Abstract
The human immunodeficiency virus type 1 (HIV-1) Gag precursor Pr55gag by itself is capable of assembling into retrovirus-like particles (VLP). In the present study, we attempted to identify the minimal Gag sequences required for the formation of VLP. Our results show that about 80% of Pr55gag can be either deleted or replaced by heterologous sequences without significantly compromising VLP production. The smallest chimeric molecule still able to efficiently form VLP was only about 16 kDa. This minimal Gag construct contained the leucine zipper domain of the yeast transcription factor GCN4 to substitute for the assembly function of nucleocapsid (NC), followed by a P-P-P-P-Y motif to provide late budding (L) domain function, and retained only the myristylation signal and the C-terminal capsid-p2 domain of Pr55gag. We also show that the L domain function of HIV-1 p6gag is not dependent on the presence of an active viral protease and that the NC domain of Pr55gag is dispensable for the incorporation of Vpr into VLP.
The gag gene is translated into a polyprotein that is sufficient to mediate the formation of retrovirus-like particles (VLP) in the absence of other viral proteins. Subsequent to the assembly of an immature virus particle, the Gag polyprotein is cleaved by the viral protease (PR) to yield the structural proteins of the mature virion. For human immunodeficiency virus type 1 (HIV-1), the major cleavage products are matrix (MA), capsid (CA), nucleocapsid (NC), and p6 (11, 26). MA remains associated with the host cell-derived lipid envelope of the virion, CA condenses into the characteristic conical core, and NC covers the genomic viral RNA within the core (11, 26).
Several regions of the 55-kDa HIV-1 Gag polyprotein (Pr55gag) have been reported to be crucial for particle formation (11, 13). The N-terminal MA domain harbors a myristylation signal which is essential for virus particle production (3, 19). Furthermore, a basic patch on the surface of the globular core of MA appears to contribute to the selective association of Pr55gag with the plasma membrane by interacting with acidic phospholipids (63). However, several studies have shown that MA can be largely deleted or even entirely replaced by a heterologous myristyl anchor without compromising the formation of extracellular particles (33, 47, 54).
CA, which follows MA in the context of Pr55gag, can by itself assemble in vitro into hollow cylindrical particles reminiscent of viral cores, but the protein concentrations required are relatively high (20, 21, 51). Interestingly, the addition of as few as four MA residues to the N terminus of CA resulted in the assembly of spherical rather than cylindrical particles, indicating that cleavage at the MA-CA junction results in conformational changes that govern the rearrangement of CA into a conical structure during virus maturation (20, 51). CA is a largely α-helical molecule with two distinct domains that can fold independently (12, 16, 38). The larger, N-terminal domain is required for core formation and virus infectivity but is dispensable for particle assembly in vivo (2, 8, 45, 46, 52). In contrast, mutations in the C-terminal third of CA, which includes the uniquely conserved major homology region (MHR), often interfere with particle assembly (2, 8, 10, 12, 37, 46). Additionally, certain substitutions or deletions which affect the N terminus of p2, the 14-amino-acid peptide that separates CA from NC, have been reported to induce grossly aberrant budding structures and to reduce the number of extracellular particles (1, 32).
The NC domain contains two copies of a conserved zinc finger-like motif which are required for the encapsidation of the genomic viral RNA (11). In the presence of RNA, NC can dramatically increase the efficiency of in vitro assembly reactions (4, 21), suggesting that NC serves to concentrate Pr55gag on viral RNA during assembly. Mutagenic analyses support this concept by showing that NC is critical for HIV-1 assembly in vivo (6, 9, 61).
The C-terminal p6 domain, which is found only in primate lentiviruses, facilitates the separation of virus particles from the cell (17, 25, 60). Additionally, p6 is required for the incorporation of the accessory viral protein Vpr (30, 35, 43) and has also been implicated in the incorporation of the viral Pol and envelope proteins (41, 59) and in the control of particle size (14, 15). Although p6 is the most variable Gag domain among primate lentiviruses, two highly conserved motifs can be discerned. One of these is located near the C terminus of the domain and, in HIV-1, is essential for the incorporation of Vpr (30, 31, 35). The second conserved motif (P-T/S-A-P-P), located near the N terminus of p6, is crucial for the role of p6 in virus release (17, 25). Domains which function at a late step of the budding process, referred to as L domains, have also been identified in other retroviruses (44, 56, 58) and, most recently, in rhabdoviruses (5). Most of these harbor a sequence which resembles the P-P-P-P-Y motif originally identified at the core of the L domain of Rous sarcoma virus (RSV) (57). However, lentivirus Gag proteins lack the P-P-P-P-Y motif but usually contain one or more copies of the P-T/S-A-P-P motif. As first shown by Parent et al. (42), L domains can act in a positionally independent manner and can be exchanged between unrelated retroviruses.
We recently reported that the C-terminal half of Pr55gag is sufficient for the efficient in vivo assembly and release of VLP (2). Interestingly, it was also recently shown that the roles of the C-terminal NC and p6 domains of Pr55gag in particle production can both be replaced by a leucine zipper domain (62), a sequence which promotes dimerization (40). In the present study, we combine these findings and demonstrate VLP formation by a small chimeric molecule which harbors a leucine zipper domain in place of NC and in which the only HIV-1 Gag components are the six-amino-acid myristylation signal and the C-terminal CA-p2 domain. Efficient VLP formation also required the presence of L domain function, but this function could be provided by a short peptide which contained the P-P-P-P-Y motif.
