Abstract
Viral protein R (Vpr) of human immunodeficiency virus type 1 is produced late in the virus life cycle and is assembled into the virion through binding to the Gag protein. It is known to play a significant role early in the viral life cycle by facilitating the nuclear import of the preintegration complex in nondividing cells. Vpr is also able to interact with nucleic acids, and we show here that it induces condensation of plasmid DNA. We have explored the possibility of using these properties in DNA transfection experiments. We report that the C-terminal half of the protein (Vpr52–96) mediates DNA transfection in a variety of human and nonhuman cell lines with efficiencies comparable to those of the best-known transfection agents. Compared with polylysine, a standard polycationic transfection reagent, Vpr52–96 was 10- to 1,000-fold more active. Vpr52–96-DNA complexes were able to reach the cell nucleus through a pH-independent mechanism. These observations possibly identify an alternate pathway for DNA transfection.
The accessory genes of lentiviruses are not essential for viral replication in tissue culture but can be critical for the establishment of a productive infection in their natural hosts. The vpr gene is found in human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus and encodes the 15-kDa viral protein R (Vpr), which is produced late in the virus life cycle and is assembled into the virion through binding to Gag (11, 38, 44). A variety of activities have been associated with Vpr. It enhances the replication of HIV-1 in lymphocytes and monocyte-derived cell lines (9, 30), it is a weak transcriptional activator of several viral promoters (1, 8, 17, 45, 50), it causes host cell arrest in the G2/M phase of the cell cycle (14, 20, 25, 41, 42), and it is a coactivator of the human glucocorticoid receptor (27). Vpr has also been implicated in transport of the viral preintegration complex (PIC) into the nucleus, a property that may help HIV-1 infect nondividing cells (18, 21, 39, 40, 48). In the absence of other HIV-1 proteins, Vpr is localized predominantly in the nucleus, although it does not contain a typical nuclear localization signal (12, 31).
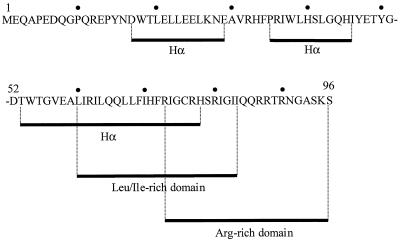
Genetic and structural studies have assigned the various functions of Vpr to overlapping domains within the molecule. Nuclear accumulation of Vpr was reported to depend on the presence of α-helices in both the N- and C-terminal halves of the protein (Fig. 1) (34, 46). The N-terminal helices are believed to be involved in incorporation of Vpr into virions. The C-terminal domain, which is rich in basic amino acids, contains elements essential for G2 arrest (33, 34) and nucleic acid binding activities (10, 55).
FIG. 1.
Vpr structure. Vpr1–96 is characterized by the presence of an α-helix (Hα)–turn–α-helix motif (residues 17 to 29 and 35 to 46) in the N-terminal region (51) and by an α-helix in the C-terminal domain (residues 53 to 78) (43). The Leu- and Ile-rich domain (residues 60 to 84) overlaps partially with both the α-helix of the C-terminal region and the arginine-rich region (residues 73 to 96). Of note, Vpr52–96 presents three negative charges and 9 to 11 positive charges, depending on the pH (histidine residues are positively charged at about pH 6).
The ability of Vpr to interact with nucleic acids and to enter the nucleus, either in the context of PICs or independent of other viral proteins, prompted us to test its ability to deliver plasmid DNA into mammalian cells. An efficient carrier of DNA for transfections must package the DNA into compact particles which can be readily taken up by cells. It must also protect DNA from cellular nucleases, allow its release from endocytic vesicles, and favor its nuclear import. While existing carriers can fulfill some of these criteria, none of them provides all of the necessary functions. The nuclear import of DNA remains the major hurdle for efficient transfection (6, 13, 53).
We report here that the C-terminal domain of Vpr (Vpr52–96), but not the N-terminal part, is able to interact with plasmid DNA and that it mediates DNA transfection with efficiencies comparable to those of the best transfection reagents.
MATERIALS AND METHODS
Plasmids.
SMD2-LucΔITR (7.6 kb) and PCIneo-Luc (7.3 kb) are expression plasmids encoding the firefly luciferase gene under the control of the human cytomegalovirus (CMV) immediate-early promoter. eGFP-C1 (4.7 kb; Clontech) encodes, under the control of the CMV promoter, a red-shifted variant of the green fluorescent protein (GFP) which has been optimized for brighter fluorescence and higher-level expression in mammalian cells. The pgk-LacZ and CMV-LacZ constructs are plasmids expressing LacZ under the control of the phosphoglycerate kinase promoter and the CMV promoter, respectively.
Peptides.
Peptide Vpr1–96 from HIV-1 strain LAI, with the sequence MEQA PEDQGPQREPYNDWTLELLEELKNEAVRHFPRIWLHSLGQHIYETYG DTWTGVEALIRILQQLLFIHFRIGCRHSRIGIIQQRRTRNGASKS, was used. The peptides were synthesized as described previously (11). Electrospray mass spectrometry was used to confirm the identities of peptides Vpr1–51 (theoretical molecular mass [MMth] = 6,165.81; calculated molecular mass [MMcalc] = 6,167), Vpr52–96 (MMth = 5,247.3; MMcalc = 5,249), Vpr1–96 (MMth = 11,394.9; MMcalc = 11,392.6), Vpr70–96 (MMth = 3,148.66; MMcalc = 3,148), Vpr80–96 (MMth = 1,941.23; MMcalc = 1,941), Vpr60–80 (MMth = 2,577.18; MMcalc = 2,578), and Arg80Vpr52–96Ala80 (MMth = 5,162.02; MMcalc = 5,162). Peptides Vpr52–75, Vpr77–96, and Vpr52–93 were synthesized by SYNTEM. The peptides were stored at −80°C as 1-mg/ml solutions in MilliQ water.
Cell culture.
