Abstract
Rubella virus (RV) genomic RNA contains two large open reading frames (ORFs): a 5′-proximal ORF encoding nonstructural proteins (NSPs) that function primarily in viral RNA replication and a 3′-proximal ORF encoding the viral structural proteins. Proteolytic processing of the RV NSP ORF translation product p200 is essential for viral replication. Processing of p200 to two mature products (p150 and p90) in the order NH2-p150-p90-COOH is carried out by an RV-encoded protease residing in the C-terminal region of p150. The RV nonstructural protease (NS-pro) belongs to a viral papain-like protease family that cleaves the polyprotein both in trans and in cis. A conserved X domain of unknown function was found from previous sequence analysis to be associated with NS-pro. To define the domains responsible for cis- and trans-cleavage activities and the function of the X domain in terms of protease activity, an in vitro translation system was employed. We demonstrated that the NSP region from residue 920 to 1296 is necessary for trans-cleavage activity. The domain from residue 920 to 1020 is not required for cis-cleavage activity. The X domain located between residues 834 and 940, outside the regions responsible for both cis- and trans-cleavage activities of NS-pro, was found to be important for NS-pro trans-cleavage activity but not for cis-cleavage activity. Analysis of sequence homology and secondary structure of the RV NS-pro catalytic region reveals a folding structure similar to that of papain.
Rubella virus (RV) is a single-strand, positive-polarity RNA virus classified in the Togaviridae family as the only member of the genus Rubivirus (12). The overall genome organization of RV (Fig. 1A) is similar to that of members of Alphavirus, the only other genus in the Togaviridae family (40), in terms of containing a nonstructural protein (NSP) open reading frame (ORF) and structural protein (SP) ORF at the 5′ and 3′ termini, respectively. In RV genomic RNA (9,762 nucleotides [nt]), the 5′-proximal NSP ORF extends from nt 41 to 6388, and the 3′-proximal SP ORF extends from nt 6512 to 9700 (10, 13, 42). Upon infection, the RV NSP is first translated from the input genomic RNA as a 2,116-amino-acid polyprotein, p200 (200 kDa), which is subsequently processed by an RV protease (NS-pro) contained within p200 into two mature protein products, p150 (residues 1 to 1301, 150 kDa) and p90 (residues 1302 to 2116, 90 kDa) (5, 7, 11, 25). The single cleavage occurs after G1301 in the sequence G1300-G1301-G1302 (7). p150 contains the proposed methyltransferase and protease sequences at its N and C termini, respectively (14, 21, 36). p90 contains the predicted helicase and RNA polymerase at its N- and C-terminal regions, respectively (15, 18, 21). Therefore, the order of conserved motifs of RV NSP, i.e., methyltransferase-protease-helicase-polymerase, differs from that of alphavirus NSP, which is methyltransferase-helicase-protease-polymerase. Along with host factors, RV NSPs form replication complexes that function to synthesize three species of viral RNAs. A full-length negative-strand RNA is first produced and serves as the template for the synthesis of 40S positive-strand genomic RNA and 24S subgenomic RNA (13). Cleavage of p200 by NS-pro plays a critical role in this process.
FIG. 1.
RV genome organization and PCR primers used for mutagenesis and amplification of different regions of RV NSP. (A) The RV genome contains an NSP ORF at its 5′ terminus and an SP ORF at its 3′ terminus. The NSPs are translated as a p200 polyprotein, which undergoes a single cleavage (indicated by a vertical arrow) into p150 and p90. The SPs are translated from a 24S subgenomic RNA accumulated during virus replication. Its starting position (nt 6436) is indicated by an arrow. Site-directed mutagenesis was by fusion PCR, which employed pBRM33 DNA as a template and two pairs of primers. For the C1152S mutation, paired primer, JSY-13–JSY-3 and JSY-2–JSY-12 were employed. For the G1301S mutation, paired primers JSY-13–JSY-5 and JSY-4–JSY-12 were used. For the G1302stop mutation, paired primers, JSY-13–YL-10 and YL-9–JSY-12 were used. All primers are shown as arrows indicating their polarities and nucleotide positions on RV genome RNA (numbered from the 5′ end of the M33 genome). Primer sequences are given in Table 1. The amplified 1.47-kb fragments containing the desired mutations were replaced back into pBR-NSP by using the NheI and EcoRV sites indicated above the genome. (B) The RV NSP ORF encodes a 200-kDa polyprotein to be processed into p150 (residues 1 to 1301) and p90 (residues 1302 to 2116). PCR primers used for making deletions from either end of p150 are shown by arrows indicating their polarities and relative amino acid positions on the NSP ORF.
Two groups of viral papain-like cysteine proteases (PCPs) have been distinguished on the basis of their ability to function in cis or in trans (14). Leader, or L-group, PCPs, located at the N terminus of the polyprotein, can function only in cis. Main, or M-group, PCPs, located in the central region of the polyprotein, can function both in cis and in trans (14). Sequence analysis also revealed an X domain (a novel conserved domain of unknown function) associated with M-group PCPs (14). On the basis of comparative sequence analysis, Gorbalenya et al. (14) proposed that RV NS-pro is an M-group PCP and identified an X domain (residues 834 to 940) adjacent to the RV NS-pro domain. Our previous studies demonstrated that RV NS-pro could function both in cis and in trans (44), a feature consistent with M-group PCPs. The catalytic dyad residues of NS-pro, C1152 and H1273, have been proposed on the basis of sequence alignment (14) supported by site-directed mutagenesis (7, 25).
Viral PCPs include members with highly variable protein size and sequence except for the close vicinity of catalytic residues (14). Only recently the tertiary structure of a viral PCP, the foot-and-mouth virus (FMDV) leader protease (Lpro), was solved as adopting a modified cellular papain fold (16). The diversity of primary sequence among PCP members prevents a direct tertiary structural prediction for other viral PCPs. Information on more PCPs is needed for a clear understanding of this protease family. RV NS-pro is located in the central region of polyprotein, and the functional proteolytic domains of RV NS-pro, in either cis- or trans-cleavage activity, have not been defined. RV NS-pro also constitutes parts of the domains mediating viral RNA synthesis. Therefore, the characterization of NS-pro functional domains contributes not only to the knowledge of the viral PCP family but also to the understanding of the biological roles of the RV NSP. In this study, we defined the regions required for cis- and trans-cleavage activities of NS-pro as well as the functional role of the X domain in both cis and trans cleavages. We have generated a panel of in-frame deletion mutants with mutations corresponding to either the N or C terminus of the p150-coding region. An in vitro translation system was used in both cis- and trans-cleavage analyses. The domain responsible for trans-cleavage activity of NS-pro was mapped to residues 920 to 1296. However, the domain for cis-cleavage activity was found to start from residue 1020, suggesting that the domain from residue 920 to 1020 is dispensable for cis cleavage. Although the X domain was found to play an important role in trans-cleavage activity, it has no effect on cis processing. We also present a predicted secondary structure for the RV NS-pro catalytic region.
MATERIALS AND METHODS
Plasmid construction.
