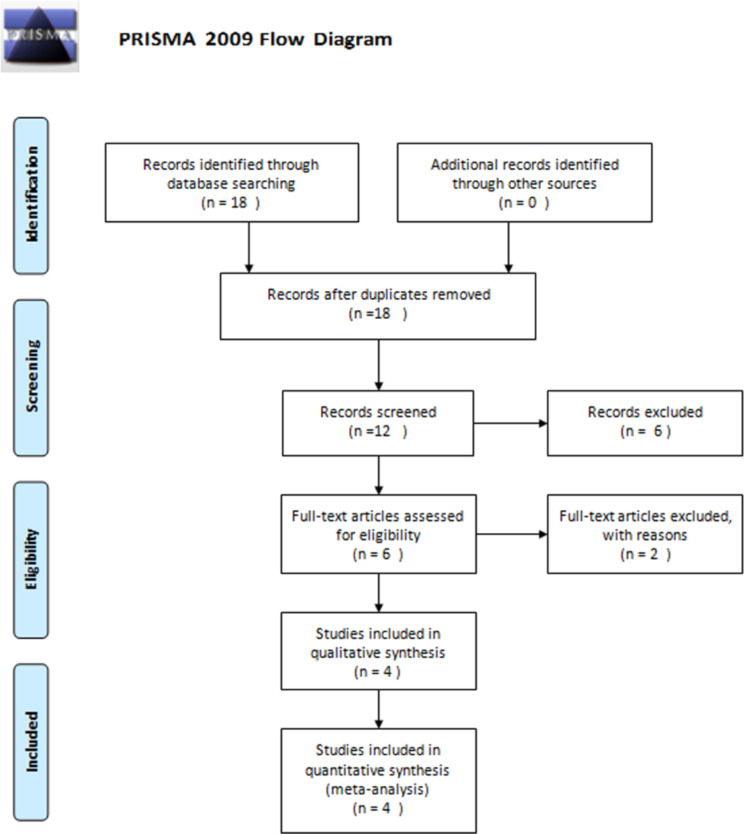
The search methodology for this meta-analysis is outlined in the PRISMA 2009 Flow Diagram (Fig. 1). Four studies in all were eventually included. A thorough summary of the most important facts and data from these investigations is shown in Table 1. Geographically, two studies from the United States, one each from France and Poland made up the included literature. Numerous genotyping techniques, such as the Masscode methodology, allele-specific assays, and Taqman assays, were used in these investigations. These studies were published between the years of 2006 and 2016, and the control groups were hospital or population-based. Every control group’s genotype frequency was in line with Hardy-Weinberg Equilibrium (HWE). These studies’ sample sizes ranged from 33 to 1120 people. The Newcastle-Ottawa Scale (NOS) scores are shown in Table 2, with an average score of 7.9 indicating a high quality across all research.
Fig. 1.
PRISMA 2009 Flow Diagram
Table 1.
General information of eligible studies enrolled in the meta-analysis
| Literature | Ethnics(Country) | Genotyping method | Control Origin | Sample capacity | Matching standard | HWE conformity | NOS |
|---|---|---|---|---|---|---|---|
| Donghui(2006a) | Caucasian(United states) | Masscode technique | HB | 332/436 | Age, sex, ethnicity | Yes | 9 |
| Donghui(2006b) | Hispanics(United states) | Masscode technique | HB | 22/20 | Age, sex, ethnicity | Yes | 8 |
| Donghui(2006c) | African(United states) | Masscode technique | HB | 19/25 | Age, sex, ethnicity | Yes | 8 |
| Jiao(2008a) | Caucasian (United states) | allele specific assay | HB | 416/436 | Age, sex, ethnicity | Yes | 9 |
| Jiao(2008b) | Hispanics (United states) | allele specific assay | HB | 26/20 | Age, sex, ethnicity | Yes | 8 |
| Jiao(2008c) | African (United states) | allele specific assay | HB | 17/16 | Age, sex, ethnicity | Yes | 8 |
| Duell(2008a) | Caucasian(France) | Taqman assay | PB | 260/860 | Age, sex, ethnicity | Yes | 8 |
| Duell(2008b) | African (France) | Taqman assay | PB | 48/104 | Age, sex, ethnicity | Yes | 7 |
| Renata(2016) | Caucasian(Poland) | Taqman assay | HB | 101/103 | Age, sex, ethnicity | Yes | 8 |
PB: Population-based; HB: Hospital-based; HWE: Hardy-Weinberg equilibrium; NOS: Newcastle-Ottawa Scale
Table 2.
