Abstract
Human Staufen (hStau), a double-stranded RNA (dsRNA)-binding protein that is involved in mRNA transport, is incorporated in human immunodeficiency virus type 1 (HIV-1) and in other retroviruses, including HIV-2 and Moloney murine leukemia virus. Sucrose and Optiprep gradient analyses reveal cosedimentation of hStau with purified HIV-1, while subtilisin assays demonstrate that it is internalized. hStau incorporation in HIV-1 is selective, is dependent on an intact functional dsRNA-binding domain, and quantitatively correlates with levels of encapsidated HIV-1 genomic RNA. By coimmunoprecipitation and reverse transcription-PCR analyses, we demonstrate that hStau is associated with HIV-1 genomic RNA in HIV-1-expressing cells and purified virus. Overexpression of hStau enhances virion incorporation levels, and a corresponding, threefold increase in HIV-1 genomic RNA encapsidation levels. This coordinated increase in hStau and genomic RNA packaging had a significant negative effect on viral infectivity. This study is the first to describe hStau within HIV-1 particles and provides evidence that hStau binds HIV-1 genomic RNA, indicating that it may be implicated in retroviral genome selection and packaging into assembling virions.
Targeting of virion components to the plasma membrane is a prerequisite for efficient human immunodeficiency virus type 1 (HIV-1) particle assembly. Precursor p55Gag, Gag/Pol polyproteins, Env glycoproteins, selected accessory proteins, and the viral RNA genome must accumulate at the plasma membrane, where the formation and release of nascent particles occur. Gag expression alone is sufficient to produce spherical virus-like particles (VLPs) which assemble and bud from the plasma membrane. To achieve this, Gag contains several signals that can promote plasma membrane targeting. These include amino-terminal myristoylation and basic amino acid signals (30). Gag targeting may also be mediated in part via Env since it has been shown that gp41 binds the Gag matrix domain (17, 31). gp41 also harbors in its cytoplasmic tail a Tyr-based trafficking motif, YXXφ, that is responsible for Env glycoprotein endocytosis and polarized virus release in epithelial and T cells (6, 21, 49, 58).
A full picture of the mechanisms involving host cell components that direct targeting of viral components to sites of viral assembly remains to be established. Several data have implicated the cytoskeleton in the targeting of viral components. The cytoskeleton forms a scaffold on which macromolecule transport is well known to occur (2), and there is now ample evidence that retroviral Gag associates with cytoskeletal proteins. For example, Gag associates with actin microfilaments (24, 63) and is colocalized with actin to the pseudopods in virus-infected cells (60), suggestive of an intracellular transport mechanism. Moreover, HIV-1 production depends on an intact actin network (65). Recent data also demonstrate that Gag directly interacts with actin (48) and can associate with a kinesin molecular motor protein, KIF-4 (70). Thus, the cytoskeleton may be directly involved in the binding and trafficking of Gag and Gag's interacting partners to sites of viral assembly. However, details about these mechanisms remain to be elucidated.
While several details regarding the targeting of virion components to the plasma membrane are becoming clearer, little is known about how the viral RNA is targeted to sites of assembly. Presumably, HIV-1 RNA-protein associations with the precursor p55Gag could come into play to influence genomic RNA localization to sites of assembly. Indeed, nucleocapsid (NC) is known to bind the psi packaging signal in genomic RNA and is principally responsible for the selection of this RNA for encapsidation (5, 18, 20, 76). However, this is likely to occur during assembly, once virion components have accumulated near the plasma membrane, consistent with evidence that indicates a role for NC during virion assembly (19, 62). Thus, host cell and/or viral proteins are likely to be uncovered to be implicated in the trafficking of viral RNA to sites of assembly during viral replication.
Staufen is a double-stranded RNA (dsRNA)-binding protein that was originally described in Drosophila melanogaster (69). In Drosophila oocytes, Staufen's principal functions are to bind RNAs and transport them to achieve, in most cases, localized translation (26, 41). It serves to transport oskar mRNA posteriorally (41) and anchors bicoid mRNA anteriorally in oocytes (26). Recently, Staufen was shown to be involved in the localization of prospero mRNA in Drosophila neuroblasts to promote an asymmetric RNA distribution during cell division (11, 47). A human homologue of Staufen (hStau) has recently been characterized by us (72) and others (50). It has a high degree of sequence and structural similarity, contains four consensus dsRNA-binding domains (dsRBD) corresponding to dsRBD2 to dsRBD5 of Drosophila Staufen (dStau), and has a tubulin-binding domain in the carboxyl terminus (72). Since its overall structure and the relative position of the dsRBDs are well conserved compared to dStau, the four dsRBDs in hStau will be referred to herein as dsRBD2 to dsRBD5. Only dsRBD3 is capable of strongly binding RNA in vitro (72). Moreover, hStau is associated with the rough endoplasmic reticulum (50, 72) and with polyribosomes (50), and it is found in ribonucleoprotein complexes in neurons (40). Recent data from studies using green fluorescent protein-hStau fusion proteins in neurons indicate that the mammalian homologue, like its Drosophila counterpart, is involved in RNA transport (43).