Standard recombinant DNA techniques were used to generate all of the plasmid constructs (37). An RV infectious cDNA clone (pBRM33) (43) was used in the plasmid construction. Plasmid pBR-NSP was derived from pBRM33 after removing the SP ORF by deleting nt 6965 to 9336 after StuI digestion and religation. Site-directed mutations and deletions were accomplished by PCR using primers that contain designed substitutions and restriction enzyme sites. All primer sequences and relative positions on the RV genome are given in Table 1.
TABLE 1.
Primers used in this work
| Primer | Polaritya | Positionb | Nucleotide sequencec |
|---|---|---|---|
| C1152S | |||
| JSY-2 | + | 3482–3508 | 5′ GACCCAAACACCAGCTGGCTCCGCGCC 3′ |
| JSY-3 | − | 3482–3508 | 5′ GGCGCGGAGCCAGCTGGTGTTTGGGTC 3′ |
| G1301S | |||
| JSY-4 | + | 3929–3955 | 5′ CTGTCTCGGGGCAGCGGCACTTGTGCC 3′ |
| JSY-5 | − | 3929–3955 | 5′ GGCACAAGTGCCGCTGCCCCGAGACAG 3′ |
| G1302stop | |||
| YL-9 | + | 3929–3961 | 5′ CTGTCTCGGGGCGGCTAAACTTGTGCCGCCACC 3′ |
| YL-10 | − | 3929–3961 | 5′ GGTGGCGGCACAAGTTTAGCCGCCCCGAGACAG 3′ |
| PCR 5′ and 3′ | |||
| JSY-13 | + | 2770–2790 | 5′ GCTGCTCGAGCGCGCCTACCG 3′ |
| JSY-12 | − | 4220–4240 | 5′ GTAGGTGGCGGCGTTCTTGAT 3′ |
| JSY-16 | − | 1737–1757 | 5′ GGTGGGCGGGGTGGCGGTAGA 3′ |
| JSY-7 | − | 3584–3600 | 5′ GCTTCGCTCAGGGCGCG 3′ |
| N-terminal deletion | |||
| YL-11 | + | 1082–1101 | 5′ ATCCATGGCGTACTATAGCGAGCGCGT 3′ |
| YL-30 | + | 2518–2533 | 5′ ATATCCATGGACCCACCGCCT 3′ |
| JSY-25 | + | 2798–2812 | 5′ ATTCCCATGGTCGCGCTAGCCGCC 3′ |
| YL-25 | + | 2960–2977 | 5′ ATTCCATGGCCACGCTGACGCACGCC 3′ |
| YL-21 | + | 3098–3115 | 5′ ATATCCATGGCGACCCCCCTCGGGGAT 3′ |
| YL-22 | + | 3344–3361 | 5′ ATATCCATGGGCATGTGCGGGAGTGAC 3′ |
| C-terminal deletion | |||
| YL-23 | − | 3893–3910 | 5′ ATTAGGCCTTAGTGGGGGCGGTCCGAGAC 3′ |
| YL-26 | − | 3908–3925 | 5′ ATTAGGCCTTAGACCGCGAGCCAAAGGTG 3′ |
| YL-28 | − | 3914–3928 | 5′ ATTAGGCCTTAGGGGACCGCGAGCCA 3′ |
| YL-27 | − | 3917–3931 | 5′ ATTAGGCCTTACAGGGGGACCGCGAG 3′ |
| YL-24 | − | 3920–3937 | 5′ ATTAGGCCTTACCGAGACAGGGGGACCGC 3′ |
+, sense primer; −, antisense primer.
Corresponding nucleotide position on the M33 genome.
Mutations are in boldface; restriction sites (NcoI site for N-terminal deletion primers and StuI site for C-terminal deletion primers) are underlined.
All PCRs were carried out using native Pfu DNA polymerase (Strategene) under the manufacturer's recommended conditions. PCR fragments were purified with the QIAquick Spin PCR purification kit (QIAgen).
(i) Plasmids pBR-200(C1152S), pBR-200(G1301S), and pBR-150.
To substitute S for catalytic C1152, to mutate G1301 to S, or to change G1302 codon GCC into the stop codon TAA, fusion PCR (43) was employed with pBRM33 DNA as a template and two pairs of primers (schematically shown in Fig. 1A). In brief, to make the C1152S mutation, two pairs of primers, JSY-13–JSY-3 and JSY-2–JSY-12, were used in two PCRs to generate two products of 738 and 758 bp, respectively; the two partially overlapping PCR products were annealed to serve as the template for amplification of the 1.47-kb fragment using JSY-13 and JSY-12. To make the G1301S mutation, two pairs of primers, JSY-13–JSY-5 and JSY-4–JSY-12, were used in two PCRs to generate products of 1.19 kb and 311 bp, respectively; the two partially overlapping PCR products were annealed to serve as the template for amplification of the 1.47-kb fragment encoding the G1301S mutation using JSY-13 and JSY-12. To make the G1302stop mutation, two paired primers, JSY-13–YL-10 and YL-9–JSY-12, were used in two PCRs to generate products of 1.19 kb and 311 bp, respectively; the two partially overlapping PCR products were annealed to serve as the template for amplification of the 1.47-kb fragment encoding the G1302stop mutation using JSY-13 and JSY-12. The resulting 1.47-kb PCR products containing the desired mutations (C1152S, G1301S, and G1302stop) were cut with NheI and EcoRV and inserted into pBR-NSP (minus the NheI/EcoRV fragment) (Fig. 1A) to produce pBR-200(C1152S), pBR-200(G1301S), and pBR-150, respectively.
(ii) Truncated plasmids.
A series of in-frame deletions corresponding to either the N or C terminus of the p150-coding region was generated by amplifying the corresponding DNA fragment from pBR-150 (containing a stop codon between p150 and p90) using available restriction sites in RV cDNA and the NcoI site containing the initiation codon of p200. The relative positions of PCR primers on the RV NSP ORF are schematically shown in Fig. 1B. To make deletions from the N terminus of p150, amplification reactions were carried out on a pBR-150 DNA template using individual N-terminal deletion primers paired with an appropriate downstream 3′-end primer (Table 1 and Fig. 1B). The N-terminal deletion primers carried the initiation codon ATG (NcoI site) followed by nucleotide sequences downstream from nt 1082, 2518, 2798, 2960, 3098, or 3344 (the respective starting positions for A348, M827, V920, A974, A1020, or G1102). In brief, to clone the construct encoding residues from A348 to G1301, primer YL-11 was paired with JSY-16 in PCR, and the resulting 675-bp product was used to replace the NcoI (nt 39)-NotI (nt 1686) fragment of pBR-150 to make pBR-A348/G1301. To clone a construct encoding residues from M827 to G1301, primer YL-30 was paired with JSY-7 in PCR, and the resulting 1.1-kb fragment was used to replace the NcoI (nt 39)-SphI (nt 3391) fragment of pBR-150 to generate pBR-M827/G1301. To make the construct encoding sequences from V920, A974, A1020, or G1102 to G1301, the primer JSY-25, YL-25, YL-21, or YL-22, respectively, was paired with JSY-12 in individual PCR, and the resulting product was used to replace the NcoI (nt 39)-NcoI (nt 4023) fragment of pBR-150 in the correct orientation. The constructed plasmids were named pBR-V920/G1301, pBR-A974/G1301, pBR-A1020/G1301, and pBR-G1102/G1301, respectively. To make deletions from the C terminus of p150, 5′ PCR primer JSY-25 was paired with each C-terminal deletion primer (YL-23, YL-24, YL-26, YL-27, or YL-28) in individual PCRs (Table 1 and Fig. 1B). The C-terminal deletion primers were complementary to nucleotide sequences consisting of sequences encoding the desired C-terminal amino acid (H1290, V1295, P1296, L1297, or R1299) plus its upstream residues, followed by stop codon TAA and the StuI restriction site. Each of the five PCR fragments was subsequently used to replace the NcoI (nt 39)-StuI (nt 6965) fragment of pBR-150 to generate plasmid pBR-V920/H1290, pBR-V920/V1295, pBR-V920/P1296, pBR-V920/L1297, or pBR-V920/R1299.