Quality assessment of the seven case–control studies according to the Newcastle-Ottawa Scale
| Literature | selection of enrolled study subjects | between-group comparability | exposure outcomes and factors | Total |
|---|---|---|---|---|
| Donghui(2006a) | 3 | 3 | 3 | 9 |
| Donghui(2006b) | 3 | 2 | 2 | 8 |
| Donghui(2006c) | 3 | 3 | 2 | 8 |
| Jiao(2008a) | 3 | 3 | 3 | 9 |
| Jiao(2008b) | 3 | 2 | 3 | 8 |
| Jiao(2008c) | 3 | 3 | 2 | 8 |
| Duell(2008a) | 3 | 3 | 2 | 8 |
| Duell(2008b) | 2 | 3 | 2 | 7 |
| Renata(2016) | 3 | 3 | 2 | 8 |
| Average | 2.9 | 2.7 | 2.3 | 7.9 |
Allele and genotype-wide meta-analysis
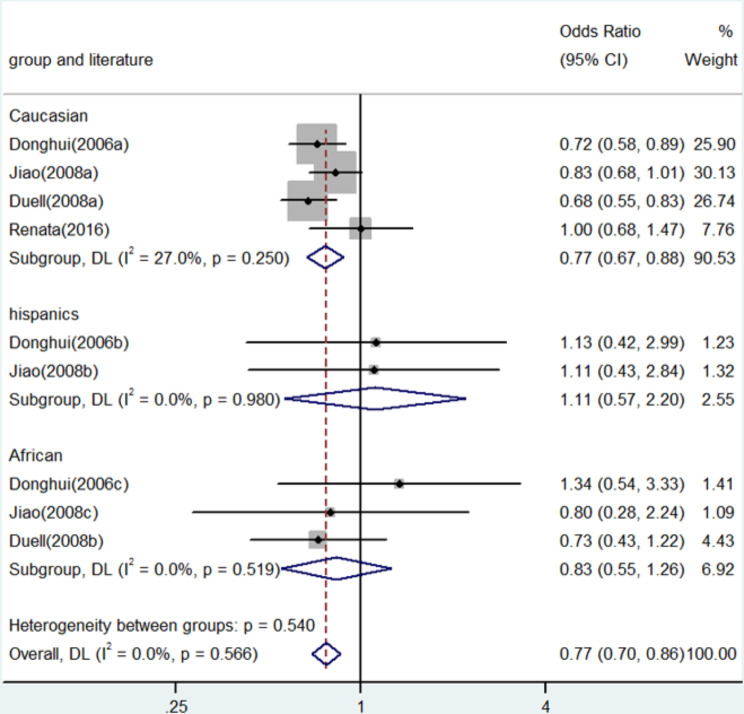
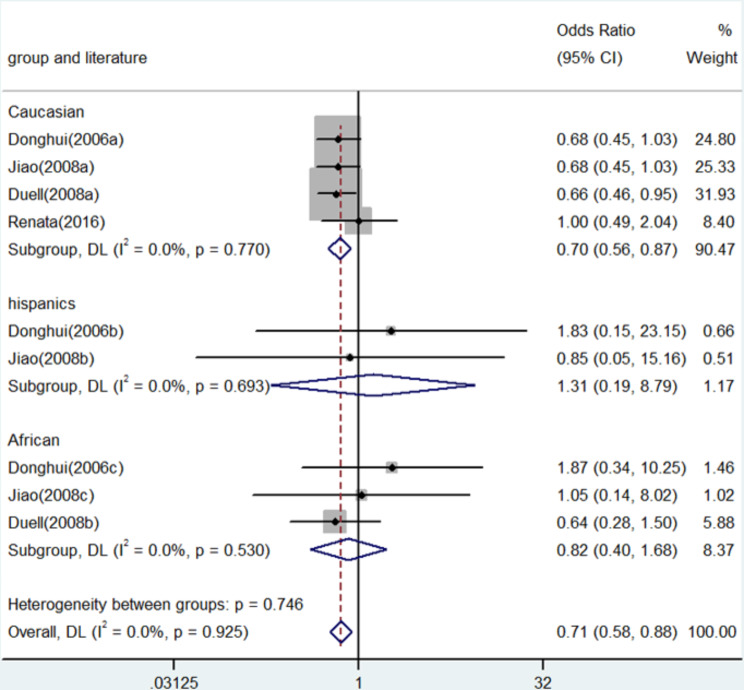
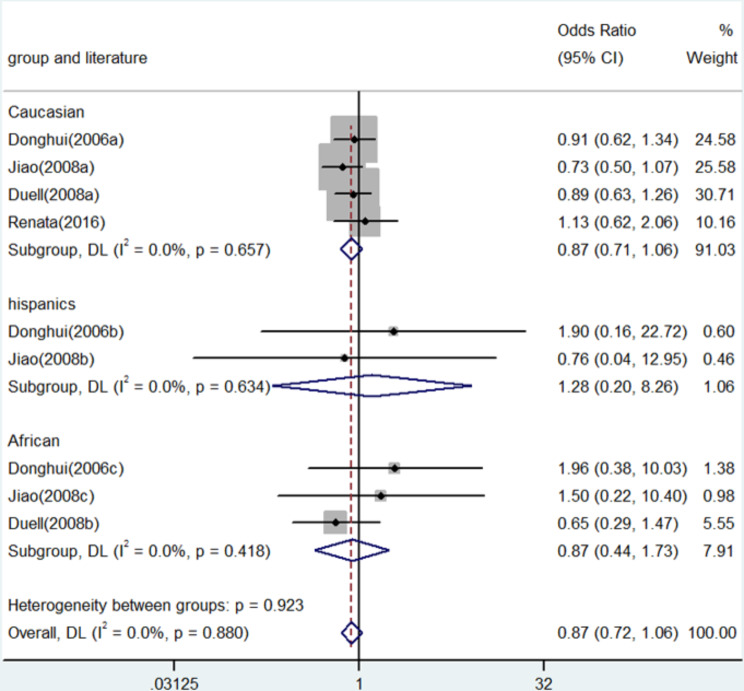
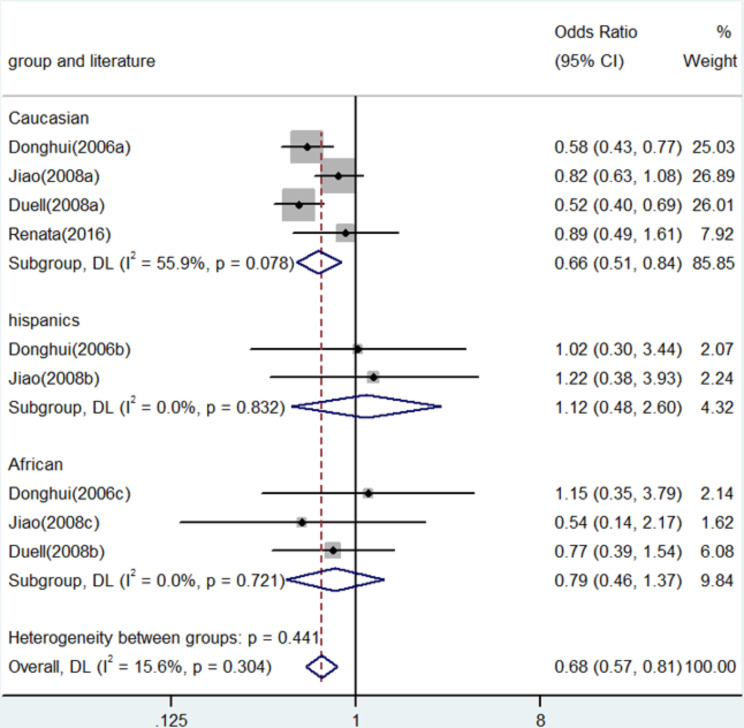
In numerous genetic studies, a strong correlation was found between the XRCC3 Thr241Met polymorphism and the risk of pancreatic cancer. To be more precise, homozygote comparison (TT vs. CC) demonstrated an OR of 0.71 (95% CI: 0.58–0.88, P = 0.001, Fig. 2), whereas allele contrast (T vs. C) revealed an OR of 0.77 (95% CI: 0.70–0.86, P < 0.001, Fig. 3). An OR of 0.67 (95% CI: 0.52–0.87, P = 0.003) was shown in the heterozygote comparison (TC vs. CC) in Fig. 4. Figure 5 show that the OR in the recessive genetic model (TT vs. CC/TC) was 0.87 (95% CI: 0.72–1.06, P = 0.167). Additionally, Fig. 6 showed an OR of 0.68 (95% CI: 0.57–0.81, P < 0.001) for the dominant genetic model (TT/TC vs. CC). A full summary of the main findings on the relationship between pancreatic cancer risk and the XRCC3 Thr241Met polymorphism was shown in Table 3
Fig. 2.
Forest plot for the associations between XRCC3 Thr241Met polymorphism and pancreatic cancer risk through homozygote comparison (TT vs. CC). OR, odds ratio; CI, confidence interval
Fig. 3.
Forest plot for the associations between XRCC3 Thr241Met polymorphism and pancreatic cancer risk through allele contrast (T vs. C). OR, odds ratio; CI, confidence interval
Fig. 4.