This report explores the involvement of Staufen in HIV-1 replication. We demonstrate here that hStau is incorporated in HIV-1 and that hStau levels correlate with the abundance of genomic RNA encapsidated in HIV-1 virions. hStau is associated with HIV-1 RNA in both cells and virus, suggesting that hStau is implicated in virus assembly at a step that controls the abundance of encapsidated RNA.
MATERIALS AND METHODS
Virus.
Cell-free preparations of DNA viruses were kindly supplied by Bernard Massie (adenovirus; Biotechnology Research Institute, Montreal, Quebec, Canada) and José Menezes and Ali Ahmad (Epstein-Barr virus and human herpesvirus 6; University of Montreal). HIV-1 clinical isolates were gifts from Mark Wainberg (McGill AIDS Centre).
Cell lines.
293T cells were maintained in Dulbecco modified Eagle medium (Gibco/BRL) and 8% fetal calf serum and transfected by the calcium phosphate coprecipitation method. MT4 and Jurkat T-cell lines were maintained in RPMI supplemented with 10% fetal calf serum and antibiotics.
Antisera.
Recombinant hStau (rhStau) was used to generate a rabbit polyclonal anti-hStau antiserum as described previously (72). Antisera to transactivation response (TAR) RNA-binding protein (TRBP) were kindly provided by Sundararajan Venkatesan (antibody 690; National Institute of Allergy and Infectious Diseases, Bethesda, Md.) and Anne Gatignol (antibodies 672 and 673; McGill University, Montreal, Quebec). Antiserum to dsRNA-activated protein kinase (PKR) was kindly provided by Antonis Koromilas (antibody 1388-F9; McGill University). An anti-Tat rabbit antiserum against the last 20 amino acids of the amino terminus was used to detect Tat. Mouse anti-gp120, anti-p17, and a rabbit anti-p24 were purchased from Intracell. Anti-HA (hemagglutinin) and anti-Myc monoclonal antisera (ascites fluid) were generated in BALB/C mice at the antibody core facility in the Department of Biochemistry, University of Montreal. Anti-IN (integrase) (36) was from the AIDS Research and Reference Reagent Program, National Institutes of Health.
Sucrose density gradient analysis of hStau in HIV-1.
A total of 108 cpm of microfiltered and ultracentrifuged pNL4.3 virus generated in 293T cells was layered onto a continuous 20 to 70% sucrose gradient and ultracentrifuged at 136,000 × g for 16 h; 14 to 15 0.7-ml fractions were collected, and reverse transcriptase (RT) activity was measured by standard assay. An aliquot from each fraction was resuspended in 2× Laemmli loading buffer was added before loading onto a 12% gel for polyacrylamide gel electrophoresis (PAGE). The proteins were transferred to nitrocellulose and probed with anti-hStau, anti-p24, anti-IN, and/or anti-NC. Antigens were visualized using luminol-based enhanced chemiluminescence (ECL) (73). An aliquot from each fraction was also taken for slot blot analysis of HIV-1 RNA using a Gibco/BRL slot blot apparatus and using a 32P-labeled cDNA probe to the 5′ leader as described previously (74).
Optiprep gradient centrifugation.
Pelleted HIV-1 (3 × 108 cpm) was loaded onto a 6 to 18% Optiprep (22) gradient and centrifuged at 183,000 × g for 1.5 h, and 17 fractions were collected from the bottom. Aliquots from each fraction were loaded onto 12% sodium dodecyl sulfate (SDS) gels for PAGE and probed for p24 and hStau by Western blot analysis. An aliquot from each gradient fraction was tested for infectious potential by syncytium formation in MT4 cells (data not shown) and in RT activity assays at 4 days postinfection. Only those fractions in the densest fractions (1 to 4) contained an appreciable amount of infectious HIV-1 particles.
Subtilisin protease resistance assay.