To make protease constructs encoding sequences from nested N termini (V920, A974, A1020, or G1102) to I1773, amplification using N-terminal deletion primers (JSY-25, YL-25, YL-21, and YL-22) and the subsequent cloning were as described above for making N-terminal deletions of p150, except that pBR-NSP (encoding RV wild-type NSP, without a stop codon between p150 and p90) rather than pBR-150 was used for both the PCR template and cloning vector. The resultant plasmids were further modified to remove the C-terminal half of the p90 sequence by BglII (nt 5355)/StuI (nt 9336) digestion, end gap filling, and religation. The derived ORFs, being terminated by a stop codon at nt 9466 (numbered according to the M33 genome), encode protein products starting from residue V920, A974, A1020, or G1102 to I1773 of the NSP sequence, followed by a 43-amino-acid sequence resulting from the shift in reading frame after deletion of nt 5355 to 9336. The plasmids were named pBR-V920/I1773, pBR-A974/I1773, pBR-A1020/I1773, and pBR-G1102/I1773. All protease constructs are shown in Fig. 2.
FIG. 2.
Plasmids and protease-encoding constructs. All protease constructs were given a two-letter name; their lengths and relative positions on the RV NSP ORF are schematically shown, labeled with starting and ending protein residues (numbered according to NSP residues).
In vitro transcription.
The cDNA clones were linearized at the unique HindIII site and transcribed with SP6 RNA polymerase (Promega) in the presence of a cap analog, 7mG5′ppp5′G (Promega), using the protocol recommended by the manufacturer. RNA transcripts were extracted once with phenol-chloroform, precipitated with ethanol, and resuspended in H2O to be used for in vitro translation.
In vitro translation.
In vitro translation was performed according to the manufacturer's (Promega) protocol in 50-μl reaction mixtures containing nuclease-treated rabbit reticulocyte lysates, an amino acid mixture minus methionine, RNasin (an RNase inhibitor), and in vitro RNA transcripts in the presence of either 400 μCi of [35S]methionine (NEN) per ml or 20 μg of cold methionine per ml. Radiolabeled proteins were visualized by fluorescence autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. In vitro translation using a TNT Quick coupled transcription-translation system was performed according to the manufacture's (Promega) protocol.
Image analysis and cleavage efficiency comparison.
Image analysis was performed on a PC computer using the Scion Image program for Windows (Beta 3b) (http://www.scioncorp.com/frames/fr_download_now.htm), the PC version of the public domain NIH Image program (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/). The cleavage ratio at certain incubation times for each protease construct was calculated as the percentage of the quantity of the cleaved products with respect to the total remaining substrate and cleaved products. The cleavage ratio was plotted against the incubation time for each construct.
Sequence analysis.
Initial alignment of primary sequences for papain (SWISS-PROT accession number P00784) and the catalytic region of RV NS-pro (M33 strain [GenBank accession number S38480 with corrections as in reference 29] and Therien strain [GenBank accession number P13889 with correction as in reference 29]) was done with the ALIGN program (27) and modified manually. Secondary-structure predictions were performed using the EMBL protein structure prediction service (http://www.embl-heidelberg.de/predictprotein/predictprotein.html). The service was described by Rost et al. (32–35).
RESULTS
Processing of RV NSP by in vitro translation.
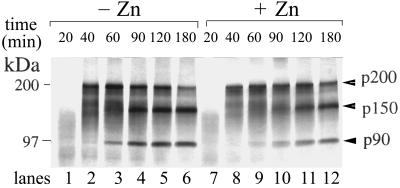
The RV NS-pro is a PCP encoded in the NSP ORF that cleaves the NSP ORF translation product (p200) at a single site to produce p150 and p90 (11). Many viral PCPs were found to be active following in vitro translation using rabbit reticulocyte lysates (4, 8, 9, 17). We therefore monitored the activity of RV-pro after translating RV NSP in vitro using the genomic-length RNA transcripts synthesized from an RV infectious cDNA clone derived from RV strain M33 (pBRM33) (43). pBRM33 was linearized with HindIII, and full-length RNA transcripts were synthesized using SP6 RNA polymerase in the presence of cap analog. In vitro translation and processing of NSP were programmed using rabbit reticulocyte lysates with synthesized RNA transcripts. A time course experiment was performed to monitor the kinetics of p200 processing. Translation of p200 was completed after 40 min (Fig. 3, lane 2); cleavage was observed at 60 min (Fig. 3, lane 3) and continued efficiently (Fig. 3, lanes 4 to 6). Liu et al. (22) suggested that activity of RV NS-pro in vitro depended on the addition of Zn2+. However, in our in vitro translation system, addition of Zn2+ was not found to be required for NS-pro activity, nor did it have a significant influence on the processing efficiency of RV NSP (Fig. 3, compare lanes 1 to 6 to lanes 7 to 12). Furthermore, using the TNT Quick coupled transcription-translation system (Promega), we also observed efficient processing of RV NSP without the addition of Zn2+ (data not shown), in contrast to the findings of Liu et al. (22) that addition of Zn2+ was essential for RV NS-pro activity in the same translation system. In addition, we found the same efficient processing of NSP from strain Therien in vitro without the addition of Zn2+, using infectious cDNA clone Robo302 (30) or its derived RNA in either the TNT transcription-translation system or rabbit reticulocyte lysate (data not shown). Liu et al. (22) used a different Therien strain cDNA construct, Robo102, and its derived subclones in their studies. However, Robo102 has a substantially lower infectivity than Robo302 (30), which might account for the observed discrepancy. Nevertheless, the exact reasons behind these contradictions need further investigation.
FIG. 3.
RV NSP processing in in vitro translation systems with or without addition of Zn2+. In vitro translation reactions with genome-length RV RNA (transcribed in vitro from pBRM33) in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of 200 μM Zn2+. Aliquots were removed at 20 (lanes 1 and 7), 40 (lanes 2 and 8), 60 (lanes 3 and 9), 90 (lanes 4 and 10), 120 (lanes 5 and 11), and 180 (lanes 6 and 12) min during incubation and subjected to SDS–8% PAGE analysis. Protein products were visualized by fluorescence autoradiography. Positions of molecular mass markers and cleavage products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
We have shown previously (44) that NS-pro can function in trans in vivo by coexpression within BHK-21 cells of a construct, p200(G1301S), that contains a cleavage site mutation (to serve as a protease) together with a construct, p200(C1152S), that contains a protease mutation (to serve as a substrate). To demonstrate the NS-pro trans-cleavage activity in vitro, three full-length mutants with alterations in the NSP ORF were constructed. pBR-200(G1301S) is a cleavage site mutant carrying a G-to-S mutation at residue 1301, pBR-200(C1152S) is a protease-inactive mutant carrying a G-to-S mutation at its catalytic C1152 residue, and pBR-150 is a mutant carrying a stop codon corresponding to residue 1302.