MATERIALS AND METHODS
Plasmids.
HXBH10-PR−, which was used to express Pr55gag, is identical to HXBH10, a vpu-positive variant of the HXB2 clone of HIV-1, except for a point mutation that inactivates PR (18). Site-directed mutagenesis was used to create the p41 mutant, which differs from HXBH10 only by a premature termination codon in gag that replaces the codon for the first residue of NC. Similarly, the p6− mutant harbors a premature termination codon in place of codon 1 of p6 (17). The ΔPTAPP mutant is a variant of HXBH10 with an in-frame deletion of codons 7 though 11 of p6 (31). The p6− and ΔPTAPP mutations were also introduced into HXBH10-PR− to prevent proteolytic processing of the mutant Gag precursor. The ΔNC-p1 mutant is a variant of HXBH10 with an in-frame deletion in gag which results in the coding regions for p2 and p6 being fused. The nucleotide sequence at the junction is 5′ TCA GCT ACC ATA ATG CTG CAG AGC AGA CCA 3′ (with p2 sequences in lightface type and p6 sequences in boldface type).
To obtain the chimeric ZWT variant of HXBH10, site-directed mutagenesis was used to introduce a PstI site into the HXBH10 gag gene immediately 3′ to the p2 coding region. By making use of a natural PstI site in the GCN4 coding sequence, we fused GCN4 codons 247 through 280 to the 3′ end of the p2 coding region. The nucleotide sequence at the fusion site is 5′ TCA GCT ACC ATA ATG CTG CAG CGT ATG AAG 3′ (with HIV-1 gag sequences in lightface type and GCN4 sequences in boldface type). In ZWT, GCN4 codon 280 is immediately followed by a PCR-generated termination codon, which in turn is followed by HIV-1 sequences, starting with the NcoI site at nucleotide (nt) 5678 of HXBH10. The ZIL construct is identical to ZWT, except that the GCN4 sequence was derived from a synthetic fragment which has both the a and the d positions of the coiled coil heptad repeats mutated to isoleucine (55). To obtain the ZWT-p6 and ZIL-p6 constructs, SacI cloning sites were generated by standard PCR methods, which allowed us to directly fuse the p6 coding sequence to the 3′ end of the GCN4 sequence in the ZWT and ZIL constructs. The nucleotide sequences at the junctions are 5′ AAG CTT GTG GGT GAG CTC CAG AGC AGA CCA 3′ for ZWT-p6 and 5′ AAA CTG ATC GGT GAG CTC CAG AGC AGA CCA 3′ for ZIL-p6 (with GCN4 sequences in boldface type and p6 sequences in lightface type). The Δ-ZWT, Δ-ZWT-p6, and Δ-ZIL-p6 constructs are variants of the previously described Δ8-277 mutant (2); they were obtained by standard cloning methods, making use of an AgeI site in the coding region for the C-terminal CA domain. The Δ-ZWT-p6(t) construct is a derivative of Δ-ZWT-p6 which harbors a premature termination codon in place of codon 15 of p6. The Δ-ZWT-p2b construct was derived from Δ-ZWT by inserting a synthetic sequence which codes for RSV p2b, followed by a stop codon, between the 3′ end of the GCN4 sequence and an NcoI site (nt 5678 of HXBH10). Vpu-negative variants of ZWT and Δ-ZWT-p2b were obtained by replacing a SalI-NheI fragment (nts 5789 to 7263 of HXBH10) with the corresponding fragment from HXB2, which harbors a defective vpu gene.
Cell culture, transfection, and viral protein analysis.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Cells (1.4 × 106) were seeded into 80-cm2 tissue culture flasks 24 h prior to transfection. The cultures were transfected with 15 μg of proviral constructs by a calcium phosphate precipitation technique and metabolically labeled with [35S]methionine (50 μCi/ml) from 48 to 60 h posttransfection. Supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm-pore-size filters. VLP released during the labeling period were spun through 20% sucrose cushions (in phosphate-buffered saline) for 2 h at 4°C and 27,000 rpm in a Beckman SW28 rotor. Pelleted VLP were lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and viral proteins were directly analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
To control for intracellular expression levels, the labeled cells were lysed in radioimmunoprecipitation assay buffer, and viral proteins were immunoprecipitated with AIDS patient serum as described previously (9). For immunoblot analysis, transfected cells were lysed directly in SDS sample buffer (156 mM Tris-HCl [pH 6.8], 2.5% SDS, 12.5% glycerol, 6% β-mercaptoethanol). The samples were then heated to 100°C for 5 min, resolved by SDS-PAGE, and electroblotted onto Hybond-C Extra membranes (Amersham Pharmacia Biotech). The membranes were incubated overnight at 4°C with rabbit anti-CA polyclonal antiserum (1:2,000; Advanced Biotechnologies) in the presence of 2% milk to block nonspecific sites, followed by incubation for 1 h at 37°C with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (1:2,000; Cappel). The blots were developed with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Sucrose density gradient fractionation.