The culture medium Dulbecco's modified Eagle medium (DMEM; GIBCO-BRL) was supplemented with 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% heat-inactivated fetal calf serum (FCS; HyClone). We used human hepatocarcinoma cells (HepG2; American Type Culture Collection [ATCC]), human epithelioid carcinoma cells (HeLa 229; ATCC), human embryonic retinoblasts (cell line 911; kindly given by Introgene), transformed human embryonic kidney cells (cell line 293; ATCC), and Swiss mouse embryo cells (NIH 3T3; ATCC).
DNA retardation assay and sensitivity of the polyplexes to DNase I.
DNA binding was studied by means of agarose gel retardation assays. Plasmid DNA (1 μg) and increasing amounts of peptide were each diluted in 25 μl of 150 mM NaCl and then mixed together. After a period of 25 min, samples (25 μl) were electrophoresed through a 1% agarose gel, using Tris-borate-EDTA buffer, and DNA was visualized after ethidium bromide staining.
To evaluate the sensitivity of the Vpr52–96-DNA complexes to DNase I digestion, naked DNA (1 μg) and preformed complexes were mixed with 10 U of DNase I in a total volume of 30 μl containing 50 mM of Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM ATP. After a 1-h incubation at 37°C, 3 μl of a 10% sodium dodecyl sulfate (SDS) solution was added to each sample. Each sample was extracted with 30 μl of phenol, and the aqueous phase was loaded onto a 1% agarose gel to examine the integrity of the plasmid DNA.
Transfection experiments.
Polylysine hydrobromide (pLys) with a degree of polymerization of about 180 was from Sigma, 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) was from Avanti Polar Lipids Inc., and polyethylenimine (PEI; 25 kDa) was from Aldrich. The PEI solution was prepared as described elsewhere (2). In some experiments, Vpr52–93 was used instead of Vpr52–96; the two peptides have indistinguishable transfection properties. Four micrograms of plasmid DNA and the desired amount of peptide, DOTAP, PEI, or pLys were added to 100 μl of 150 mM NaCl and gently mixed. After 25 min of incubation, the mixture was diluted with serum-free medium to a final volume of 1 ml; 0.5 ml of the transfection mixture was then put in each well of duplicate plates. A total of 100,000 to 300,000 cells, depending on the cell type, were plated in each well of 24-well plates (Nunc) 1 or 2 days before transfection. For all experiments, the final transfection volume was 0.5 ml per well. After about 3 h, the transfection medium was replaced with fresh medium containing 10% FCS. Each experiment was carried out several times; within a series, experiments were done in duplicate. The transfection experiments involving chloroquine (Sigma; final concentration, 100 μM) and bafilomycin A1 (Sigma; final concentration, 50 to 175 nM) were done as described above except that the drug was added after dilution of the complexes with DMEM, just prior to the addition of the transfection medium to the cells.
For luciferase activity determinations, cells were harvested after 24 to 48 h in 250 μl of lysis buffer (8 mM MgCl2, 1 mM dithiothreitol, 1 mM EDTA, 1% Triton X-100, 15% glycerol, and 25 mM Tris-phosphate buffer [pH 7.8]). The cell lysate was then transferred into Eppendorf tubes and centrifuged for 5 min at 10,000 × g to pellet debris. From an aliquot of the supernatant (50 μl), luciferase activity (in light units) was measured in a 96-well plate format with a PhL luminometer (Mediators Diagnostika) with 10-s integration after automatic injection of 100 μl of assay buffer (lysis buffer without Triton X-100 but supplemented with 2 mM ATP) and 100 μl of a luciferin solution (167 μM, in water; Molecular Probes). Background luciferase activity (300 light units) was subtracted from each value, and the transfection efficiency was expressed as light units per 10 s per well (with 1 light unit being equivalent to 10 counts); the values presented are means of duplicate determinations.
β-Galactosidase activity was measured by a chemiluminescence method (Tropix). For GFP expression measurements, the cells were trypsinized 24 to 48 h after transfection and analyzed by flow cytometry (FACSCalibur; Becton Dickinson).
Association and/or endocytosis of Vpr52–96-DNA complexes.
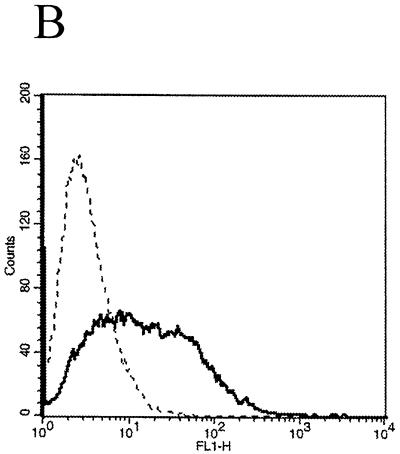
Plasmid DNA was incubated with the fluorescent intercalating dye YOYO-1 (Molecular Probes; about one dye molecule/300 bp), and Vpr52–96–DNA complexes were prepared as described above. The fluorescent complexes were added to 293 cells, which were then incubated at 37°C for 4 h. The cells were washed twice with cold phosphate-buffered saline (PBS) and harvested in PBS–1 mM EDTA. Cells were then analyzed by flow cytometry.
Cell proliferation assay, protein quantification, and cell cycle study.
One day after transfection, the cell culture medium was removed and replaced by serum-free DMEM containing 0.5 mg of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT; Sigma)/ml. The insoluble blue formazan crystals that formed after 3 to 4 h of incubation were dissolved in 240 μl of an acidified isopropanol-SDS mixture (200 μl of isopropanol-HCl and 40 μl of 3% SDS in water), and the absorbance at 570 nm was measured. Untreated cells were used as a control (100% absorbance). The protein content of the transfected cells was measured by the Bradford dye-binding method, using the Bio-Rad protein assay. For cell cycle analysis, cells were washed, trypsinized, centrifuged, and resuspended in 1 ml of PBS containing 30 μg of propidium iodide (Sigma), 0.02% saponin, and RNase A (10 μl of a 10-mg/ml solution). The cellular DNA content was analyzed by flow cytometry after 30 min of incubation in the dark at room temperature.