To assay for trans-cleavage activity of NS-pro, two separate translation reactions were carried out. Radiolabeled p200(C1152S) was synthesized in vitro in the presence of [35S]methionine to serve as a source of substrate for protease, and p200(G1301S) or p150 was synthesized in the absence of [35S]methionine to serve as a source of protease. After a 1-h incubation at 30°C, both in vitro translation reactions were terminated by the addition of RNase A and cycloheximide. When the radiolabeled p200(C1152S) was added to the unlabeled p200(G1301S) or p150 translation reaction mixture, the cleavage products of p150 and p90 were detected after a 1-h incubation (Fig. 4A and B), indicating that trans cleavage of p200(C1152S) catalyzed by p200(G1301S) or p150 protease had occurred. No cleavage product was observed when p200(C1152S) was incubated alone for 5 h (Fig. 4C). For a control, p200(G1301S) or p150 was synthesized in the presence of [35S]methionine at 30°C and incubated alone for 5 h (Fig. 4A and B, lanes 1).
FIG. 4.
RV p200(G1301S) and p150 cleave substrate protein in trans. In vitro translation was carried out as described in the legend to Fig. 3 with slight modifications. Protein products were labeled by synthesis in the presence of [35S]methionine or were unlabeled. After incubation at 30°C for 1 h, translation was terminated by the addition of RNase A and cycloheximide to final concentrations of 1 and 0.6 mg/ml, respectively. Protein product p200(G1301S) or p150 (synthesized in the absence of [35S]methionine) was mixed with radiolabeled substrate p200(C1152S) and incubated at 30°C for up to 5 h. Samples removed at various times were subjected to SDS-PAGE analysis. (A) RV p200(G1301S) was labeled in the presence of [35S]methionine and incubated alone for 5 h (lane 1). After translation reactions were terminated, unlabeled p200(G1301S) was mixed with 35S-labeled substrate p200(C1152S) and incubated at 30°C. Aliquots were removed at intervals from 0 to 5 h and subjected to SDS-PAGE analysis (lanes 2 to 7). (B) RV p150 was labeled in the presence of [35S]methionine and incubated alone for 5 h (lane 1). After translation reactions were terminated, unlabeled p150 was mixed with 35S-labeled substrate p200(C1152S) and further incubated at 30°C. Aliquots were removed from 0 to 5 h (lanes 2 to 7) and subjected to SDS-PAGE analysis. (C) In vitro transcription and translation of p200(C1152S) were carried out as described in the legend to Fig. 3. Samples were taken at the indicated times and subjected to SDS-PAGE analysis. Positions of molecular mass markers and cleavage products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Construction of truncated NS-pro cDNA clones.
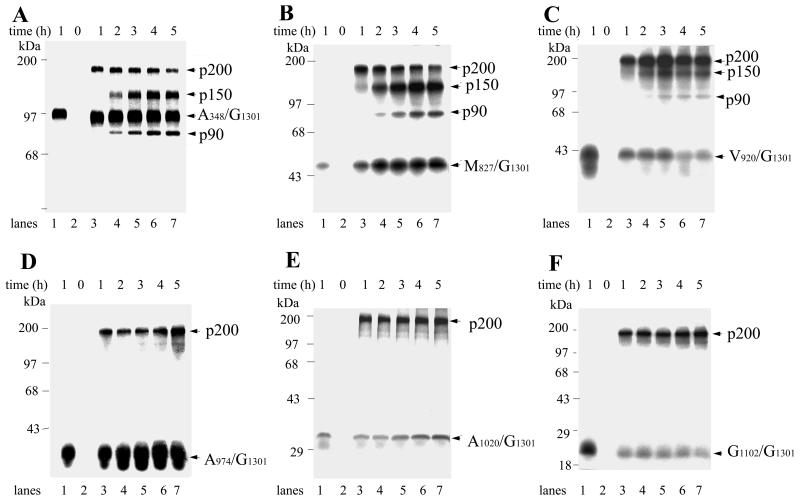
In order to identify the minimal domains required for functional protease activities, a panel of protease constructs was generated and expressed in vitro for protease activity analysis. The cDNA fragments encoding NS protease constructs were generated by PCR as described in Materials and Methods. The constructed plasmids were inserted downstream of the SP6 RNA polymerase promoter and RV 5′ untranslated region. Sequences from nt 9336 to poly(A) sequences of the RV full-length cDNA clone (pBRM33) were also preserved to provide poly(A) addition sites in these cDNA clones. The plasmids, protease products, and their relative positions on the RV NSP ORF are shown in Fig. 2. All of the protease constructs are designated by a two-letter name indicating starting and ending amino acid positions. A348/G1301, M827/G1301, V920/G1301, A974/G1301, A1020/G1301, and G1102/G1301 are protein fragments starting from A348, M827, V920, A974, A1020, and G1102, respectively, and extending to G1301, the end of p150. Fragments V920/H1290, V920/V1295, V920/P1296, V920/L1297, and V920/R1299 extend from V920 to positions H1290, V1295, P1296, L1297, and R1299, respectively. These fragments did not contain a cleavage site and were examined for their trans-cleavage capacity. Fragments V920/I1773, A974/I1773, A1020/I1773, and G1102/I1773 extend from A348, V920, A974, A1020, and G1102, respectively, to I1773 of the NSP sequence. They contain cleavage sites and C-terminal tails to be used for cis-cleavage analysis.
Defining the NS-pro domain required for trans cleavage.
To determine the trans-cleavage activity of the generated protease constructs, six protease constructs with nested N-terminal deletions (A348/G1301, M827/G1301, V920/G1301, A974/G1301, A1020/G1301, and G1102/G1301) were translated in vitro separately, producing protein products with apparent molecular masses of 102, 50, 41, 35, 30, and 21 kDa, respectively (Fig. 5, lanes 1). Each was examined for trans protease activity against substrate by cotranslation with p200(C1152S). In the cases of A348/G1301, M827/G1301, and V920/G1301, cleavage products p150 and p90 were detected after a 1-h incubation and increased with incubation time, suggesting that these constructs possess trans-cleavage activity (Fig. 5A, B, and C, lanes 2 to 6). No detectable cleavage products (p150 or p90) could be observed in the reactions of A974/G1301, A1020/G1301, and G1102/G1301 (Fig. 5D, E, and F, lanes 2 to 6), suggesting that they could not form active protease to cleave in trans. Our data suggested that the domain containing active trans protease starts at V920 or after, but at least upstream of A974.
FIG. 5.