Forest plot for the associations between XRCC3 Thr241Met polymorphism and pancreatic cancer risk through heterozygosis comparison (TC vs. CC). OR, odds ratio; CI, confidence interval
Fig. 5.
Forest plot for the associations between XRCC3 Thr241Met polymorphism and pancreatic cancer risk through recessive genetic model (TT vs. TC/CC). OR, odds ratio; CI, confidence interval
Fig. 6.
Forest plot for the associations between XRCC3 Thr241Met polymorphism and pancreatic cancer risk through dominate genetic model (TC/TT vs. CC). OR, odds ratio; CI, confidence interval
Table 3.
Meta-analysis of the XRCC3 C18067T polymorphism and pancreatic cancer risk
| Comparison | Population | N | Test of association | Mode | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | χ2 | P | I2 | ||||
| T versus. C | Overall | 9 | 0.77 | 0.70-0.86 | 0 | Fixed | 6.73 | 0.566 | 0 |
| Caucasian | 4 | 0.77 | 0.67-0.88 | 0 | Fixed | 4.11 | 0.250 | 27.0 | |
| Hispanics | 2 | 1.11 | 0.57-2.20 | 0.753 | Fixed | 0 | 0.980 | 0 | |
| African | 3 | 0.83 | 0.55-1.26 | 0.506 | Fixed | 1.31 | 0.519 | 0 | |
| TT versus. CC | Overall | 9 | 0.71 | 0.58-0.88 | 0.001 | Fixed | 3.14 | 0.925 | 0 |
| Caucasian | 4 | 0.70 | 0.56-0.87 | 0.001 | Fixed | 1.13 | 0.770 | 0 | |
| Hispanics | 2 | 1.31 | 0.19-8.79 | 0.782 | Fixed | 0.16 | 0.693 | 0 | |
| African | 3 | 0.82 | 0.40 -1.68 | 0.590 | Fixed | 1.27 | 0.530 | 0 | |
| TC versus. CC | Overall | 9 | 0.67 | 0.52-0.87 | 0.003 | Random | 13.43 | 0.098 | 40.5 |
| Caucasian | 4 | 0.63 | 0.44-0.89 | 0.008 | Random | 10.47 | 0.015 | 71.4 | |
| Hispanics | 2 | 1.09 | 0.45-2.61 | 0.848 | Fixed | 0.13 | 0.716 | 0 | |
| African | 3 | 0.80 | 0.41-1.57 | 0.517 | Fixed | 0.94 | 0.624 | 0 | |
| TT versus. TC/CC | Overall | 9 | 0.87 | 0.72-1.06 | 0.167 | Fixed | 3.74 | 0.923 | 0 |
| Caucasian | 4 | 0.87 | 0.71-1.06 | 0.173 | Fixed | 1.61 | 0.657 | 0 | |
| Hispanics | 2 | 1.28 | 0.20-8.26 | 0.797 | Fixed | 0.34 | 0.634 | 0 | |
| African | 3 | 0.87 | 0.44-1.73 | 0.697 | Fixed | 1.74 | 0.418 | 0 | |
| TT/TC versus. CC | Overall | 9 | 0.68 | 0.57-0.81 | 0 | Fixed | 9.48 | 0.441 | 15.6 |