[35S]methionine-labeled VLP obtained from transfected HeLa cells were concentrated by centrifugation through 20% sucrose cushions, resuspended in Dulbecco's modified Eagle's medium, and layered on top of a preformed 20 to 60% sucrose gradient (in phosphate-buffered saline). Following centrifugation at 40,000 rpm in an SW41 rotor for 19 h at 4°C, 16 0.5-ml fractions were collected. The fractions were precipitated with 10% trichloroacetic acid and analyzed by SDS-PAGE. The density of each fraction was determined on an Abbe MARK II refractometer.
RESULTS
Dimerization and trimerization domains differ in their ability to substitute for NC-p1-p6 in particle formation.
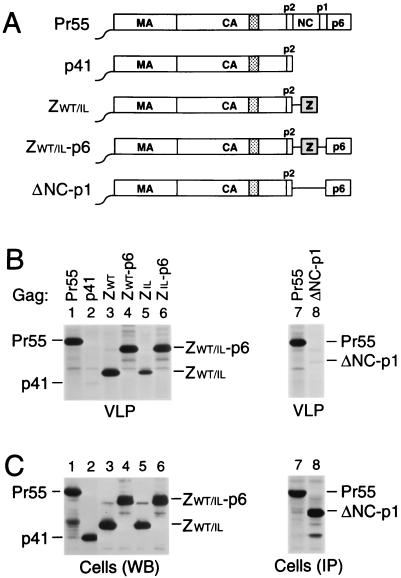
Several studies have demonstrated that the C-terminal NC and p6 domains of the HIV-1 Gag polyprotein Pr55gag are crucial for particle assembly and release. Interestingly, Zhang et al. (62) recently reported that particle production can be rescued by replacing the NC-p1-p6 region of Pr55gag with a coiled coil sequence capable of forming parallel homodimers. To explore whether the ability of the coiled coil to assume a particular oligomerization state is crucial, we chose to use versions of the GCN4 leucine zipper domain which form either dimers or trimers in solution and in crystal structures (22, 23, 39). In one case, the coding region for NC-p1-p6 was replaced with the 33 codons for the wild-type GCN4 dimerization domain, yielding the ZWT construct (Fig. 1A). A second construct, ZIL, harbors a GCN4 zipper variant with isoleucine substitutions at four a and four d positions of its heptad repeats. It has been shown that the presence of isoleucine residues at these positions directs trimer formation (22, 23, 55).
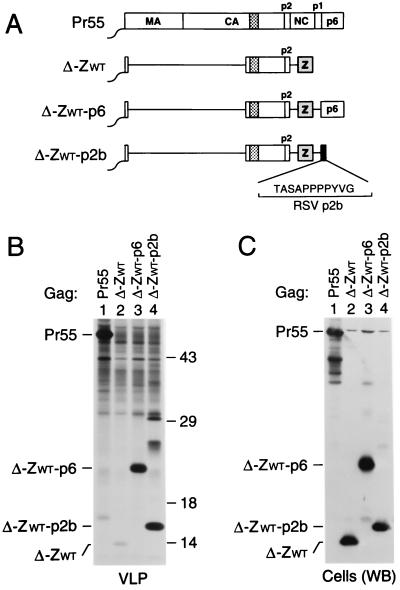
FIG. 1.
Replacement of the assembly functions of NC and p6 by GCN4 zipper sequences. (A) Comparison of the domain organizations of the HIV-1 Gag precursor Pr55gag and of mutant Gag molecules. The wavy line at the N termini indicates the presence of a myristylation signal. The position of the MHR within the CA domain is indicated by a cross-hatched box. Horizontal lines denote in-frame deletions, and wild-type or mutant GCN4 zipper (Z) domains inserted in place of Gag sequences are represented by a gray box. (B) VLP formation. HeLa cells were transfected with a PR-defective HIV-1 provirus expressing wild-type Pr55gag or with the indicated Gag mutants, followed by metabolic labeling with [35S]methionine. VLP released during the labeling period were pelleted through sucrose and analyzed by SDS-PAGE. The migration positions of the wild-type and mutant Gag precursors are indicated. (C) Comparison of cell-associated Gag protein levels by Western blotting (WB) with anti-CA antiserum (lanes 1 to 6) or by immunoprecipitation (IP) with patient serum from [35S]methionine-labeled cell lysates (lanes 7 and 8).
To examine whether these modifications affect particle formation, proviral constructs harboring the chimeric gag genes were transfected into HeLa cells. After metabolic labeling with [35S]methionine, extracellular particles were pelleted through sucrose cushions, and their protein content was then directly analyzed by SDS-PAGE. As expected, a mutant which expressed a C-terminally truncated Gag precursor that corresponded precisely to the authentic Gag cleavage intermediate p41 (MA-CA-p2) (Fig. 1A) produced virtually no particles (Fig. 1B, lane 2), despite efficient expression of the truncated Gag product (Fig. 1C). In accordance with the finding that the CREB leucine zipper domain can replace the assembly function of NC-p1-p6 (62), fusing the wild-type GCN4 dimerization domain to the C terminus of p41 dramatically improved particle formation (Fig. 1B, lane 3). Taking into account the number of methionine residues present in each Gag construct, the release levels of ZWT protein in the experiment shown in Fig. 1B, as quantified by PhosphorImager analysis, were about two-thirds those obtained for Pr55gag and were even closer to wild-type levels in other experiments. In contrast, the ZIL construct produced only about 10% the amount of particulate Gag protein seen with the provirus expressing wild-type Pr55gag (Fig. 1B, compare lanes 1 and 5).