Subtilisin assays were performed essentially according to Ott et al. (56, 57), with minor modifications. Pelleted virus preparations (pNL4.3; 2 × 108 cpm) were mock treated or treated with 0.001, 0.1, and 1 mg of subtilisin (Boehringer Mannheim) per ml in 10 mM Tris-HCl (pH 8)–1 mM CaCl2 containing bovine serum albumin (1.5 mg/ml; ICN) for 16 to 24 h at 37°C. Subtilisin was inactivated by phenylmethylsulfonyl fluoride. Virus was then repelleted as described above, resuspended in phosphate-buffered saline (PBS), made to 1× Laemmli buffer, and loaded onto PAGE followed by Western blotting. Blots were sequentially probed with anti-gp120, a human AIDS patient serum (serum 162), or anti-p24 to reveal p24, anti-p17, and anti-hStau.
rhStau.
rhStau was produced in bacteria as described previously (72) and concentrated in 10 mM Tris (pH. 7), using Centricon filters as described by the manufacturer (Millipore).
DNA and transfections.
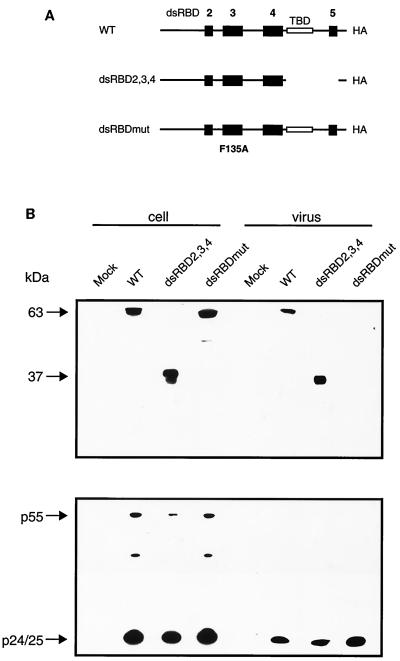
Proviral constructs used in these studies are pNL4.3 (AIDS Research and Reference Reagent Program), HIV-2 (ROD) (AIDS Research and Reference Reagent Program), and HxBru (74). Murine leukemia virus (MLV) provirus was kindly provided by Guy Lemay (University of Montreal). HA-tagged hStau constructs driven by the Rous sarcoma virus (RSV) promoter (WT [wild type], dsRBD2,3,4 and dsRBDmut) have been described elsewhere (72; M. Luo and L. DesGroseillers, submitted for publication). WT, dsRBD2,3,4, and dsRBDmut express HA-tagged proteins of 63, 37, and 63 kDa, respectively. 293T cells were transfected with 10 μg of HxBru and 5 μg of each hStau mutant expressor. Cell lysates and 1.5 × 108 cpm of virus were run in parallel and probed with an anti-HA monoclonal antibody to reveal the hStau molecules. Anti-p24 was used to detect Gag in cell and viral lysates.
dsRNA-binding protein expression plasmids.
For selectivity and overexpression studies, 293T cells were transfected with 10 μg of pNL4.3 and 5 μg of the following mammalian expression plasmids: pcDNA3/RSV-hStau, pcDNA3-TRBP2 (from Anne Gatignol, McGill AIDS Centre), pcDNA3-PKR (from Eliane Meurs, Institut Pasteur), or pcDNA3 (Invitrogen) vector control. At 48 h posttransfection, cells were washed and lysed in PBS containing detergents. The supernatants were harvested, precleared, and microfiltered, and virus was purified through a 20% sucrose cushion. An equal quantity of protein (40,000 cell equivalents) and 2 × 108 cpm of virus were run in parallel in each of four blots. Antigens were identified by Western blot and ECL analyses (73).
Rev-independent Gag expression.
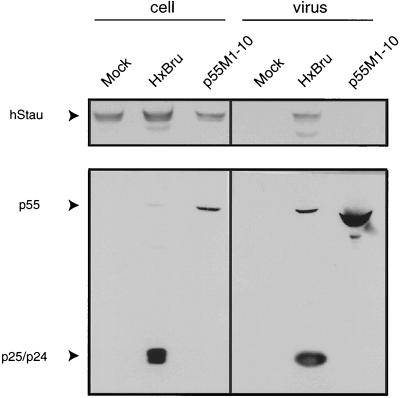
To examine hStau incorporation in VLPs, 293T cells were mock transfected or transfected with 10 μg of HxBru provirus or p55M1-10 Gag expressor (generously provided by George Pavlakis [66]). pcDNA3 was included in the mock transfections. At 48 h posttransfection, cells were washed and lysed in radioimmunoprecipitation (RIPA) buffer. Supernatants were harvested, precleared, and microfiltered (0.45-μm pore size). Virus was then purified by ultracentrifugation and collected in PBS. Equal quantities of protein from the cell lysates were loaded in parallel with 50% of the virus particle preparations onto 12% SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were sequentially probed for hStau and p24 (to identify p55 and p24).