| Caucasian | 4 | 0.66 | 0.51-0.84 | 0.001 | Random | 6.80 | 0.078 | 55.9 | |
| Hispanics | 2 | 1.12 | 0.48-2.60 | 0.793 | Fixed | 0.04 | 0.832 | 0 | |
| African | 3 | 0.79 | 0.46-1.37 | 0.409 | Random | 0.65 | 0.721 | 81.7 | |
OR, odds ratio; CI, confidence interval
Evaluation of between-study heterogeneity
Overall, the analysis revealed minimal heterogeneity across most genetic models. Notable exceptions were observed in the heterozygote comparison (χ² = 10.47, P = 0.015, I² = 71.4, Table 3) and the dominant genetic model (χ² = 6.80, P = 0.078, I² = 55.9, Table 3), where significant heterogeneity was detected. Meta-regression analysis pinpointed sample size as a critical factor contributing to this heterogeneity. Specifically, the study conducted by Renata et al. introduced substantial heterogeneity, attributable primarily to its relatively small sample size.
Subgroup analysis and publication bias
Subgroup analysis was meticulously performed based on ethnicity, revealing significant findings particularly within the Caucasian population. Comprehensive details of these findings are available in Table 3. The funnel plot analysis showed no apparent asymmetry, and the reliability of these results was further supported by Egger’s test, which yielded a non-significant bias (P = 0.560).
Discussion
The etiology of pancreatic cancer remains a topic of ongoing research, with current consensus suggesting a multifactorial influence spanning environmental, hereditary, demographic, lifestyle, genetic, and psychosocial factors. Environmental elements, particularly lifestyle choices such as smoking, exposure to secondhand smoke, and excessive alcohol consumption, are notably impactful in the incidence and progression of pancreatic cancer.
Moreover, psychological and demographic factors are acknowledged to modulate the risk of pancreatic cancer. The varying susceptibility within populations, despite similar environmental risk exposures, underscores the significance of genetic predisposition in the disease’s onset and progression. The role of genetic factors, especially at