Southern blot analysis.
293 cells were transfected with 3 μg of plasmid SMD2-LucΔITR in six-well plates, washed once with PBS, trypsinized, centrifuged, and washed again with PBS. Selective low-molecular-weight DNA extraction was performed according to the method of Hirt (22). After linearization of the recovered DNA, the samples were electrophoresed on a 1% agarose gel. After transfer of the samples to a nylon membrane (Biodyne B; Pall), hybridization was carried out with the linearized SMD2-LucΔITR construct labeled with alkaline phosphatase (AlkPhos kit; Amersham). The specific bands were detected after addition of the enhanced chemifluorescent substrate by chemifluorescent imaging (Storm; Molecular Dynamics).
Electron microscopy.
Vpr peptides at a final concentration of 0.08 μg/μl (11.6 μM for the Vpr52–96 and 25 μM for Vpr77–96) were mixed with 0.02 μg of plasmid DNA (3.4 nM)/ml in a final volume of 50 μl of 150 mM NaCl. Five microliters of the mixture was deposited onto an electron microscope grid covered with a thin carbon film previously activated by a glow discharge in the presence of pentylamine. The grids were then stained with 2% aqueous uranyl acetate, drained, and blotted. The observations were done in the annular dark-field mode with a Zeiss model 902 electron microscope, filtering out inelastically scattered electrons for enhanced contrast and resolution (28).
RESULTS
Interactions between Vpr derivatives and plasmid DNA.
Full-length Vpr (Vpr1–96) and several Vpr subfragments (Table 1 and Fig. 1) were synthesized and tested for their capacity to interact with plasmid DNA at a physiological ionic strength (150 mM NaCl). Peptide-DNA interactions can be measured by determining the amount of peptide required to retard the migration of plasmid DNA toward the cathode during agarose gel electrophoresis. Table 1 shows that all peptides except Vpr1–51 and Vpr80–96 were able to retard plasmid DNA. Vpr1–96, Vpr52–96, and Vpr52–93 were active at plus/minus charge ratios below 1, whereas the other peptides required a charge ratio above 1. Vpr77–96 behaved unexpectedly in that 5 μg of peptide retarded most, but not all, of the DNA. Even with 30 μg of Vpr77–96 there was still a smear of DNA, indicating partial or unstable complex formation (data not shown). Interestingly, Vpr52–75, which possesses three negative charges and three positive charges at pH 7 (Fig. 1), completely retarded 1 μg of DNA while Vpr80–96 (six positive charges and one negative charge) was inactive even in large quantities (30 μg). Thus, the ability of Vpr derivatives to bind DNA was not simply linked to their positive charges, suggesting that structural features are probably involved.
TABLE 1.
Capacity of Vpr derivatives to retard plasmid DNA migration
| Peptide | Amt of peptide needed for complete DNA retardation (μg)a | Plus/minus charge ratio of the peptide-DNA complex |
|---|---|---|
| Vpr1–96 | 4 | 0.5 |
| Vpr1–51 | >30b | NAd |
| Vpr52–96 | 2 | 0.8 |
| Vpr52–93 | 2 | 0.77 |
| Vpr52–75 | 20 | 2.1 |
| Vpr60–80 | 2.5 | 1.5 |
| Vpr70–96 | 2 | 1.4 |
| Vpr77–96 | ∼5c | 3.6 |
| Vpr80–96 | >30b | NA |
The indicated amounts of peptides are the minimal amounts required to retard completely 1 μg of plasmid DNA.
The largest amount of peptide tested was 30 μg.
With 5 μg of peptide, most of the DNA was retarded, but complete retardation was not attained even with a significantly larger amount of peptide (i.e., 30 μg).
NA, not applicable.
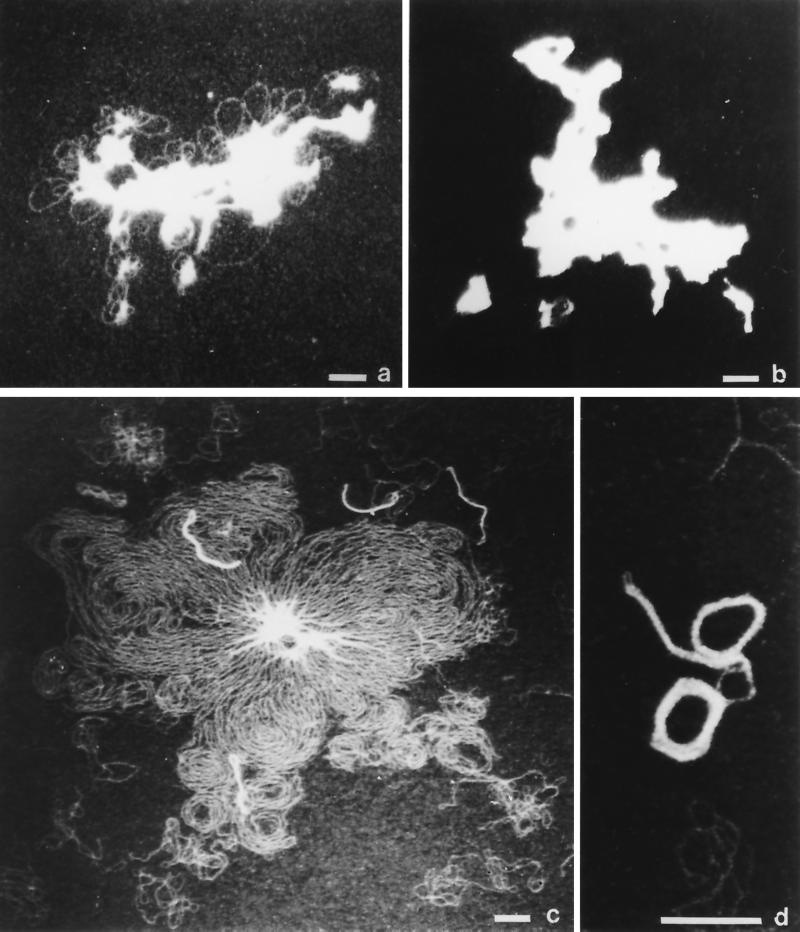
The complexes formed between selected Vpr fragments and plasmid DNA were analyzed by electron microscopy. Figure 2a and b show that Vpr52–96 induced a high level of compaction of DNA molecules. DNA condensation results from intermolecular interactions by which several molecules are incorporated in the condensed structure, leading to the formation of aggregates with irregular sizes and shapes. In contrast, Vpr77–96, which was less active in the retardation assay, induced an ordered and reversible condensation without collapse of the DNA molecule. Figure 2 shows that in a single Vpr77–96-DNA preparation, different steps of compaction were evident (lamellar structures and rods [Fig. 2c], as well as toroids [Fig. 2d]).