Dimerization and trimerization domains are equally capable of substituting for NC-p1.
The chimeric constructs described above lacked both NC and the C-terminal p6 domain of Pr55gag. The p6 domain is found only in primate lentiviruses and has been implicated in the final release of assembled particles from the cell surface (17, 25, 42, 60). To determine whether the absence of a p6 domain contributed to the relatively low particle yields obtained with the ZIL construct, the p6 coding sequence was fused in frame to the 3′ terminus of the variant GCN4 zipper sequence, yielding ZIL-p6 (Fig. 1A). An analogous construct (ZWT-p6) harboring the wild-type GCN4 leucine zipper sequence in place of NC-p1 was also made. Of note, the MA, CA, p2, and p6 regions encoded by the ZWT-p6 and ZIL-p6 constructs are predicted to be identical to those encoded by the parental HXBH10 provirus.
In contrast to ZWT and ZIL, the ZWT-p6 and ZIL-p6 constructs retained the coding sequence for PR. However, because of the absence of the frameshift site required for the expression of PR (27), the chimeric Gag precursors produced by ZWT-p6 and ZIL-p6 were expected to remain unprocessed. Therefore, the ability of the chimeric constructs to produce particles was again compared to that of a full-length HIV-1 provirus which encodes an inactive PR. After transfection into HeLa cells, the ZWT-p6 and ZIL-p6 constructs both produced amounts of particles similar to those produced by the control expressing wild-type Pr55gag (Fig. 1B, compare lanes 1, 4, and 6). Cells transfected with the ΔNC-p1 construct (Fig. 1A) efficiently expressed a shortened Gag polyprotein but released about 40-fold less particulate Gag protein into the medium than cells expressing Pr55gag (Fig. 1B and C, lanes 7 and 8), confirming the critical role of NC in VLP formation. We conclude that the wild-type and mutant GCN4 zipper domains were both fully capable of replacing the assembly function of NC.
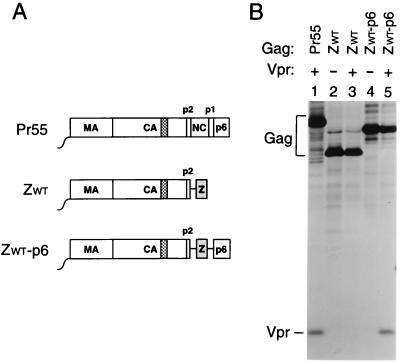
The NC domain is dispensable for Vpr incorporation.
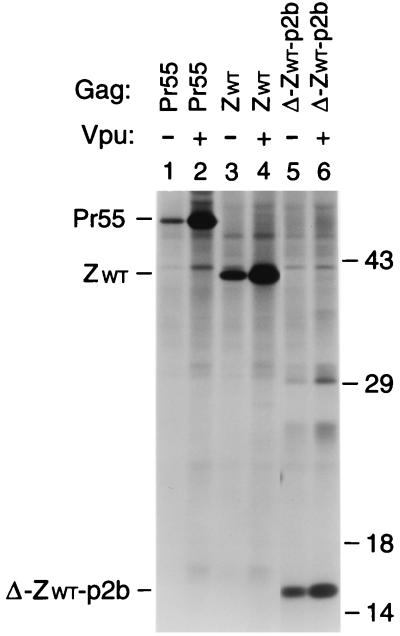
Vpr is a small accessory protein of primate lentiviruses which is specifically incorporated into virions. Mutagenic analyses have shown that the incorporation of HIV-1 Vpr into VLP depends on the presence of a C-terminal region of p6 (30, 31, 35). However, in in vitro binding studies, HIV-1 Vpr displayed a significantly higher affinity for NC than for p6, prompting the suggestion that NC cooperates with p6 in the encapsidation of Vpr (7, 34, 49). The availability of the ZWT and ZWT-p6 constructs allowed us to test this model, because they efficiently form VLP in the absence of NC. Because the parental provirus expressing Pr55gag and the ZWT and ZWT-p6 chimeras were all vpr negative, each of these constructs was cotransfected with a plasmid providing HIV-1 Vpr in trans. As a control, the two Gag chimeras were transfected alone. As expected, VLP produced by the ZWT chimera, which lacks a p6 domain, failed to incorporate Vpr (Fig. 2, lane 3). In contrast, VLP formed by the ZWT-p6 chimera incorporated at least as much Vpr as particles produced by full-length Pr55gag (Fig. 2, compare lanes 1 and 5). These results confirm the central role of p6 in the incorporation of Vpr and demonstrate that NC is dispensable.
FIG. 2.
Vpr incorporation in the absence of NC. (A) Schematic representation of the Gag constructs used. See the legend to Fig. 1 for details. (B) Analysis of Vpr incorporation. HeLa cells were transfected with vpr-negative proviral constructs expressing wild-type or mutant Gag precursors. Where indicated, a construct expressing a hemagglutinin epitope-tagged version of HIV-1 Vpr was cotransfected. [35S]methionine-labeled particulate material released into the supernatant was pelleted through sucrose and analyzed directly by SDS-PAGE.