the single nucleotide polymorphism (SNP) level, is increasingly recognized in the pathogenesis of pancreatic cancer. Susceptibility genes of interest primarily include oncogenes, tumor suppressor genes, and DNA repair genes, with their polymorphisms accounting for individual variations in disease response and progression due to environmental factors.
XRCC3, a crucial DNA repair gene, functions as a molecular scaffold, facilitating single-strand break repair and base excision repair by binding to repair-related proteins. Epidemiological data increasingly suggest a link between repair gene polymorphisms and cancer risk, with XRCC3 implicated in various malignancies, including lymphoma, lung, esophageal, salivary gland, colorectal, cervical, breast, and stomach cancers [21–28]. Our findings indicate a decreased risk in Caucasian populations, marking this as potentially the first meta-analysis to focus on this aspect. However, the current results indicate that XRCC3 Thr241Met polymorphism is not associated with the risk of pancreatic cancer in Hispanics population or African population. We think the different results from different population are not surprising, because the results of genetic polymorphism are influenced by many aspects such as geography, ethnicity and environment. And even within the same continent or within the same country, there can still be racial differences. China, Iran, India, Saudi Arabia and other countries are all in Asia, but there are obvious differences in their race. Furthermore, even if different studies study the same race, they may get different results due to the difference in sample size, which is also the reason for our meta-analysis. By conducting Between-Study Heterogeneity and meta-regression, we successfully identified the source of heterogeneity and reduced the impact of heterogeneity to a very low level through subgroup analysis, thus ensuring the reliability of the meta-analysis results. Finally, the sensitivity analysis and publication bias also show that our results are very convincing.