FIG. 2.
Electron microscopy of Vpr52–96-DNA and Vpr77–96-DNA complexes. The visualization of Vpr-DNA complexes was performed by positive staining and annular dark-field electron microscopy. The Vpr52–96-DNA and Vpr77–96-DNA complexes were prepared in 150 mM NaCl as described in Materials and Methods. (a and b) Vpr52–96 induces the formation of aggregates of plasmid DNA. The aggregates in panel b, which represent a large majority of the events that can be observed on the grid, result from the incorporation of several DNA molecules. (c and d) Vpr77–96 induces a compaction of the DNA molecules, without collapse and effective aggregations, but an ordered condensation with lamellar structures. Such condensation can lead to the formation of rods or toroids. (d) Illustration of toroidal compaction of plasmid DNA mediated by Vpr77–96. Scale bars, 100 nm.
DNA transfection activity of Vpr derivatives.
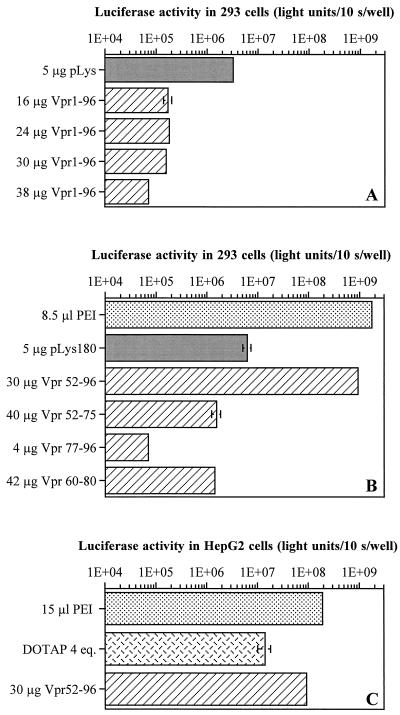
Full-length Vpr and Vpr subfragments with complete or partial DNA binding activity were tested for their ability to transfect DNA into human 293 cells. The peptides were complexed to a luciferase expression plasmid (CMV-Luc) and incubated with the cells for 3 h. Luciferase activity was measured about 30 h later. The controls included polyplexes formed with PEI and pLys. PEI is a potent transfection reagent even in the absence of endosomolytic agents (2, 3), whereas pLys cannot efficiently escape the endocytic vesicles (26). Figure 3A indicates that Vpr1–96 mediated significantly less gene transfer than did pLys. In contrast, Vpr52–96 (Fig. 3B) and Vpr52–93 (data not shown) were up to 500-fold more efficient than pLys and only 2- to 20-fold less efficient than PEI was under its best conditions. In spite of their DNA binding activities, Vpr52–75, Vpr77–96, and Vpr60–80 displayed poor transfection efficiencies (Fig. 3B).
FIG. 3.
Transfection efficiencies of Vpr derivatives. Increasing amounts of transfection agents were mixed with a constant amount of reporter plasmid (4 μg per duplicate), and the mixtures were incubated for 3 h with cells in a serum-free medium. Luciferase activity was measured 30 h posttransfection. The transfection efficiency was expressed as total light units per 10 s per well, and the means of duplicate determinations are shown. Only the results obtained under the best conditions are shown. (A) Comparison of transfection efficiencies of Vpr1–96 and pLys on 293 cells. (B) Comparison of transfection efficiencies of Vpr derivatives pLys and PEI on 293 cells. (C) Comparison of transfection efficiencies of Vpr52–96, DOTAP, and PEI on HepG2 cells. Of note, 1 eq. of DOTAP represents the amount of lipid needed to neutralize the negative charges carried by 4 μg of DNA.
Using another CMV-luciferase construct (PCIneoLuc) or an expression plasmid encoding the lacZ gene under the control of the human phosphoglycerate kinase promoter, we also observed a higher transfection efficiency with Vpr52–96 than with pLys (data not shown). We then tested whether the number of cells expressing the reporter gene was higher with Vpr52–96 than with pLys. We transfected a GFP expression construct and measured the percentage of transfected cells by flow cytometry 40 h posttransfection. While less than 1% of the 293 cells expressed GFP when transfected with pLys under optimized conditions, 26% and 37% were GFP positive when Vpr52–96 and the monocationic lipid DOTAP, respectively, were used as the carriers (data not shown).
Following transfection, the toxicity of Vpr52–96-DNA complexes for 293 cells was monitored by measuring the total amount of protein per well, by counting the cells, and by measuring cell proliferation by the MTT colorimetric assay (37). Cell survival after transfection with Vpr52–96 was between 70 and 90% of that of nontreated cells. This cytotoxicity was comparable to that observed with other transfection reagents. Our data are in good agreement with the recent findings of Jacotot and coworkers, who showed that the cytocidal effect of Vpr52–96 was abolished when agents such as RNA or DNA interacted with the H(S)RIG region of the peptide (23).