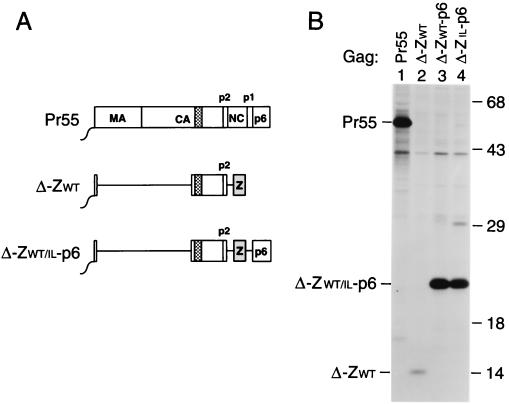
Efficient VLP formation in the absence of MA, the N-terminal CA domain, and NC.
We recently reported that the HIV-1 gag sequences coding for MA and for the N-terminal domain of CA are not required for efficient particle assembly or release (2, 47). Taken together with the observation that Pr55gag sequences distal to p2 can be replaced by a coiled coil, these results raised the possibility that the C-terminal domain of CA may be the only component of Pr55gag necessary to obtain particle formation.
To test this hypothesis, we deleted gag codons 8 through 277 from ZWT. The resulting Δ-ZWT construct codes for a 14-kDa protein composed of the six-residue Gag myristylation signal, the C-terminal domain of CA-p2, and the wild-type GCN4 leucine zipper domain (Fig. 3A). Additionally, the Δ8-277 mutation was introduced into the ZWT-p6 and ZIL-p6 constructs, yielding Δ-ZWT-p6 and Δ-ZIL-p6 (Fig. 3A). We demonstrated previously that the Δ8-277 deletion, when introduced into a wild-type HIV-1 gag gene, had only moderate effects on particle yields (2). However, in the context of the ZWT construct, the Δ8-277 deletion reduced VLP production more than 15-fold (Fig. 3B, lane 2). Interestingly, fusing the p6 domain to the C terminus of the ZWT protein increased VLP production to levels which approached those obtained with wild-type Pr55gag (Fig. 3B, lanes 1 to 3). Comparable levels of VLP production were also obtained with the Δ-ZIL-p6 construct (Fig. 3B, lane 4), demonstrating that the oligomerization preference of the coiled coil replacing NC was not crucial. All quantitations take into account the fact that Pr55gag has 15 methionine residues, whereas the mutant Gag forms shown in Fig. 3 contained only 6 methionines. While the intensities of Pr55gag and of the deletion mutants containing p6 appeared similar in Fig. 3B, the mutant bands in fact yielded about threefold fewer counts, as determined by PhosphorImager analysis.
FIG. 3.
Efficient VLP formation by minimal HIV-1 Gag constructs. (A) Schematic drawing illustrating the Gag regions retained and the position of the GCN4 zipper domain used to replace the assembly function of NC. See the legend to Fig. 1 for details. (B) VLP formation by transfected HeLa cells. VLP released during metabolic labeling were pelleted through sucrose and analyzed by SDS-PAGE. The positions of wild-type Pr55gag and of chimeric Gag molecules are indicated on the left. The migration positions of molecular mass markers (in kilodaltons) are indicated on the right.
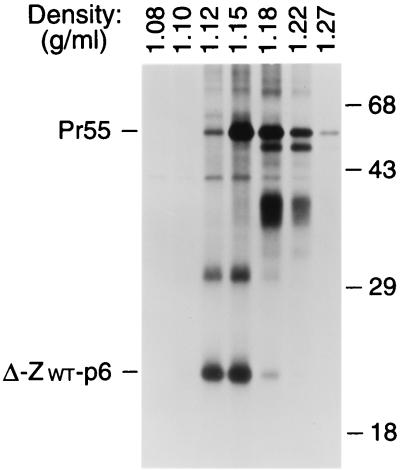
To confirm that the Δ-ZWT-p6 molecule was released in the form of VLP, we compared the density of the particulate structures produced by the chimeric construct to that of authentic immature HIV-1 virions. HeLa cells transfected with the Δ-ZWT-p6 construct or with a provirus expressing full-length Pr55gag were metabolically labeled with [35S]methionine, and particulate material released into the supernatant was pelleted through 20% sucrose. The pelleted material was pooled and fractionated by sucrose density gradient centrifugation. Labeled proteins were then recovered from each fraction by trichloroacetic acid precipitation and separated by SDS-PAGE. As shown in Fig. 4, the Δ-ZWT-p6 protein was released in particles of uniform density, with a peak at 1.15 g/ml. On average, particles formed by the Δ-ZWT-p6 construct had a somewhat lower density than authentic HIV-1 virions produced by the HXBH10-PR− provirus, consistent with a lower ratio of protein to lipid mass. Taken together, these results show that the C-terminal domain of CA-p2, in combination with a membrane anchor, a protein-protein interaction domain, and p6, can direct the efficient formation of VLP.
FIG. 4.
Comparison of particle densities. [35S]methionine-labeled VLP formed by wild-type Pr55gag and by the chimeric Δ-ZWT-p6 molecule were pooled and fractionated in a 20 to 60% sucrose gradient. Sixteen fractions were collected, precipitated with 10% trichloroacetic acid, and analyzed by SDS-PAGE. The peak Gag protein fractions are shown, and the densities of individual fractions are indicated above each lane. Numbers at right are in kilodaltons.