However, several limitations warrant consideration. Primarily, the inclusion of more diverse studies, particularly from Asian, African, and other ethnic groups, would enhance the meta-analysis. Additionally, the influence of various confounders could not be fully assessed due to insufficient data, and the overall participant number in our study is relatively small.
In summary, our findings suggest that the XRCC3 Thr241Met polymorphism is associated with a reduced risk of pancreatic cancer in Caucasian populations. Further research is imperative to corroborate these findings.
Data sharing statement
All data generated or analyzed during this study are included in this published article.
Acknowledgements
We sincerely acknowledge the staff in Clinical Laboratories for their support.
Author contributions
Mangmang He and Tianyu Liang conceived study design and conceived the content concept; Wenjing Wu and Sen Xu performed the data collection, extraction and analyzed the data. Lingzhi Chen and Chaomin Ji interpreted and reviewed the data and drafts. All authors were involved in literature search, writing the paper and had final approval of the submitted and published versions. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethical approval
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Competing interests
The authors declare no competing interests.
Disclosure
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenjing Wu and Sen Xu contributed equally to this work.
Contributor Information
Tianyu Liang, Email: Liangtianyu1111@163.com.
Mangmang He, Email: hmmzjsrmyy0121@163.com.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Davis FG, Dolecek TA, McCarthy BJ, Villano JL. Toward determining the lifetime occurrence of metastatic brain tumors estimated from 2007 United States cancer incidence data. Neuro Oncol. 2012;14:1171–7. doi: 10.1093/neuonc/nos152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Yang H, Wang SY, Ou W, Sun HB, Fang Q. [Clinical characteristics and prognosis of very young patients with breast cancer in the southern of China] Ai Zheng. 2009;28:1310–6. doi: 10.5732/cjc.009.10230. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Wang Z, Yan Y, et al. XRCC3 C18067T polymorphism contributes a decreased risk to both basal cell carcinoma and squamous cell carcinoma: evidence from a meta-analysis. PLoS ONE. 2014;9:e84195. doi: 10.1371/journal.pone.0084195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin LY, Chen X, Li P, Yang Z, Mo WN. Association between the XRCC3 Thr241Met polymorphism and cervical cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2014;14:6703–7. doi: 10.7314/APJCP.2013.14.11.6703. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Chen X, Liu B, et al. Quantitative assessment of the associations between DNA repair gene XRCC3 Thr241Met polymorphism and gastric cancer. Tumour Biol. 2014;35:1589–98. doi: 10.1007/s13277-013-1219-8. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, Chen X, Li T, Li M, Liang H. Association of OGG1 Ser326Cys polymorphism and pancreatic cancer susceptibility: evidence from a meta-analysis. Tumour Biol. 2014;35:2397–402. doi: 10.1007/s13277-013-1317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Yan Y, Li P, et al. Association of GSTP1 -313A/G polymorphisms and endometriosis risk: a meta-analysis of case-control studies. Eur J Obstet Gynecol Reprod Biol. 2013;171:362–7. doi: 10.1016/j.ejogrb.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Si D, Yao Y, Chen X, Qiu J. Ethnicity-stratified analysis of the association between P53 rs1042522 polymorphism and women HPV infection: a meta-analysis. Microb Pathog. 2021;161:105099. doi: 10.1016/j.micpath.2021.105099. [DOI] [PubMed] [Google Scholar]