Vpr52–96 was also able to mediate DNA transfection in other cell lines. The best Vpr52–96 and pLys conditions were determined for each cell line by testing several concentrations of both reagents. Table 2 shows the ratios of the highest levels of luciferase activity obtained with Vpr52–96 and pLys. As with 293 cells, Vpr52–96-mediated transfection resulted in up to 3 orders of magnitude higher luciferase activities on 911 and HepG2 cells than did pLys under optimal conditions, thus reaching efficiencies comparable to those obtained with PEI or DOTAP (Fig. 3C). For transfections into NIH 3T3 cells and HeLa cells, Vpr52–96 was only 3 to 10 times more efficient than pLys.
TABLE 2.
Transfection efficiency of Vpr52–96 versus that of pLys
| Cell line | Vpr52–96/pLys ratioa
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| 293 | 67 | 112 | 225b |
| 911 | 30 | 110 | NDc |
| HepG2 | 500 | 720 | 1,400 |
| HeLa | 7 | 10 | ND |
| NIH 3T3 | 3 | 5 | ND |
The values are the ratios of the highest levels of luciferase activity obtained with Vpr52–96 to those obtained with pLys. Data from two to three independent experiments are shown.
Data are shown in Fig. 3B.
ND, not done.
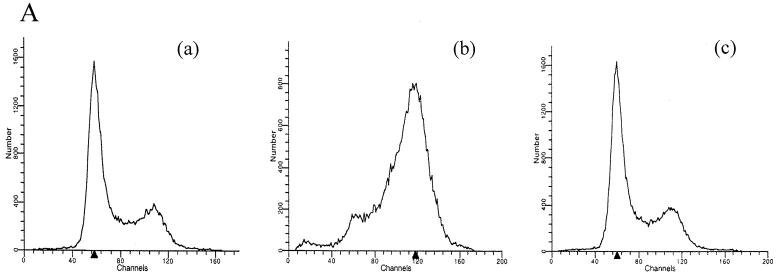
The leucine-rich region and the C terminus of Vpr can stimulate reporter gene expression directed from the HIV-1 long terminal repeat as well as from heterologous viral promoters (17, 19, 50). Importantly, this stimulation is usually under 10-fold and is linked to G2 arrest activity (16, 17, 19). To rule out the possibility that Vpr52–96 transactivated reporter gene expression in our experiments, we performed a series of control studies. First, we determined whether the externally added peptide Vpr52–96, alone or mixed with plasmid DNA, was able to induce cell cycle arrest. Although less than 30% of the cells expressed the reporter gene following transfection, most of the cells took up polyplexes, as shown by flow cytometry after incubation of the cells with fluorescent Vpr52–96-DNA complexes (Fig. 4B). The cell cycle was analyzed after propidium iodide staining of DNA 24 and 30 h after transfection (see Materials and Methods). Control cells and cells incubated with Vpr52–93 (data not shown) or with Vpr52–93–CMV-Luc complexes showed normal cell cycle profiles, with about 50% of the cells being in G0/G1. The experiment was controlled by treatment of 293 cells with thymidine and nocodazole, which resulted in accumulation of cells in the G2 phase (to >70%) (Fig. 4A). We concluded that Vpr52–96 alone or complexed with DNA did not mediate G2 arrest in our system. In a second control experiment, we used the mutant Arg80Ala, which is unable to arrest cells in G2 (16, 17). We compared the mutated peptide Arg80Vpr52–96Ala80 to wild-type Vpr52–96 with regard to the ability to form complexes with plasmid DNA and to transfect 293 cells. The mutated peptide behaved like Vpr52–96 in the DNA retardation assay and was as active as in DNA transfections (data not shown).
FIG. 4.
Study of possible Vpr52–96-induced G2/M phase arrest. (A) The cell cycles of untreated (a), thymidine- and nocodazole-treated (b), and Vpr52–96-transfected (c) 293 cells were analyzed by flow cytometry as described in Materials and Methods. (B) Evaluation of the association and/or endocytosis of fluorescent DNA-Vpr52–96 complexes on 293 cells after 4 h of incubation at 37°C. The autofluorescence of the cells is shown by the broken line. The x axis represents the relative fluorescence intensity.
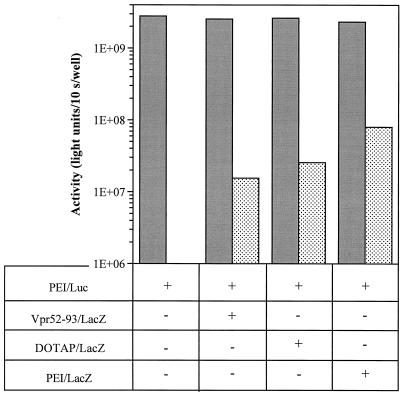
Finally, we looked at a possible transactivation effect of Vpr52–93 by applying complexes to 293 cells previously transfected with a CMV-driven reporter gene. Cells first received PEI–CMV-Luc complexes, and after 43 h they were transfected again with either Vpr52–93–CMV-LacZ, DOTAP–CMV-LacZ, or PEI–CMV-LacZ. Thirty hours after the second transfection, the luciferase and β-galactosidase activities were measured. Figure 5 shows that none of the secondary transfections led to enhanced luciferase expression. Thus, we concluded that the high levels of reporter gene expression obtained with Vpr52–96 cannot be explained by transactivation of the CMV promoter.
FIG. 5.
Does Vpr52–96 transactivate the CMV promoter of the reporter plasmid? 293 cells transfected with PEI 25 kDa–CMV-Luc were transfected again (except the control) 43 h later with (+) or without (−) the following polyplexes: Vpr52–93–CMV-LacZ, DOTAP–CMV-LacZ, and PEI 25 kDa–CMV-LacZ. Thirty hours after the second transfection, the luciferase (solid gray bars) and the β-galactosidase (stippled bars) activities were determined by measuring luminescence.
Intracellular fate of the transfection complexes.