Requirement for L domain function.
Previous studies have shown that p6 facilitates the final separation of HIV-1 virions from the cell surface (17, 25, 60). This so-called L domain function is critically dependent on the presence of a highly conserved P-T/S-A-P-P motif near the N terminus of p6 (17, 25). Furthermore, the first 12 residues of p6, which include the P-T/S-A-P-P motif, can to some extent replace the L domain of RSV (42). To examine whether this N-terminal region of p6 is capable of rescuing VLP production by the Δ-ZWT chimera, the first 14 residues of p6 were fused in frame to the C terminus of the GCN4 zipper sequence. In contrast to full-length p6, the N-terminal p6 fragment only modestly increased VLP production in repeated experiments (data not shown), indicating that the P-T/S-A-P-P motif is not sufficient for full L domain function.
The potent enhancement of VLP production by full-length p6 suggested that the Δ-ZWT chimera is defective at a late stage of budding. However, it also remained possible that the full-length p6 domain contributed additional Gag-Gag contact sites that became critical for early assembly steps in the absence of large portions of Pr55gag. In an effort to distinguish between these possibilities, we examined whether the defect of the Δ-ZWT chimera could be corrected by the entirely unrelated L domain of RSV. The RSV L domain has been fine mapped to a conserved P-P-P-P-Y motif in the p2b region of the Gag precursor and has been shown to function in a positionally independent manner (42, 57). Remarkably, when fused to the C terminus of the Δ-ZWT molecule (Fig. 5A), the 11-amino-acid RSV p2b peptide enhanced VLP formation as effectively as the entire HIV-1 p6 domain, causing an approximately 20-fold increase in the release of particulate Gag protein (Fig. 5B, lane 4). As expected, this result was not due to increased expression levels, because the intracellular steady-state levels of the Δ-ZWT and Δ-ZWT-p2b molecules, as detected by immunoblotting with anti-CA antiserum, were comparable (Fig. 5C). In addition to the predicted 16-kDa product, VLP formed by the Δ-ZWT-p2b construct contained several slower-migrating species (Fig. 5B, lane 4) whose identities are currently under investigation. Taken together, our results support the notion that the 14-kDa Δ-ZWT Gag protein is assembly competent but requires an L domain for efficient VLP release.
FIG. 5.
Rescue of VLP formation by the L domain of RSV. (A) Schematic presentation of the Gag constructs used. The RSV p2b peptide attached to the C terminus of the wild-type GCN4 leucine zipper (Z) is represented by a black box, and the amino acid sequence of p2b is given in the one-letter code. See the legend to Fig. 1 for other details. (B) VLP formation by transfected HeLa cells. [35S]methionine-labeled particulate material released into the medium was pelleted through sucrose and analyzed directly by SDS-PAGE. Numbers at right are in kilodaltons. (C) Cell-associated Gag detected by Western blotting (WB) with anti-CA antiserum.
Requirement for Vpu.
Vpu, a small accessory protein unique to HIV-1, can significantly enhance the release of virus particles from the cell surface (18, 29, 50). To determine whether the effect of Vpu depends on specific Gag domains, variants of the ZWT and Δ-ZWT-p2b constructs lacking a vpu initiation codon were made. Consistent with previous work (18), HeLa cells transfected with a vpu-negative variant expressing full-length Pr55gag released 25-fold less particulate Gag protein than cells transfected with the vpu-positive version (Fig. 6, lanes 1 and 2). In the absence of Vpu, the levels of the ZWT and Δ-ZWT-p2b proteins in the particulate fractions were higher than those of full-length Pr55gag (Fig. 6, lanes 1, 3, and 5), indicating that the presence of the GCN4 leucine zipper reduced the requirement for Vpu for efficient VLP release. However, the ZWT and Δ-ZWT-p2b constructs both remained responsive to Vpu (Fig. 6, lanes 3 to 6), indicating that neither MA, the N-terminal domain of CA, NC, nor p6 is required.
FIG. 6.
Effect of Vpu on VLP release. HeLa cells were transfected with proviral constructs harboring a wild-type or mutated gag gene and either an intact or a defective vpu gene. Particulate material released during metabolic labeling was pelleted and analyzed directly by SDS-PAGE. Numbers at right are in kilodaltons.
A role for p6 in particle release by full-length proviruses in the absence of PR.