- 12.Niu K, Chen X, Lu Y. COL3A1 rs1800255 polymorphism is associated with pelvic organ prolapse susceptibility in caucasian individuals: evidence from a meta-analysis. PLoS ONE. 2021;16:e0250943. doi: 10.1371/journal.pone.0250943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin X, Wu Y, Yin S, Chen X, Zhang Y. Association between the IL-10 and IL-6 polymorphisms and brucellosis susceptibility: a meta-analysis. BMC Med Genet. 2020;21:63. doi: 10.1186/s12881-020-01006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin X, Yin S, Zhang Y, Chen X. Association between TLR2 Arg677Trp polymorphism and tuberculosis susceptibility: a meta-analysis. Microb Pathog. 2020;144:104173. doi: 10.1016/j.micpath.2020.104173. [DOI] [PubMed] [Google Scholar]
- 15.Yuanyuan G, Xue Y, Yachao L, Xiao F, Xu C. Association between IL-18 -607 C/A polymorphism and the risk of prostate Cancer: a Meta-analysis of case-control studies. Asian Pac J Cancer Prev. 2019;20:1595–602. doi: 10.31557/APJCP.2019.20.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin X, Yin S, Zhang Y, Chen X. Quantitative assessment of the association between IL-10 -592 A/C polymorphism and Kawasaki disease risk in Chinese population: evidence from a meta-analysis. Cardiol Young. 2018;28:811–15. doi: 10.1017/S1047951118000380. [DOI] [PubMed] [Google Scholar]
- 17.Jin X, Yin S, Zhang Y, Chen X. Association between TLR2 + 2477G/A polymorphism and bacterial meningitis: a meta-analysis. Epidemiol Infect. 2018;146:642–47. doi: 10.1017/S0950268818000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Jiang M, Zhao RK, Gu GH. Quantitative Assessment of the Association between ABC Polymorphisms and Osteosarcoma Response: a Meta-analysis. Asian Pac J Cancer Prev. 2015;16:4659–64. doi: 10.7314/APJCP.2015.16.11.4659. [DOI] [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 20.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 21.Cheng S, Wang L, Wang L, Wang Z. Association of XRCC3 gene rs861539 polymorphism with gastric cancer risk: evidence from a case-control study and a meta-analysis. Int J Clin Exp Pathol. 2015;8:1911–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, You HH, Jia YJ, Guo JD, Du HL. Association of C722T polymorphism in XRCC3 gene with larynx cancer: a meta-analysis. Tumour Biol. 2014;35:5427–30. doi: 10.1007/s13277-014-1707-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Zhong Z, Zhang X, et al. DNA repair gene XRCC3 T241M polymorphism and bladder cancer risk in a Chinese population. Genet Test Mol Biomarkers. 2012;16:640–3. doi: 10.1089/gtmb.2011.0334. [DOI] [PubMed] [Google Scholar]
- 24.Hu S, Jing Y, Liu F, Han F. Association between XRCC3 rs861539 Polymorphism and the Risk of Ovarian Cancer: Meta-Analysis and Trial Sequential Analysis. Biomed Res Int. 2022; 2022:3915402. [DOI] [PMC free article] [PubMed]
- 25.Sarwar R, Mahjabeen I, Bashir K, Saeed S, Kayani MA. Haplotype based analysis of XRCC3 gene polymorphisms in thyroid Cancer. Cell Physiol Biochem. 2017;42:22–33. doi: 10.1159/000477109. [DOI] [PubMed] [Google Scholar]
- 26.Bashir H, Majid S, Hamid R, et al. Polymorphism of the XRCC3 gene and risk of gastric cancer in a Kashmiri population: a case-control study. Eur J Cancer Prev. 2015;24:167–75. doi: 10.1097/CEJ.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 27.Peng Q, Mo C, Tang W, et al. DNA repair gene XRCC3 polymorphisms and bladder cancer risk: a meta-analysis. Tumour Biol. 2014;35:1933–44. doi: 10.1007/s13277-013-1259-0. [DOI] [PubMed] [Google Scholar]
- 28.Fan J, Fan Y, Kang X, Zhao L. XRCC3 T241M polymorphism and melanoma skin cancer risk: a meta-analysis. Oncol Lett. 2015;9:2425–29. doi: 10.3892/ol.2015.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.