NC and psi mutant proviruses.
The wild-type and mutant proviruses used were pNL4.3, HxBru, 28C/49C-S NC, 15C/18C-S NC, 36C/39C-S NC, delta 14K-50T NC, and psi mutant proviruses (15, 39, 44, 45, 71); 15 μg of each was transfected into 293T cells, and virus was harvested at 48 h posttransfection. Virus was assayed for RT activity using multiple dilutions as well as by p24 by enzyme-linked immunosorbent assay (ELISA). Equal quantities of virus were taken for Northern analysis as described previously (74). p17 was used to control for loading.
Infectivity assays.
For infectivity assays, equal quantities of p24 were used to infect MAGI indicator cells (42) and monocytic indicator BF-24 cells (28), and infectivity was quantitated by colorimetric and chloramphenicol acetyltransferase (CAT) activity assays, respectively. BF-24 cells were washed extensively and lysed by freeze-thaw in 0.25 M Tris (pH 7.5), followed by heat inactivation. CAT activity in cells was determined by standard assay by thin-layer chromatography (54), and quantitation was performed by phosphorimager analysis.
Immunoprecipitation and RT-PCR analyses.
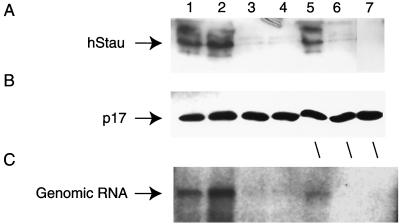
293T cells (8 × 105) were transfected with RSV-hStau-HA, HxBru, both HxBru and hStau-HA, or both HxBru and dsRBDmut. Virus was harvested and purified as described above at 48 h posttransfection. Cells were washed extensively with diethyl pyrocarbonate (DEPC)-treated PBS. One half of the cells was taken for RNA analysis for both HIV-1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNA. An NP-40 lysis extraction method (74) was used to extract cytosolic RNA, and equal quantities of RNA were slot blotted and subsequently probed for HIV-1 and GAPDH RNA (74). The other half of the cells and virus were lysed in DEPC-treated RIPA buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 0.5% NP-40, 0.01% sodium deoxycholate). After removal of DNA by centrifugation, protein was quantitated and input hStau was assessed by Western blot analysis using anti-HA. For immunoprecipitations, the protein content in each of the lysates was normalized. The lysates were precleared using preimmune sera from their respective hosts (for hStau-HA, we used a monoclonal to c-Myc of approximate equal immunoglobulin G content; for TRBP, rabbit preimmune serum was used). After 2 h, 100 μl of a 50:50 slurry of protein G-Sepharose (Pharmacia) was added and incubated for 2 h. Brief centrifugation followed to remove the Sepharose beads. Each sample was split into two tubes; 5 μl of either preimmune or immune serum, was added, followed by addition of protein G-Sepharose as above. Following five washes with RIPA buffer (minus detergents) and two with water, pelleted samples were treated with 10 μg of RQ1 DNase (Gibco/BRL) for 60 min followed by two phenol and one phenol-chloroform-isoamyl alcohol (50-49-1) extraction. The RNA was precipitated with ethanol and glycogen carrier (Boehringer Mannheim). The RNA pellets were resuspended in DEPC-treated water and used in RT-PCR analysis essentially as instructed by the manufacturer (Perkin-Elmer RT-PCR Kit) (46). The RNA was reverse transcribed with random hexamers (Gibco/BRL), and the PCR amplification using 1 U Taq DNA polymerase (Sigma) used two sets of primers to amplify HIV-1 RNA. Set a consisted of a sense oligomer containing an XhoI site to the TAR (positions +1 to 22) and an antisense oligomer immediately upstream of the major splice donor site (52). This set amplifies a 285-bp fragment from both genomic and subgenomic HIV-1 RNAs. Set b was chosen to identify the presence of unspliced genomic RNA in immunoprecipitates and included the TAR oligomer above and an antisense oligomer to the coding region of Gag to generate a PCR fragment of 462 bp (52). Controls for RT-PCR included the exclusion of RT from the RT reaction and treatment of the immunoprecipitation complexes with RNase A before DNase I treatment above (data not shown).