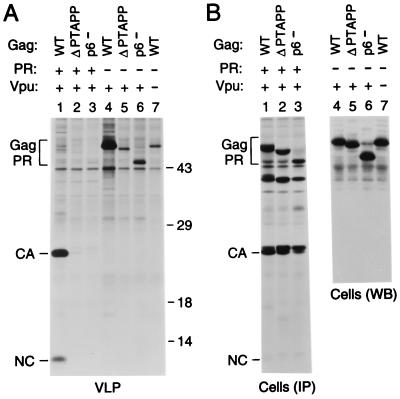
It has been reported that p6 is required for efficient particle formation by full-length HIV-1 proviruses but becomes dispensable in the absence of PR activity (25). However, the results described above show that, at least under certain circumstances, p6 can significantly enhance particle production in the absence of PR. To determine how this PR-independent effect of p6 compares to its role in a full-length provirus, we used variants of the infectious HXBH10 molecular clone and of its PR-negative variant HXBH10-PR− which harbor a premature termination codon that prevents the synthesis of p6 (17). We also made versions of HXBH10 and of HXBH10-PR− that lack the codons for the highly conserved P-T-A-P-P motif in p6, which is required for L domain function (17, 25). Consistent with previous reports (17, 25), particle formation by full-length proviruses encoding active PR decreased dramatically in the absence of the P-T-A-P-P motif or if the p6 domain was missing entirely (Fig. 7A, lanes 1 to 3). However, the p6 mutants also exhibited significant defects in particle formation if PR was inactive (Fig. 7A, lanes 4 to 6), in spite of efficient expression of the mutant Gag proteins (Fig. 7B). Indeed, in the PR-negative context, the absence of the P-T-A-P-P motif reduced particle production by about as much as a lack of Vpu (Fig. 7A, compare lanes 5 and 7), whose effect is independent of PR (18). Thus, at least in our experimental conditions, p6 was required for efficient particle production by full-length proviruses lacking active PR.
FIG. 7.
PR-independent requirement for p6 in the context of a full-length provirus. (A) VLP formation. HeLa cells were transfected with the replication-competent HXBH10 molecular clone (lane 1), with the PR-deficient HXBH10-PR− provirus (lane 4), with a vpu-deficient variant of HXBH10 (lane 7), or with versions of HXBH10 and HXBH10-PR− harboring the indicated mutations in p6 (lanes 2, 3, 5, and 6). [35S]methionine-labeled particulate material released by the transfected cells was sedimented through sucrose and analyzed by SDS-PAGE. Numbers at right are in kilodaltons. (B) Cell-associated Gag protein levels determined by immunoprecipitation (IP) from [35S]methionine-labeled cell lysates with patient serum (lanes 1 to 3) or by Western blotting (WB) with anti-CA antiserum (lanes 4 to 7). WT, wild type; Gag PR, Gag precursor.
DISCUSSION
This study demonstrates efficient VLP production by minimal HIV-1 Gag constructs which have NC replaced by a leucine zipper domain and retain only the myristyl anchor, the C-terminal third of CA-p2, and a late assembly domain. In a previous study, the smallest HIV-1 gag gene product capable of forming VLP was a 28-kDa protein which contained the entire CA-p2 region (53). However, the efficiency of VLP formation was low. In the present study, the use of heterologous sequences to replace NC and p6 allowed efficient VLP formation even by a 16-kDa protein that retained only the 6-amino-acid myristylation signal, CA residues 146 to 231, and the 14-amino-acid p2 peptide from the HIV-1 Gag precursor.
The crystal structure of CA residues 146 to 231 shows a globular domain composed of four helices which form a dimer, and equilibrium sedimentation analyses indicate that CA residues 146 to 231 dimerize with an affinity similar to that of the full-length CA protein (12). The MHR, which lies at the N terminus of the domain, does not contribute to the dimer interface (12) but is nevertheless essential for HIV-1 particle assembly (37). In a recent study, we found that all of the MHR had to be retained for efficient VLP formation, even though MA and the entire N-terminal CA domain were dispensable (2). We also retained the p2 “spacer” peptide in our minimal Gag constructs, because it has been shown that mutations in p2 can cause severe defects in the assembly of uniformly curved buds and, as a consequence, in the formation of extracellular particles (1, 32). In the context of Pr55gag, the CA-p2 boundary is predicted to form an additional α helix which is not present in mature CA, and our recent mutagenic analysis supports the view that the propensity of the N terminus of p2 to adopt an α-helical conformation is crucial for its role in assembly (1).
The present study confirms that the NC and p6 domains of Pr55gag are both critical for particle formation. However, in accord with previous observations (62), the entire C-terminal NC-p1-p6 region of Pr55gag became dispensable for particle production if a leucine zipper domain was inserted in its place. A mutant zipper domain which forms trimers rather than dimers was also fully capable of replacing NC-p1, suggesting that the role of NC in assembly is mainly to induce proximity and that the oligomerization preference of the region is not crucial. Interestingly, the mutant GCN4 zipper form was clearly less efficient than the wild-type form in replacing the role of p6 in particle formation. A requirement for p6 was even more apparent with Gag constructs which lacked the N-terminal half of Pr55gag, except for six amino acids that provided a myristyl anchor for membrane attachment. In the latter context, even the wild-type leucine zipper domain was unable to replace the role of NC-p1-p6 in assembly or release. However, a dramatic increase in VLP production was obtained if p6, which includes the L domain of Pr55gag, was fused to the C terminus of the zipper sequence. Furthermore, the unrelated 11-amino-acid p2b peptide from the RSV Gag precursor, which also functions as an L domain (42, 56, 57), was as effective as the entire HIV-1 p6 domain in rescuing VLP formation. Taken together, these results indicate that our minimal Gag construct, which retains only the myristyl anchor and the C-terminal CA-p2 domain of Pr55gag, is assembly competent but highly dependent on the presence of an L domain for the final release of VLP.