In vitro translation of protease constructs (A348/G1301, M827/G1301, V920/G1301, A974/G1301, A1020/G1301, and G1102/G1301) and examination of their trans-cleavage activities. In vitro transcription and translation were carried out as described in Materials and Methods. A348/G1301 (A), M827/G1301 (B), V920/G1301 (C), A974/G1301 (D), A1020/G1301 (E), and G1102/G1301 (F) were translated individually to give 102-, 50-, 41-, 35-, 30-, and 21-kDa products, respectively (lanes 1). Cotranslation of each construct with substrate p200(C1152S) was carried out at 30°C for 0 to 5 h. Samples were removed at each time point and subjected to SDS-PAGE analysis (lanes 2 to 7). Positions of molecular mass markers and cleavage products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Five protease constructs (V920/H1290, V920/V1295, V920/P1296, V920/L1297, and V920/R1299) extending from V920 to different C termini were of the same molecular mass, 41 kDa, when expressed in vitro (Fig. 6, lanes 1). As described above, each of them was examined for potential trans-cleavage activity by cotranslation with p200(C1152S) for up to 5 h (Fig. 6). The appearance of cleavage products (p150 and p90) in the reactions of V920/P1296, V920/L1297, and V920/R1299 demonstrated that they preserve protease activity (Fig. 6C, D, and E, lanes 2 to 6). In contrast, neither V920/H1290 nor V920/V1295 was able to cleave substrate p200 (Fig. 6A and B, lanes 2 to 6). We therefore mapped the C terminus of the active RV NS-pro domain exactly to P1296. The weak trans-cleavage activity observed in V920/P1296, V920/L1297, and V920/R1299 could be due to the absence of the X domain in these constructs (see “Effect of N-terminal regions on cleavage efficiency” below).
FIG. 6.
In vitro translation of protease constructs (V920/H1290, V920/V1295, V920/P1296, V920/L1297, and V920/R1299) and examination of their trans-cleavage activities. In vitro transcription and translation were as described in Materials and Methods. V920/H1290 (A), V920/V1295 (B), V920/P1296 (C), V920/L1297 (D), and V920/R1299 (E) were translated in vitro separately to yield a 40-kDa product (lanes 1). Cotranslation with substrate p200(C1152S) was carried out for 0 to 5 h (lanes 2 to 7), and samples were subjected to SDS-PAGE analysis. Positions of molecular mass markers and cleavage products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Domains required for cis cleavage.
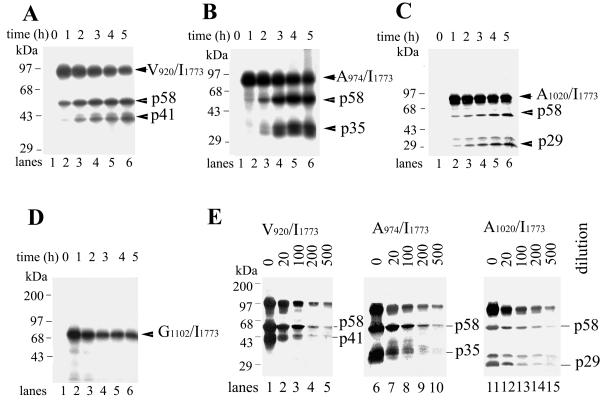
RV NS-pro is known to possess both trans- and cis-cleavage activities (44). The cotranslation experiments described above identified the NS-pro domain required for trans cleavage within the fragment from V920 to P1296. This domain may or may not be the exact domain required for cis cleavage. Since the C terminus of the cis protease construct must extend beyond the cleavage site, we therefore examined the N-terminal domain requirement for cis cleavage by analysis of in vitro translation of the protease constructs V920/I1773, A974/I1773, A1020/I1773, and G1102/I1773. Processing of these protein constructs would accumulate a cleavage product with an apparent molecular mass of 58 kDa (p58, the C-terminal fragment extending from residue 1302 to 1773) throughout the incubation time. We found that V920/I1773 and A974/I1773 underwent efficient processing. V920/I1773 was translated as a 98-kDa polyprotein, which was then cleaved into 58- and 41-kDa products within a 1-h incubation (Fig. 7A). A974/I1773 was translated as a 92 kDa product and cleaved into 58- and 35-kDa products efficiently (Fig. 7B). Self-cleavage of A1020/I1773 (87 kDa) into 58- and 29-kDa products was less efficient (Fig. 7C). The band above p29 seems to be a translation by-product rather than a cleavage product, since its amount did not increase with time as the p29 band did. No cleavage could be observed in the case of G1102/I1773 (78 kDa) (Fig. 7D). Of these constructs, only V920/I1773 contains a protease domain (V920 to G1301) that can cleave in trans at low efficiency (Fig. 5C). Therefore, the highly efficient processing of V920/I1773 represents cis-cleavage activity rather than trans-cleavage activity. The other two constructs, A974/I1773 and A1020/I1773, do not contain the necessary domain (V920 to R973) for trans cleavage and can function only in cis. To confirm that constructs V920/I1773, A974/I1773, and A1020/I1773 function in cis, a dilution experiment was performed (Fig. 7E). Translation reactions with V920/I1773 (Fig. 7E, lanes 1 to 5), A974/I1773 (Fig. 7E, lanes 6 to 10), and A1020/I1773 (Fig. 7E, lanes 11 to 15) were carried out using serial dilutions (0, 1:20, 1:100, 1:200, and 1:500) of each RNA transcript. For each of them, the total translation products decreased correspondingly when more-diluted RNA was added. However, cleavage products were clearly demonstrated, and the cleavage ratio (determined as described in Materials and Methods) remained roughly unchanged from that for to nondiluted samples, suggesting that these cleavages occur in cis. Our results suggest that a construct starting from A1020 to a residue after the cleavage site such as to include the N terminus of p90 is sufficient for cis processing. Comparison between domains required for cis- and trans-cleavage activities of NS-pro indicated that the domains involved in cis- and trans-cleavage activities are different. Obviously, the cis protease domain must contain a cleavage site and C-terminal tail for cis cleavage to occur. This is not the case for the trans protease domain. However, it is interesting that the N-terminal domains are different between cis and trans cleavage for RV NS-pro. The domain from V920 to A1020 is required for trans cleavage but is dispensable for cis cleavage. It will be of interest to examine the functions of the domain from V920 to A1020 in trans cleavage.
FIG. 7.
Autolytic processing of protease constructs V920/I1773, A974/I1773, A1020/I1773, and G1102/I1773. V920/I1773 (A), A974/I1773 (B), A1020/I1773 (C) and G1102/I1773 (D) were transcribed and translated as described in Materials and Methods for 5 h in the presence of [35S]methionine. Samples were removed at the indicated times and subjected to SDS-PAGE analysis. (A) V920/I1773 was translated as a 98-kDa protein and subsequently processed into 58- and 41-kDa fragments. Positions of V920/I1773, p58, and p41 are indicated by arrows. (B) A974/I1773 gave a 92-kDa product, which was autocleaved into p58 and p35, as indicated by arrows. (C) A1020/I1773 was translated into an 87-kDa product, whose processing generated p58 and p29 as indicated. (D) G1102/I1773 gave a 78-kDa protein, whose autolytic processing was undetectable. (E) Translation reactions of V920/I1773, A974/I1773, and A1020/I1773 were each programmed with input RNA at 0, 1:20, 1:100, 1:200, and 1:500 dilutions, and mixtures were incubated for 4 h. Positions of molecular mass markers and cleavage products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Effect of N-terminal regions on cleavage efficiency.