RT-PCR analyses using Roche Biotechnology's Amplicor RT-PCR kit were performed according to the manufacturer's instructions, using a model 9600 Perkin-Elmer PCR Thermocycler. RNA from equal quantities of virus (as determined by p24 ELISA) was quantitated with this method. The number of copies was related to the number of virions to obtain a per-virion count. This analysis was performed twice using multiple dilutions, and similar results were obtained.
RESULTS
hStau incorporation in HIV-1.
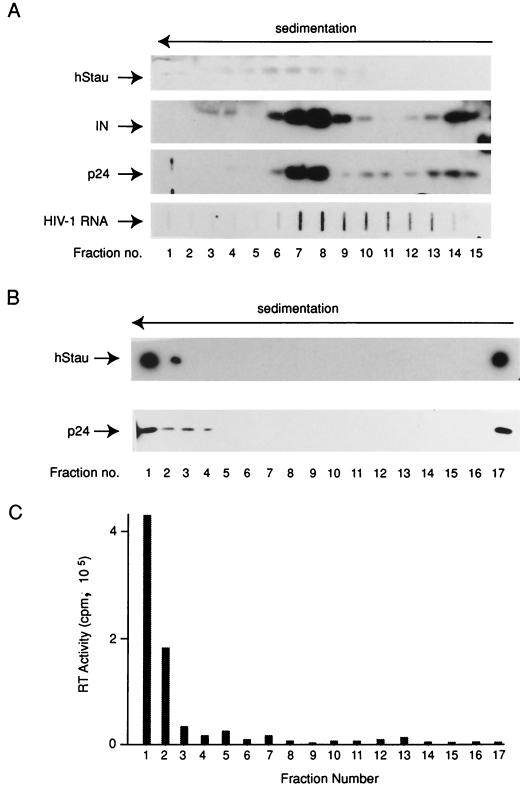
We investigated hStau's association with HIV-1 by determining whether hStau was incorporated in HIV-1 particles, a possible result of its dsRNA-binding capacity. Using a polyclonal antiserum generated to purified rhStau, we identified hStau in purified viral preparations of laboratory strains of HIV-1 (15, 71). To substantiate hStau virion incorporation, sucrose density gradient analyses were performed. pNL4.3 virus was generated from 293T cells. Pelleted virus was fractionated through a 20 to 70% sucrose gradient, and the presence of hStau in each fraction (bottom is fraction 1) was evaluated by Western blot analysis. hStau was found to cosediment with peak p24, IN, and HIV-1 RNA levels (Fig. 1A) and with peak RT activity (data not shown), strongly indicating association with viral particles.
FIG. 1.
hStau is incorporated in HIV-1. (A) Sucrose density gradient analysis of HIV-1 (pNL4.3). hStau, IN, and p24 were assessed in each fraction by Western analysis. HIV-1 RNA in each fraction was assessed by slot blot analysis as described in Materials and Methods. (B) Optiprep analysis. Pelleted HIV-1 particles were fractionated in an Optiprep gradient; 17 fractions were collected and assessed for hStau and p24 content. (C) An aliquot from each Optiprep fraction was taken and used to infect MT4 cells. Five days postinfection, an aliquot of the supernatant from each infection was assayed for RT activity to assess infectious potential.
A recently developed velocity gradient virion purification method (22) was described to separate virus particles and microvesicular contaminants typically found in pelleted virus particles (7). This method uses Optiprep, which forms iso-osmotic solutions at all densities, allowing better separation than sucrose gradients. Sucrose cushion-purified virus was fractionated through a continuous 6 to 18% Optiprep gradient, and 17 fractions were collected. An aliquot from each gradient fraction was assessed for hStau and p24 content by Western blot analysis. Figure 1B shows the results of this analysis. As shown previously (22), p24 is predominantly found in the higher-density fractions, corresponding to the four bottom-most fractions. Only these bottom-most fractions contain infectious HIV-1 particles, as determined by infecting MT4 cells with aliquots from each fraction (Fig. 1C) and by syncytium formation (not shown). Most of the hStau cosediments with p24 in this gradient, suggesting strong association with infectious HIV-1 particles. Both p24 and hStau immunoreactivity was found in the top-most gradient fraction (Fig. 1B). These fractions did not contain infectious particles, suggesting that hStau and p24 in this fraction may be derived from lysed virus particles that did not enter the gradient. Alternatively, a fraction of these proteins is present in 293T microvesicles.