The role of p6 in virus release has been subject to controversy, as several groups failed to detect a clear requirement for p6 (24, 28, 36, 43, 48). One study appeared to reconcile these observations by showing that p6 is required for particle production in the context of a full-length HIV-1 molecular clone but becomes dispensable if proteolytic processing of the Gag precursor is prevented (25). However, our results with minimal Gag constructs demonstrated that p6 and the unrelated L domain of RSV can both significantly enhance VLP formation in the absence of proteolytic processing. Because of these observations, we reexamined the requirement for p6 in a full-length HIV-1 provirus. In the absence of the p6 domain or of the conserved P-T-A-P-P motif near the N terminus of the domain, substantial defects in particle production remained, even after PR was inactivated. One possible explanation for this discrepancy between our results and those of Huang et al. (25) is that the requirement for p6 depends on Gag expression levels. In support of this possibility, we previously observed that deletions in MA can significantly enhance particle production but that this effect becomes less evident at high Gag expression levels (47).
Intriguingly, while the use of L domains appears widespread, if not universal, among retroviruses, efficient virus release can evidently be achieved in other ways, as exemplified by the ability of leucine zipper domains to replace p6. However, the use of an L domain may allow Gag-Gag interactions to remain sufficiently flexible to permit not only virus assembly and release but also the subsequent rearrangements required for virus maturation and uncoating. Because of their exquisite dependence on the presence of an L domain, the minimal Gag constructs described here should be useful tools for the identification of viral sequences with L domain function.
ACKNOWLEDGMENTS
We thank Winfried Weissenhorn for generously providing DNA encoding wild-type and mutant GCN4 zipper domains.
This work was supported by National Institutes of Health grants AI29873 and AI28691 (Center for AIDS Research) and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Accola M A, Höglund S, Göttlinger H G. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borsetti A, Öhagen A, Göttlinger H G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson L, Yu X-F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 7.De Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix J L, Roques B P. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 8.Dorfman T, Bukovsky A, Öhagen A, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman T, Luban J, Goff S P, Haseltine W A, Göttlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 11.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 12.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 13.Garnier L, Bowzard J B, Wills J W. Recent advances and remaining problems in HIV assembly. AIDS. 1998;12:S5–S16. [PubMed] [Google Scholar]
- 14.Garnier L, Parent L J, Rovinski B, Cao S-X, Wills J W. Identification of retroviral late domains as determinants of particle size. J Virol. 1999;73:2309–2320. doi: 10.1128/jvi.73.3.2309-2320.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier L, Ratner L, Rovinski B, Cao S-X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 17.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc Natl Acad Sci USA. 1993;88:3195–3199. doi: 10.1073/pnas.90.15.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross I, Hohenberg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 22.Harbury P B, Kim P S, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 23.Harbury P B, Zhang T, Kim P S, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 24.Hoshikawa N, Kojima A, Yasuda A, Takayashiki E, Masuko S, Chiba J, Sata T, Kurata T. Role of the gag and pol genes of human immunodeficiency virus in morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J Gen Virol. 1991;72:2509–2517. doi: 10.1099/0022-1317-72-10-2509. [DOI] [PubMed] [Google Scholar]
- 25.Huang M, Orenstein J M, Martin M A, Freed E O. p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 27.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 28.Jowett J B M, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 29.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo E, Mammano F, Cohen E A, Göttlinger H G. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo E, Göttlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M S, Garcia A G, Bhattacharyya U, Mascagni P, Austen B M, Roberts M M. The Vpr protein of human immunodeficiency virus type 1 binds to the nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun. 1996;218:352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y-L, Bennett R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mammano F, Öhagen Å, Höglund S, Göttlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 39.O'Shea E K, Klemm J D, Kim P S, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 40.O'Shea E K, Rutkowski R, Kim P S. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 41.Ott D E, Chertova E N, Busch L K, Coren L V, Gagliardi T D, Johnson D G. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6gag protein produces a mutant that fails to package its envelope protein. J Virol. 1999;73:19–28. doi: 10.1128/jvi.73.1.19-28.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reicin A S, Öhagen A, Yin L, Höglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reil H, Bukovsky A A, Gelderblom H R, Göttlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Royer M, Cerutti M, Gay B, Hong S-S, Devauchelle G, Boulanger P. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 49.Selig L, Pages J-C, Tanchou V, Preveral S, Berlioz-Torrent C, Liu L X, Erdtmann L, Darlix J-L, Benarous R, Benichou S. Interaction with the p6 domain of the Gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol. 1999;73:592–600. doi: 10.1128/jvi.73.1.592-600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger E F, Cohen E A, Lu Y, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C-T, Lai H-Y, Li J-J. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C-T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weissenhorn W, Calder L J, Dessen A, Laue T, Skehel J J, Wiley D C. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6065–6069. doi: 10.1073/pnas.94.12.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu X-F, Dawson L, Tian C-J, Flexner C, Dettenhofer M. Mutations of the human immunodeficiency virus type 1 p6gag domain result in reduced retention of Pol proteins during virus assembly. J Virol. 1998;72:3412–3417. doi: 10.1128/jvi.72.4.3412-3417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu X-F, Matsuda Z, Yu Q-C, Lee T-H, Essex M. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J Gen Virol. 1995;76:3171–3179. doi: 10.1099/0022-1317-76-12-3171. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]