The protease constructs examined in this study were found to have variable cleavage efficiencies, depending on the region and length deleted. Cleavage efficiency was compared among different protease constructs by using the percentage of cleaved products with respect to total proteins, expressed as a cleavage ratio. The cleavage ratio for each protease construct at certain incubation time was calculated as described in Materials and Methods and plotted against time.
Figure 8A compares the trans-cleavage efficiency among positive controls [p200(G1301S) and p150], A348/G1301, M827/G1301, V920/G1301, V920/P1296, V920/L1297, and V920/R1299. For p200(G1301S) and p150, the function time began when the protease and substrate were mixed. However, for A348/G1301, M827/G1301, V920/G1301, V920/P1296, V920/L1297, and V920/R1299, the protease functioned only after it had been translated, which took about 40 to 60 min. Therefore, the effective function time was taken as the real incubation time minus 60 min. The eight protease constructs could be separated into two groups according to their proteolytic activity: one group with high cleavage ratios (70 to 90%), including p200(G1301S), p150, A348/G1301, and M827/G1301, and the other group with low cleavage ratios (5 to 17%), including V920/G1301, V920/P1296, V920/L1297, and V920/R1299. A348/G1301 and M827/G1301 had cleavage efficiencies comparable to those of p200(G1301S) and p150, the positive controls for trans-cleavage activity, whereas V920/G1301, differing from M827/G1301 in lacking an X domain, had a substantially lower cleavage ratio (17%) than M827/G1301 (82%). These results suggest an important role of the X domain in trans cleavage.
FIG. 8.
Cleavage efficiencies of protease constructs. Protein bands of substrate and cleavage products on SDS-PAGE gel were quantitated using the Scion image program. The cleavage ratio for each protease construct was calculated as described in Materials and Methods and plotted against incubation time. (A) trans-cleavage efficiency comparisons. ◊, p200(G1301S); ⧫, p150; ○, A348/G1301; ×, M827/G1301; ●, V920/G1301; ▵, V920/P1296; ▴, V920/L1297; ■, V920/R1299. (B) cis-cleavage efficiency comparisons. ⧫ and ○, wild-type NSP in the absence or presence, respectively, of Zn2+; ▴, V920/I1773; ∗, A974/I1773; □, A1020/I1773.
As discussed above, the processing of V920/I1773 is largely by cis cleavage, and the processing of A974/I1773 or A1020/I1773 is the consequence of cis cleavage only. Therefore, the processing efficiencies of V920/I1773, A974/I1773, and A1020/I1773 (Fig. 8B) reflected their respective cis-cleavage abilities. The constructs compared in Fig. 8B can be classified into two groups: one with high processing ratios (60 to 70%), including wild-type NSP, V920/I1773, and A974/I1773, and the other (A1020/I1773) with a processing ratio as low as 35%. The fact that V920/I1773 and A974/I1773 had as efficient cis cleavage as wild-type NSP suggested that the lack of and X domain in V920/I1773 and A974/I1773 had no significant influence on their self-processing. However, the domain from residue 974 to 1020, although not required absolutely, had a substantial effect on cis cleavage.
Secondary-structure prediction for RV NS-pro.
Gorbalenya et al. (14) reported sequence similarity between papain, a cellular cysteine protease, and RV NS-pro in the vicinity of catalytic C and H residues as determined through local alignment. The catalytic C and H residues are separated by 133 residues in papain and 120 residues in RV NS-pro (14). There are 24 residues upstream of the catalytic C residue in papain. The active RV NS-pro domain identified in this work was larger than papain, with about 230 residues upstream of the catalytic C1152 that are required for trans cleavage or about 130 residues upstream of C1152 that are required for cis cleavage. It has been reported that through sequence alignment and secondary-structure comparison to known protein structures, topologic prediction of uncharacterized proteins is possible (38). Skern et al. (38) proposed a papain-like fold for the FMDV Lpro, a viral cysteine protease, from the analyses of predicted secondary structure. This prediction was confirmed by a recent crystallographic analysis of FMDV Lpro showing a globular papain-like catalytic domain with adaptation for the specific requirements of the virus (16). In the hope of obtaining initial structural information on RV NS-pro, we compared the primary and secondary structures of RV NS-pro to those of papain. The analyses were performed with strains M33 and Therien, both of which are wild-type isolates of RV and differ from each other by two residues (A1140V and R1201W, M33 versus Therien) within the examined NS-pro catalytic region (from residue 1128 to 1301), with identical results. Only the result for RV strain M33 is presented here (Fig. 9).
FIG. 9.
Comparison of primary and secondary structures between RV NS-pro and papain. The protein sequence of papain (SWISS-PROT accession number P00784) was aligned to that of NS-pro (strain M33; GenBank accession number S38480 with corrections as in reference 29) using ALIGN software with manual adjustment. The papain sequence is numbered as for mature protease, and the RV-NS-pro sequence is numbered as for the NSP ORF. :, identical residues; ., conserved amino acids. Cys and His at the catalytic sites are in boldface and shaded. Features of the secondary structure of papain (20) are illustrated above the sequence. Secondary structure for RV NS-pro was calculated by the EMBL protein prediction server (32–35) and is illustrated below the NS-pro sequence. α-helices are shown as curves, and β-sheets are shown as arrows. α-helices are named according to the nomenclature of Kamphuis et al. (20); β-sheets are named as described by Skern et al. (38).
To determine the global similarity between papain and RV NS-pro with respect to their catalytic sites, sequence alignment between papain and the RV NS-pro catalytic region (residues 1128 to 1296) was made using the ALIGN program (27) with manual modification (Fig. 9). The alignment gave an identity of 18.1%, and, as expected, the two sequences exhibited most similarity around C and H, the catalytic sites. The derived alignment was further supported by the analysis of the predicted secondary structure of RV NS-pro (Fig. 9). RV NS-pro was predicted to have the α-β structural organization found in cellular PCPs. This prediction has three α-helices (αL1, αL2/3, and αR1) and six β-sheets (A to F) in RV NS-pro. Most of these were present in the papain structure at corresponding, positions including βA, αL1, αR1, βC, βD, βE, and βF (20, 21). The match was highest in the catalytic C and H regions, with differences occurring in the linker regions and the C end. In NS-pro, an α-helix, αL2/3, took the place where two helices, αL2 and αL3, occurred in papain. βB1 and βB2 for NS-pro did not match the position of βB for papain well. Furthermore, the αR2 in the linker region and βG at the C end were missing in the NS-pro prediction. RV NS-pro had a shorter linker region (120 residues) between catalytic C and H residues than papain (133 residues) and a shorter tail after the catalytic H residue (23 residues) than papain (53 residues), which explained the discrepancies. It was proposed (38) that loops between the secondary elements that define the papain topology can be modified, by insertions or deletions, without interfering with the overall folding of the molecule. Therefore, the global similarity of their secondary structures suggested that RV NS-pro might maintain a papain-like topology for its catalytic region, whereas those differences may come from the adaptive changes for the viral specificity. Crystallographic data are necessary for precise structure determination for RV NS-pro.