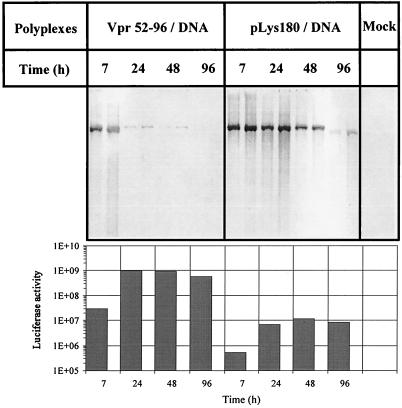
To determine whether the higher transfection levels obtained with Vpr52–96 compared to pLys were due to more-efficient DNA delivery, we harvested transfected 293 cells 7, 24, 48, and 96 h after transfection and isolated low-molecular-weight DNA. Southern blot analysis indicated that significant amounts of plasmid DNA were present at 7 h posttransfection in both Vpr52–96- and pLys-transfected cells (Fig. 6). One day after Vpr52–96-mediated transfection, the amount of plasmid present in the 293 cells was dramatically decreased whereas cells transfected with pLys still contained large amounts of DNA. After 4 days, no DNA was detectable in Vpr52–96-transfected cells whereas the signal remained in the case of pLys. Although much less DNA accumulated following Vpr52–96 transfection, reporter gene activity was superior to that of pLys at all time points (Fig. 6, lower panel). Of note, the high level of luciferase activity observed at day 4 cannot be explained by the accumulation of enzyme during the first hours because the half-life of this enzyme in mammalian cells is only 3 h (47).
FIG. 6.
Efficiency of delivery of plasmid DNA by Vpr52–96. 293 cells plated in six-well plates were transfected, or not (mock), with either Vpr52–96–CMV-Luc or pLys–CMV-Luc complexes. At different time points after transfection, cells were harvested and lysed and the low-molecular-weight DNA was recovered. At the same time, the luciferase activity was measured. For each set of conditions, two different volumes of the Hirt extracts (i.e., one-fourth and one-half of the volume of one well) were used. The graph below the blot presents the levels of luciferase activity obtained at different time points after transfection with Vpr52–96 or pLys.
This experiment indicates that most of the delivered DNA does not contribute to transgene expression. This DNA is trapped either in vesicles or in the cytosol and is eventually degraded at a rate that depends on the capacity of its carrier to provide protection against nucleases (29). To evaluate the capacity of Vpr52–96 to protect DNA against degradation, we incubated Vpr52–96-DNA complexes with DNase I for 1 h at 37°C, and after elimination of Vpr by SDS treatment, DNA was analyzed by agarose gel electrophoresis. Vpr52–96 was not able to preserve the integrity of the plasmid (data not shown), in contrast to pLys (7). Thus, the rapid disappearance of the transfected DNA (Fig. 6) may reflect the sensitivity of Vpr52–96- DNA complexes to nucleases.
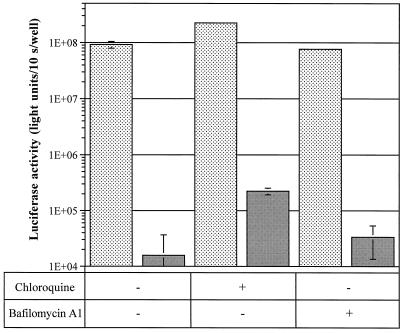
Ligand-pLys-mediated gene transfer can be significantly enhanced in the presence of chloroquine (15, 54), a weak lysosomotropic base which accumulates in acidic endocytic vesicles, neutralizes their pH, and destabilizes their membranes. Another beneficial effect of chloroquine in pLys-mediated transfection experiments may be the dissociation of polyplexes (15). Figure 7 shows that Vpr52–96-mediated transfection of HepG2 cells was 2- to 10-fold more efficient in the presence of 100 μM chloroquine. This enhancement is comparable to that observed with pLys (Fig. 7). We concluded that at least some of the Vpr52–96-DNA complexes are localized in chloroquine-sensitive endocytic vesicles.
FIG. 7.
Transfection of HepG2 cells with Vpr52–96–CMV-Luc in the presence of either chloroquine or bafilomycin A1. HepG2 cells plated in 24-well plates were transfected with 24 μg of Vpr52–96 and 4 μg of CMV-Luc (stippled bars) or 4 μg of pLys and 4 μg of CMV-Luc (solid gray bars) in the presence (+) or absence (−) of either 100 μM chloroquine or 175 nM bafilomycin A1. After 2.5 h of incubation, the transfection medium was removed and replaced with DMEM containing 10% FCS. The luciferase assay was performed 2 days after transfection.
Several peptides known to enhance pLys-mediated DNA transfection are believed to act by disrupting the endosome membrane at low pH values (35, 49). Their fusogenic activity is triggered when protonation of their side-chain carboxy groups induces a change from a random conformation to an α-helix. This process is inhibited by bafilomycin A1, a specific inhibitor of the vacuolar proton pump (4, 52). To test the possibility that Vpr52–96 was acting along a similar pathway, we transfected HepG2 cells in the presence of increasing amounts of bafilomycin A1. Figure 7 shows that gene transfer was not significatively altered under these conditions. This indicates that if Vpr52–96 complexes are released from endosomes, it happens via a pH-independent mechanism.
In summary, our study of the intracellular fate of Vpr52–96-DNA complexes indicates that only a minority of them efficiently find their way to the nucleus whereas most are trapped in vesicles or in the cytosol and are quickly degraded. The active fraction of complexes ending up in the nucleus either avoids the endocytic route or can be released from endosomes by a pH-independent mechanism.
DISCUSSION
Vpr is incorporated into HIV virions through interactions with at least two gag-encoded proteins, p6 and Ncp7. It is known to bind RNA and DNA (10, 55) and therefore may be an integral component of the PIC following reverse transcription. The presence of Vpr has been shown to facilitate nuclear import of the complex, possibly through interactions with karyopherins (40). Considering these properties, we tested the possibility of using Vpr directly as a DNA transfection agent.