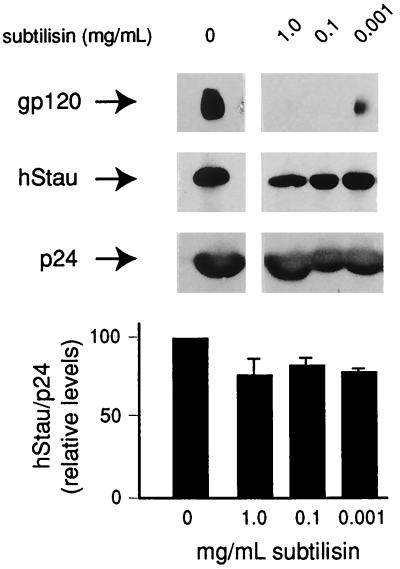
Virus preparations produced from 293T cells contain negligible amounts of microvesicles (29) and therefore are useful for identification of virion-incorporated host cell proteins. Nevertheless, a subtilisin protease assay (56) was performed to substantiate hStau virion incorporation. Purified virus was mock digested or digested with a range of subtilisin concentrations (Fig. 2). While envelope glycoprotein, gp120, was completely degraded with 1.0 and 0.1 mg of subtilisin per ml as expected, p24 and p17 (data not shown) remained protected from protease action since they are found within the virus (Fig. 2). hStau remained intact, though a fraction (approximately 20 to 25%) was sensitive to the subtilisin treatment at the highest subtilisin concentration tested, reflecting what we observed in the Optiprep analyses (Fig. 1B). This phenomenon has previously been observed in virus preparations generated from H9 and CEMss cells, where subtilisin treatment reduced the apparent amount of HIV-1-incorporated actin (57), suggesting that a fraction of this protein is not within virus particles. On the other hand, we could not detect hStau in 200 μg of purified microvesicles from these CEMss cells (a gift from David Ott, NCI-FCRDC, Frederick, Md. [data not shown]), indicating that vesicle-associated hStau is not necessarily observed in all cells. gp120 is incompletely degraded by subtilisin at 0.001 mg/ml, indicating that this concentration of protease is insufficient to remove extravirion—and hence unincorporated—proteins. hStau is not inherently resistant to subtilisin treatment, since 25 μg of recombinant hStau is completely degraded by 0.01 mg of subtilisin per ml (data not shown).
FIG. 2.
Subtilisin protease resistance assay. Subtilisin assays were performed as described in Materials and Methods. Following subtilisin treatment, virus was repelleted and loaded onto SDS-polyacrylamide gels. Subtilisin resistance of gp120, p24, and hStau was assessed by Western analysis. The histogram shows the average from three experiments (±SEM) for each treatment. The signal of hStau was quantitated by phosphorimager analysis and was related to that of p24.
hStau in other retroviruses.
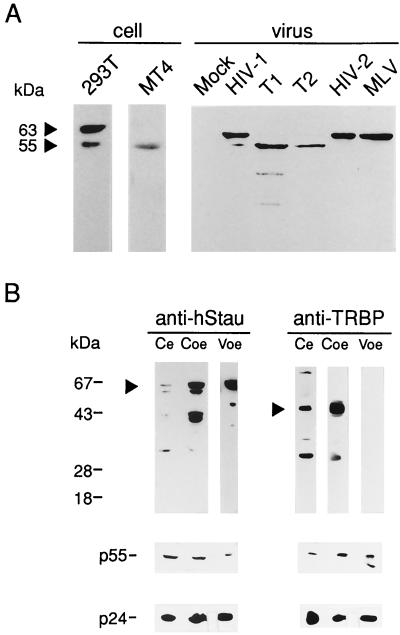
We next examined hStau incorporation in two T-tropic viral clinical isolates passaged in MT4 cells and in the retroviruses HIV-2 and MLV generated in 293T cells. These RNA viruses incorporated hStau (Fig. 3A). hStau was not detectable in mock-transfected 293T cell supernatants and importantly was undetectable in purified, cell-free preparations of the DNA viruses, adenovirus, Epstein-Barr virus, and human herpesvirus 6 (data not shown), suggesting a role in RNA virus biology. While we have characterized at least two isoforms of hStau in human cells (72), we note in this report a preferential cell-type-dependent expression and HIV-1 incorporation of hStau in epithelial 293T and MT4 T lymphocytes (Fig. 3A). Upon longer exposure of this blot, both hStau isoforms do become visible, however, in virus generated in both 293T (HIV-1, MLV, and HIV-2) and MT4 (T1 and T2) cells.