DISCUSSION
We have used an in vitro translation system to identify domains important for cis- and trans-cleavage activities of RV NS-pro. The results are summarized in Fig. 10. Through analysis of protease activity using RNA transcripts from cloned material with serial deletions from either end of p150, we have demonstrated that RV NS-pro requires a region from residue 920 to 1296 to perform functional trans cleavage (Fig. 5 and 6). The N-terminal region of NS-pro was roughly determined to reside between residues 920 and 974 (Fig. 5C and D). The C end was precisely determined to be P1296 (Fig. 6B and C). However, the minimal NS-pro domain (residues 920 to 1296) for trans cleavage processed only 5 to 17% of substrate after 4 h of incubation, compared to the 70 to 96% for the positive controls, p200(G1301S) and p150 (Fig. 8A). The minimal NS-pro domain that maintains as high trans-cleavage ability as the positive controls is found in the construct M827/G1301 (80% cleavage at 4 h), starting from around residue M827 (Fig. 8A).
FIG. 10.
Functional domains of RV NS-pro and comparison with complete papain sequence. Residues of RV NS-pro are numbered as for the RV NSP ORF. The X domain (residues 834 to 940) is shown by the dotted line, positioned outside the essential protease domains of NS-pro shown as a solid heavy line. The essential domains for either trans- or cis-cleavage activity are indicated by the arrows starting from residue 920 or 1020. The region from residue 1128 to 1301 is compared to the mature papain protease in this work, and the boundary is indicated by vertical dotted lines. The predicted catalytic residues Cys and His of RV NS-pro and catalytic residues Cys and His of papain are represented by open squares. For a complete papain sequence, residues 1 to 18 encode the signal peptide shown as a heavy line, residues 19 to 133 are the pro region, and residues 134 to 345 contain the mature papain sequence.
RV NS-pro possesses both cis and trans activities (44). Both use the same cysteine protease cleavage mechanism and thus should employ the same core catalytic structure (19). They may vary in other external domain requirements. In addition to the protease domain, the cis protease must include intact cleavage site and substrate regions. The most significant difference between cis and trans cleavage for RV NS-pro lies at the N-terminal domains. When a panel of protease constructs with nested N-terminal deletions (V920/I1773, A974/I1773, A1020/I1773, and G1102/I1773), with weak or no trans-cleavage activity, were examined for autoprocessing, V920/I1773, A974/I1773, and A1020/I1773 were active in cis, while G1102/I1773 was not (Fig. 7). Thus, protease active in trans starts after residue 920 but before residue 974, while that in cis begins after residue 1020. These results suggested that the core protease domain (required for both cis and trans activity) ranges from around residue 1020 to residue 1296, and that the fragment from residue 920 to 1020 is required only for trans cleavage while being dispensable for cis processing (Fig. 10). Our data are the first to show that RV NS-pro uses different domains for cis and trans cleavage. Since trans cleavage is a bimolecular interaction, it is likely that the domain of residues 920 to 1020 is involved in protein-protein interaction, required to position substrate protein into the trans protease catalytic site. For cis cleavage, this protein-protein interaction is not necessary in order to hold substrate and enzyme together. Identification of the trans-specific domain facilitates future studies on the biological significance of trans-cleavage activity.
Sequence analysis on M-group PCPs of several virus families (such as alphavirus and coronavirus, etc.) had identified a conserved X domain near the protease domain (14, 39). In RV, this X domain lies N-terminal to the protease domain, ranging from residue 834 to 940 (Fig. 10) (14). Functions of the X domain remain to be characterized. Association of the X domain with M-group PCPs (possessing both cis and trans activity) rather than L-group PCPs (containing only cis activity) encouraged the speculation that the X domain might be involved in the regulation of polyprotein processing (14). Elimination of the X domain from PLP-1 of mouse hepatitis virus reduced cleavage by 22 to 63% (3, 4, 41). In our experiments, RV NS-pro remained enzymatically active after all or most of the X domain had been removed. V920/G1301 cleaved substrate in trans, and A974/I1773 processed itself efficiently (Fig. 5C and 7B). However, cleavage efficiencies differed considerably. In trans cleavage, the absence of the X domain in V920/G1301 caused a substantially decreased cleavage ratio (17%) compared to that for M827/G1301 (82%), which contains the X domain (Fig. 8A). In contrast, cis cleavage was not affected significantly by the presence of the X domain, since V920/I1773 and A974/I1773 (both missing the X domain) processed themselves almost as efficiently as the positive control, wild-type NSP (Fig. 8B). Our results demonstrate the importance of the X domain in trans cleavage. Although it is unclear at present what function it could play in RV NS-pro trans cleavage, we speculate that this proline-rich region might provide a protein-protein interaction domain that enhances the opportunity for protease to meet its trans cleavage substrate and thus decreases the Km of protease. Further studies of the biologic significance and functional mechanism of the X domain in NSP processing and virus replication are indicated.
The PCP family include a group of cellular and viral proteases which employ the catalytic C and H dyad. The distant relationship between viral and cellular PCPs was suggested from many primary sequence comparisons (2, 14) and from the crystal structure of FMDV Lpro, the only structure determined on a viral PCP (16). Sequence alignment showed that the catalytic region of RV NS-pro (from residue 1128 to 1296) has global sequence similarity with papain (Fig. 9). Secondary-structure comparison also supported their topologic relationship (Fig. 9). It is possible that the catalytic region of RV NS-pro exhibits a papain-like folding with adapted modifications. To obtain the full tertiary structural information of RV NS-pro will require crystallographic analysis.
The additional N-terminal region of the RV NS-pro core domain (from A1020 to A1127) has no corresponding sequence in papain and was excluded from alignment with it. It is likely that this N domain may not contain sequences directly required for protease activity. Rather, it may serve other subsidiary functions, such as folding assistance, conformational stability, and/or protein-protein interactions. Papain, as well as other proteases, is translated as a proprotease with an additional N-terminal region (115 residues of pro region for papain) (SWISS-PROT accession number P00784) (Fig. 10). In many proteases, the pro region plays an active role in protein folding. Subtilisin (6), α-lytic protease (1), carboxypeptidase A1 (28), and carboxypeptidase Y (31) do not fold into active conformations in the absence of their pro regions. The pro region is essential for folding of at least one PCP (cathepsin L) (40). The pro regions of PCPs can also perform other biologic roles, such as stabilization (23, 40) and subcellular targeting (24, 26). It would be interesting to determine whether the region from A1020 to A1127 of RV NS-pro serves subsidiary roles similar to those of the pro regions for many other proteases.
In summary, our work identifies the domains required by RV NS-pro for trans and cis cleavage (Fig. 10). Both cleavages require a core catalytic domain from A1020 to P1296, while containing different N and C ends. Protease cleaving in trans needs an additional domain (V920 to A1020) at the N end, while cis protease contains a four-residue linker, the cleavage site G1301, and the substrate region at the C end. We also demonstrated that the X domain is important in trans cleavage of RV NS-pro but has no significant influence on cis cleavage. Defining the regions and roles of protease-related domains of RV NS-pro clarifies our understanding of this specific viral PCP and provides a basis for comparison with other protease members. A remote homology to papain is noticed from primary sequence analysis and predicted secondary structure, suggesting that the RV NS-pro catalytic region has a papain-like topology. Crystallographic data are needed for a precise three-dimensional structure determination.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada. Y.L. is supported by a studentship from the British Columbia Children's Hospital Foundation. S.G. is an investigator of the British Columbia Children's Hospital Foundation.