We first analyzed the DNA binding activities of a variety of Vpr-derived peptides in gel retardation assays. Our study confirmed that peptide fragments from the C-terminal domain (residues 52 to 96), but not those from the N terminus (residues 1 to 51), are able to form complexes with DNA. We showed that Vpr52–96 induces the compaction of plasmid DNA, leading to the formation of aggregates with irregular sizes and shapes. Rather unexpectedly, the positively charged peptide Vpr80–96 was found to be unable to bind DNA efficiently. Instead, residues between positions 70 and 80 appear to play a key role in the formation of a DNA binding domain.
Transfection experiments identified Vpr52–96 and Vpr52–93 as agents with higher transfection efficiencies than pLys. Surprisingly, full-length Vpr, which also binds DNA, was always significantly less efficient than pLys in these experiments. Recent observations indicate that Vpr1–51 can partially hinder nucleic acid recognition by Vpr52–96 (10). Our results suggest that the transfection activity of the Vpr52–96 region is not available in the context of the whole Vpr protein. It is not known whether an active configuration related to the transfection activity described here is unveiled at any step of HIV infection. Yet, it is tempting to speculate that the ability of Vpr52–96 to act as a DNA carrier is related to the role of Vpr as an enhancer of the nuclear import of viral DNA.
Multiple activities have been associated with Vpr, including cell cycle arrest in G2 and transactivation of viral promoters. We believe that the expression of the luciferase reporter gene which we used in our experiments to measure the transfection efficiency was not influenced by these activities, for the following reasons: (i) no cell cycle arrest was observed following transfection with Vpr52–96-DNA complexes, (ii) a Vpr52–96 peptide containing a mutation that eliminates G2 arrest was as active as the wild-type peptide, and (iii) Vpr52–93-mediated transfection did not transactivate a CMV promoter already present in the transfected cells (Fig. 5).
The mechanism of cellular uptake of Vpr-DNA complexes still needs to be elucidated. Positively charged DNA complexes can enter cells after binding to membrane-associated sulfated proteoglycans (36). Under our experimental conditions, with a plus/minus charge ratio of around 2.5, the complexes could follow this pathway. We have observed that although higher levels of gene expression are reached when Vpr52–96 is used, smaller amounts of DNA are found within the cells than when pLys is employed. This can be related to the higher susceptibility to DNase displayed by the Vpr-DNA complexes. It also indicates that a large number of DNA molecules do not participate in the expression of the reporter gene. Most likely, these molecules are trapped in acidic vesicles, and we showed that a proportion are released from these vesicles upon chloroquine treatment. It is not clear, however, whether endocytosis is an obligatory pathway used by Vpr-DNA complexes for access to the cytoplasm. Alternatively, the active complexes may enter the cell directly after membrane permeabilization. Indeed, Macreadie et al. have reported that the C-terminal part of Vpr causes permeabilization in different yeast strains when added extracellularly (32, 33).
Once released in the cytosol, Vpr-DNA complexes may also be more efficiently transported to the nucleus than pLys/DNA polyplexes due to interactions with the nuclear import machinery (24, 40). In this regard, it is interesting that the advantage of Vpr over pLys was less conspicuous in HeLa cells, in which Vpr-like activities were detected (5, 40). To the best of our knowledge, Vpr52–96 represents a unique example of a natural peptide capable of binding DNA and transporting it into cells as efficiently as the best-known transfection agents. It is now important to identify the cellular partners involved in the transfection pathway. This may allow the design of a new class of peptides for gene transfer.
ACKNOWLEDGMENTS
We thank scientists at Genethon for helpful discussions and critical reading of the manuscript. We thank Valérie Allo for excellent assistance. The Development and Production department of Genethon helped by providing us with plasmid DNA.
This work was performed with the financial support of the Association Française contre les Myopathies (AFM).
REFERENCES
- 1.Agostini I, Navarro J M, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J Mol Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 2.Boussif O, Lezoualc'h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boussif O, Zanta M A, Behr J P. Optimized galenics improve in vitro gene transfer with cationic molecules up to 1000-fold. Gene Ther. 1996;3:1074–1080. [PubMed] [Google Scholar]
- 4.Bowman E J, Siebers A, Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M, Adzhubei A. Viral protein R of HIV-1. Rev Med Virol. 1999;9:39–49. doi: 10.1002/(sici)1099-1654(199901/03)9:1<39::aid-rmv235>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Capecchi M R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- 7.Chiou H C, Tangco M V, Levine S M, Robertson D, Kormis K, Wu C H, Wu G Y. Enhanced resistance to nuclease degradation of nucleic acids complexed to asialoglycoprotein-polylysine carriers. Nucleic Acids Res. 1994;22:5439–5446. doi: 10.1093/nar/22.24.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E A, Terwilliger E F, Jalinoos Y, Proulx J, Sodroski J G, Haseltine W A. Identification of HIV-1 vpr product and function. J Acquir Immune Defic Syndr. 1990;3:11–18. [PubMed] [Google Scholar]
- 9.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 10.de Rocquigny, H., A. Caneparo, T. Delaunay, J. Bischerour, J.-F. Mouscadet, and B. P. Roques. The interaction of the Vpr C-terminus with nucleic acids is modulated by its N-terminus. Eur. J. Biochem., in press. [DOI] [PubMed]
- 11.de Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix J-L, Roques B P. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 12.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowty M E, Williams P, Zhang G, Hagstrom J E, Wolff J A. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 15.Erbacher P, Roche A C, Monsigny M, Midoux P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA/lactosylated polylysine. Exp Cell Res. 1996;225:186–194. doi: 10.1006/excr.1996.0169. [DOI] [PubMed] [Google Scholar]