FIG. 3.
hStau is incorporated in HIV-1, HIV-2, and MLV. (A) HIV-1 (pNL4.3), HIV-2 (ROD), and MLV were produced in 293T cells, and T-tropic HIV-1 (T1 and T2) was harvested from MT4 cells; 5 × 107 RT cpm of pelleted virus was loaded onto gels, and virion-associated hStau was assessed by Western blotting. Cell lysates from 293T and MT4 cells (left) show preferential cell-specific expression of a hStau isoform (55 or 63 kDa) due to translation initiation from alternatively spliced transcripts (72). Longer exposures reveal the presence of both hStau species in all virus extracts. (B) HIV-1 selectively incorporates in HIV-1. 293T cells were transfected with pNL4.3 and 5 μg of the expression plasmid for hStau-HA, TRBP, or PKR. Ce, cell extract from pNL4.3-pcDNA3-transfected cells (endogenous levels of antigen included in each panel); Coe and Voe, cell and virus extract from dsRNA-binding protein-overexpressing cells, respectively. Blots were probed with anti-hStau, anti-TRBP, anti-PKR (not shown), or anti-Tat (not shown). p24 was used to reveal p24 and p55 in virus and cell lysates (bottom). Antigens were revealed by ECL and are indicated by arrowheads. Revelation of TRBP in Ce required an exposure time 30 times longer than that for Coe. PKR overexpression completely inhibited HIV-1 production (4); therefore, Ve virus was assessed for the inclusion of PKR. TRBP, PKR, and Tat were undetectable in longer exposures (data not shown). This analysis was performed twice with identical results.
hStau HIV-1 incorporation is selective.
We addressed the selectivity of hStau incorporation by examining the presence of related proteins of hStau in purified viral preparations. hStau can bind HIV-1 TAR, as shown in Northwestern analysis (L. Wickham and L. DesGroseillers, unpublished data), like TRBP, PKR, and HIV-1 Tat. TRBP, PKR, and hStau also share similar dsRNA-binding motifs and thus belong to the same RNA-binding protein family (12). We were not able to detect any other TAR RNA-binding protein, including Tat, PKR, and TRBP, in highly concentrated viral preparations. In data presented in Fig. 3B, hStau is readily detectable endogenously in cells and in overexpression conditions. The appearance of the ≈40-kDa bands in hStau-overexpressing cells is likely due to degradation products during overexpression; these bands are not always observed, however. In virus from hStau-overexpressing cells, hStau content is enhanced (see Fig. 8A) and is immediately apparent. TRBP, on the other hand, remained undetectable in virus preparations derived from HIV-1-transfected 293T cells, despite an approximate 150-fold increase in cellular TRBP levels in overexpression conditions (Fig. 3B). Three different TRBP antisera (kind gifts from Sundararajan Venkatesan and Anne Gatignol) were used in these analyses, and all three yielded identical results. Thus, hStau is unique among these dsRNA-and TAR RNA-binding proteins since it is the only protein from this family that is detectably incorporated in HIV-1.
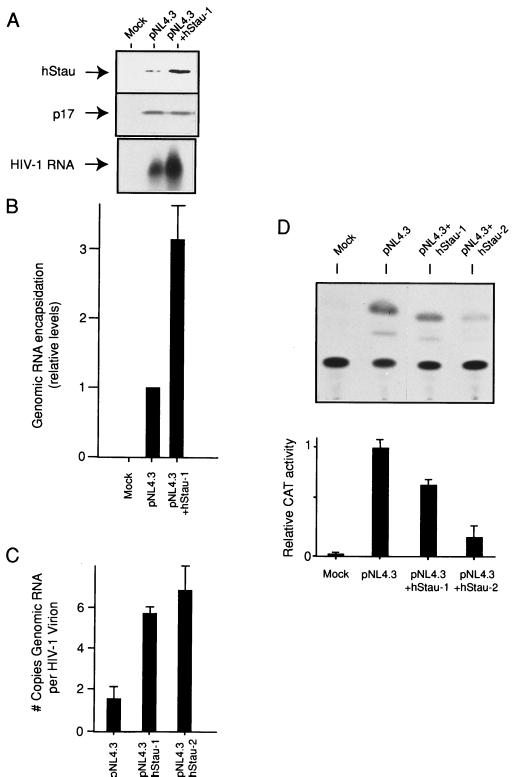
FIG. 8.