We thank T. K. Frey for providing Therien strain infectious clone Robo302 and for helpful discussions.
REFERENCES
- 1.Baker D, Sohl J L, Agard D A. A protein-folding reaction under kinetic control. Nature. 1992;356:263–265. doi: 10.1038/356263a0. [DOI] [PubMed] [Google Scholar]
- 2.Berti P J, Storer A C. Alignment/phylogeny of the papain superfamily of cysteine proteases. J Mol Biol. 1995;246:273–283. doi: 10.1006/jmbi.1994.0083. [DOI] [PubMed] [Google Scholar]
- 3.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like protease of the murine coronavirus mouse hepatitis virus A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonilla P J, Hughes S A, Pinon J F, Weiss S R. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 5.Bowden D S, Westaway E G. Rubella virus structural and nonstructural proteins. J Gen Virol. 1984;65:933–943. doi: 10.1099/0022-1317-65-5-933. [DOI] [PubMed] [Google Scholar]
- 6.Bryan P, Wang L, Hoskins J, Ruvinov S, Strausberg S, Alexander P, Almog O, Gilliland G, Gallagher T. Catalysis of a protein reaction: mechanistic implications of the 2.0 A structure of the subtilisin-prodomain complex. Biochemistry. 1995;34:10310–10318. doi: 10.1021/bi00032a026. [DOI] [PubMed] [Google Scholar]
- 7.Chen J-P, Strauss J H, Strauss E G, Frey T K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol. 1996;70:4707–4713. doi: 10.1128/jvi.70.7.4707-4713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi G H, Pawlyk D M, Nuss D L. The autocatalytic protease p29 encoded by hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology. 1991;183:747–752. doi: 10.1016/0042-6822(91)91004-z. [DOI] [PubMed] [Google Scholar]
- 9.Den Boon J A, Faaberg K S, Meulenberg J J M, Wassenaar A I M, Plagemann P G W, Gornalenya A E, Snijder E J. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papain-like cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez G, Wang C Y, Frey T K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during Togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forng R-Y, Frey T K. Identification of the rubella virus nonstructural proteins. Virology. 1995;206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- 12.Francki R I B, Fauquet C M, Knudson D L, Brown F, editors. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. Suppl. 2. Vienna, Austria: Springer-Verlag; 1991. [Google Scholar]
- 13.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorbalenya A E, Koonin E V, Lai M M-C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 16.Guarne A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy W R, Strauss J H. Processing the nonstructural polyproteins of Sindbis virus: nonstructural protease is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:998–1007. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerase from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamphuis I G, Drenth J, Baker E N. Thiol proteases. Comparative studies based on the high-resolution structures of papain and actinidin, and on amino acid sequence information for cathepsins B and H, and stem bromelain. J Mol Biol. 1985;182:317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- 20.Kamphuis I G, Kalk K H, Swarte M B A, Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984;179:233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- 21.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Ropp S L, Jackson R J, Frey T K. The rubella virus nonstructural protease requires divalent cations for activity and functions in trans. J Virol. 1998;72:4463–4466. doi: 10.1128/jvi.72.5.4463-4466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mach L, Mort J S, Glossl J. Noncovalent complexes between the lysosomal proteinase cathepsin B and its propeptide account for stable, extracellular, high molecular mass forms of the enzyme. J Biol Chem. 1994;269:13036–13040. [PubMed] [Google Scholar]
- 24.Mao P L, Jiang Y, Wee B Y, Porter A G. Activation of caspase-1 in the nucleus requires nuclear translocation of pro-caspase-1 mediated by its prodomain. J Biol Chem. 1998;273:23621–23624. doi: 10.1074/jbc.273.37.23621. [DOI] [PubMed] [Google Scholar]
- 25.Marr L D, Wang C-Y, Frey T K. Expression of the rubella virus nonstructural protein ORF and demonstration of proteolytic processing. Virology. 1994;198:1–7. doi: 10.1006/viro.1994.1070. [DOI] [PubMed] [Google Scholar]
- 26.McIntyre G F, Erickson A H. The lysosomal proenzyme receptor that binds procathepsin L to microsomal membranes at pH5 is a 43-kDa integral membrane protein. Proc Natl Acad Sci USA. 1993;90:10588–10592. doi: 10.1073/pnas.90.22.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers E, Miller W. Optimal alignments in linear space. CABIOS. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 28.Phillips M A, Rutter W J. Role of the prodomain in folding and secretion of rat pancreatic carboxypeptidase A1. Biochemistry. 1996;35:6771–6776. doi: 10.1021/bi960113o. [DOI] [PubMed] [Google Scholar]
- 29.Pugachev K V, Abernathy E S, Frey T K. Genomic sequence of the RA27/3 vaccine strain of rubella virus. Arch Virol. 1997;142:1165–1180. doi: 10.1007/s007050050150. [DOI] [PubMed] [Google Scholar]
- 30.Pugachev K V, Abernathy E S, Frey T K. Improvements of the specific infectivity of the rubella virus (RUB) infectious clone: determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J Virol. 1997;71:562–568. doi: 10.1128/jvi.71.1.562-568.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos C, Winther J R, Kielland-Brandt M C. Requirement of the propeptide for in vivo formation of active yeast carboxypeptidase Y. J Biol Chem. 1994;269:7006–7012. [PubMed] [Google Scholar]
- 32.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 35.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. CABIOS. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 36.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the “Sindbis-like” supergroup of positive-strand RNA virus. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Skern T, Fita I, Guarne A. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J Gen Virol. 1998;79:301–307. doi: 10.1099/0022-1317-79-2-301. [DOI] [PubMed] [Google Scholar]
- 39.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao K, Stearns N A, Dong J, Wu Q L, Sahagian G G. The proregion of cathepsin L is required for proper folding stability, and ER exit. Arch Biochem Biophys. 1994;311:19–27. doi: 10.1006/abbi.1994.1203. [DOI] [PubMed] [Google Scholar]
- 41.Teng H, Pinon J D, Weiss S R. Expression of murine coronavirus recombinant papain-like proteinase: efficient cleavage is dependent on the lengths of both the substrate and the proteinase polypeptides. J Virol. 1999;73:2658–2666. doi: 10.1128/jvi.73.4.2658-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolinsky J S. Rubella. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lipincott-Raven Publishers; 1996. pp. 899–921. [Google Scholar]
- 43.Yao J, Gillam S. Mutational analysis, using a full-length rubella virus cDNA clone, of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J Virol. 1999;73:4622–4630. doi: 10.1128/jvi.73.6.4622-4630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao J, Yang D, Chong P, Hwang D, Liang Y, Gillam S. Proteolytic processing of rubella virus nonstructural proteins. Virology. 1998;246:74–82. doi: 10.1006/viro.1998.9179. [DOI] [PubMed] [Google Scholar]