- 16.Felzien L K, Woffendin C, Hottiger M O, Subbramanian R A, Cohen E A, Nabel G J. HIV transcriptional activation by the accessory protein, VPR, is mediated by the p300 co-activator. Proc Natl Acad Sci USA. 1998;95:5281–5286. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forget J, Yao X J, Mercier J, Cohen E A. Human immunodeficiency virus type 1 vpr protein transactivation function: mechanism and identification of domains involved. J Mol Biol. 1998;284:915–923. doi: 10.1006/jmbi.1998.2206. [DOI] [PubMed] [Google Scholar]
- 18.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Albright A V, González-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh W C, Rogel M E, Kinsey C M, Michael S F, Fultz P N, Nowak M A, Hahn B H, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 20.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzinger N K, Bukinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 23.Jacotot E, Ravagnan L, Loeffler M, Ferri K F, Vieira H L, Zamzami N, Costantini P, Druillennec S, Hoebeke J, Briand J P, Irinopoulou T, Daugas E, Susin S A, Cointe D, Xie Z H, Reed J C, Roques B P, Kroemer G. The HIV-1 viral protein R induces apoptosis via a direct effect on the mitochondrial permeability transition pore. J Exp Med. 2000;191:33–46. doi: 10.1084/jem.191.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kichler A, Zauner W, Morrison C, Wagner E. Ligand-polylysine mediated gene transfer. In: Felgner P L, et al., editors. Artificial self-assembling systems for gene delivery. Washington, D.C.: American Chemical Society; 1996. pp. 120–128. [Google Scholar]
- 27.Kino T, Gragerov A, Kopp J B, Stauber R H, Pavlakis G N, Chrousos G P. The HIV-1 virion-associated protein vpr is a coactivator of the human glucocorticoid receptor. J Exp Med. 1999;189:51–62. doi: 10.1084/jem.189.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Cam E, Delain E. Nucleic acids-ligand interactions. In: Morel G, editor. Visualization of nucleic acids. Boca Raton, Fla: CRC Press; 1995. pp. 331–356. [Google Scholar]
- 29.Lechardeur D, Sohn K J, Haardt M, Joshi P B, Monck M, Graham R W, Beatty B, Squire J, O'Brodovich H, Lukacs G L. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 30.Levy D N, Refaeli Y, MacGregor R R, Weiner D B. Serum Vpr regulates productive infection and latency of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:10873–10877. doi: 10.1073/pnas.91.23.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macreadie I G, Arunagiri C K, Hewish D R, White J F, Azad A A. Extracellular addition of a domain of HIV-1 Vpr containing the amino acid sequence motif H(S/F)RIG causes cell membrane permeabilization and death. Mol Microbiol. 1996;19:1185–1192. doi: 10.1111/j.1365-2958.1996.tb02464.x. [DOI] [PubMed] [Google Scholar]
- 33.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midoux P, Mendes C, Legrand A, Raimond J, Mayer R, Monsigny M, Roche A C. Specific gene transfer mediated by lactosylated poly-l-lysine into hepatoma cells. Nucleic Acids Res. 1993;21:871–878. doi: 10.1093/nar/21.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mislick K A, Baldeschwieler J D. Evidence for the role of proteoglycans in cation-mediated gene transfer. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 38.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J Biol Chem. 1998;273:13347–13352. doi: 10.1074/jbc.273.21.13347. [DOI] [PubMed] [Google Scholar]
- 40.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Re F, Luban J. HIV-1 Vpr: G2 cell cycle arrest, macrophages and nuclear transport. Prog Cell Cycle Res. 1997;3:21–27. doi: 10.1007/978-1-4615-5371-7_2. [DOI] [PubMed] [Google Scholar]
- 43.Schüler W, Wecker K, de Rocquigny H, Baudat Y, Sire J, Roques B P. NMR structure of the (52–96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions. J Mol Biol. 1999;285:2105–2117. doi: 10.1006/jmbi.1998.2381. [DOI] [PubMed] [Google Scholar]
- 44.Selig L, Pages J-C, Tanchou V, Prévéral S, Berlioz-Torrent C, Liu L X, Erdtmann L, Darlix J-L, Benarous R, Benichou S. Interaction with the p6 domain of the Gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol. 1999;73:592–600. doi: 10.1128/jvi.73.1.592-600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subbramanian R A, Kessous-Elbaz A, Lodge R, Forget J, Yao X J, Bergeron D, Cohen E A. Human immunodeficiency virus type 1 Vpr is a positive regulator of viral transcription and infectivity in primary human macrophages. J Exp Med. 1998;187:1103–1111. doi: 10.1084/jem.187.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subbramanian R A, Yao X J, Dilhuydy H, Rougeau N, Bergeron D, Robitaille Y, Cohen E A. Human immunodeficiency virus type 1 Vpr localization: nuclear transport of a viral protein modulated by a putative amphipathic helical structure and its relevance to biological activity. J Mol Biol. 1998;278:13–30. doi: 10.1006/jmbi.1998.1685. [DOI] [PubMed] [Google Scholar]
- 47.Thompson J F, Hayes L S, Lloyd D B. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 48.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel M L. Influenza virus hemagglutinin HA-2 N-terminal fusogenic peptides augment gene transfer by transferrin-polylysine-DNA complexes: toward a synthetic virus-like gene-transfer vehicle. Proc Natl Acad Sci USA. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 51.Wecker K, Roques B P. NMR structure of the (1–51) N-terminal domain of the HIV-1 regulatory protein Vpr. Eur J Biochem. 1999;266:1–12. doi: 10.1046/j.1432-1327.1999.00858.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 53.Zabner J, Fasbender A J, Moninger T, Poellinger K A, Welsh M J. Cellular and molecular barriers to gene transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 54.Zenke M, Steinlein P, Wagner E, Cotten M, Beug H, Birnstiel M L. Receptor-mediated endocytosis of transferrin-polycation conjugates: an efficient way to introduce DNA into hematopoietic cells. Proc Natl Acad Sci USA. 1990;87:3655–3659. doi: 10.1073/pnas.87.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S, Pointer D, Singer G, Feng Y, Park K, Zhao L J. Direct binding to nucleic acids by Vpr of human immunodeficiency virus type 1. Gene. 1998;212:157–166. doi: 10.1016/s0378-1119(98)00178-4. [DOI] [PubMed] [Google Scholar]