hStau overexpression results in enhanced genomic RNA content in virions and dramatically reduces infectivity. 293T cells were transfected with 10 μg of pNL4.3 (plus vector control) or pNL4.3 with hStau-HA cDNA expressor, pNL4.3/hStau-HA (hStau-1, 1 μg; hStau-2, 5 μg). (A) At 48 h posttransfection, virus was harvested and hStau and p17 were identified by Western analysis. Genomic RNA levels were determined by Northern blot analysis, and the results from three experiments (± standard deviation) are presented in panel B. RNA levels in pNL4.3 were arbitrarily set to 1. Only one concentration of hStau-HA was tested (hStau-1). (C) A sensitive RT-PCR method was used to quantitate the number of copies of HIV-1 genomic RNA per virion from hStau-overexpressing cells as described in Materials and Methods. The average (± standard deviation) from two independent determinations is shown. (D) Results from infectivity assays in hStau overexpression studies. BF-24 monocytic indicator cells were infected with virus from mock-, pNL4.3- or pNL4.3-hStau-HA-transfected cells. A representative result from a CAT assay is shown; the bottom panel shows the average virus infectivity levels (±SEM) from three separate experiments. These effects were dose dependent.
hStau incorporation determinant.
It was of interest to determine the molecular determinant within hStau that mediated HIV-1 incorporation. We therefore attempted to identify the region of hStau that conferred incorporation by supplying hStau mutant proteins in trans with HxBru proviral DNA (74). With a series of HA-tagged hStau expressors (Fig. 4A) we localized the major incorporation determinant within the first three dsRBDs (Fig. 4B). This region includes the functional dsRBD (dsRBD3). We therefore tested dsRBDmut, a full-length hStau dsRNA-binding mutant. This protein possesses a single point mutation (F135A) in the dsRBD3 and does not bind RNA, as shown in in vitro Northwestern analyses (Luo and DesGroseillers, submitted). Upon coexpression with HIV-1, dsRBDmut virion incorporation was undetectable, suggesting that hStau incorporation is a direct or indirect result of dsRNA binding via this domain. The protein stability was estimated for each of these mutants in pulse-chase experiments. The WT and dsRBDmut proteins have identical half-lives (approximately 15 h), while the dsRBD2,3,4 hStau mutant has a half-life of approximately 8 h (data not shown).
FIG. 4.
hStau incorporation is mediated through the dsRBD3. (A) WT and hStau mutant constructs were tested for the ability to be incorporated in trans with HxBru as described in Materials and Methods. dsRBD2,3,4 contains the first three dsRBDs, including the major functional domain (dsRBD3). An RNA-binding mutant (dsRBDmut; F135AhStau) was also tested to test the importance of this domain. TBD, tubulin-binding domain. (B) Results from Western analyses of hStau proteins (using anti-HA in Western analyses) and Gag (using anti-p24) in cell and virus extracts. p24 is used to normalize for virus loading. Arrows indicate expected sizes of the expressed hStau proteins.
hStau is not incorporated in VLPs.
VLPs were generated using a recently described Rev-independent Gag-expressor, p55M1-10 (66). Upon transfection of this plasmid, p55 is synthesized and VLPs bud from the plasma membrane (66). Particles are immature and noninfectious. hStau content in these VLPs was assessed. 293T cells were mock transfected or transfected with HxBru or p55M1-10; 48 h posttransfection, virus was harvested and pelleted, and cells were lysed with detergents. hStau was found in all cell lysates (Fig. 5, left). However, when hStau content was assessed in the pelleted virus or VLP preparations, we observed hStau associated only with HxBru virus and not in the mock or p55M1-10 lanes (Fig. 5, right). These data demonstrate that the empty VLPs do not contain hStau and, in addition, indicate that p55Gag is not a major determinant for hStau incorporation, at least in the absence of viral RNA.
FIG. 5.
hStau is not incorporated in VLPs. 293T cells were mock transfected or transfected with 10 μg of HxBru or p55M1-10. Cell and viral lysates were prepared, and hStau (top) and p55 and p25/p24 (bottom) were identified by Western analyses. hStau was only found in purified HxBru virus, not in VLPs generated with p55M1-10.
hStau incorporation correlates with genomic RNA content in HIV-1.
We then attempted to correlate genomic RNA encapsidation with hStau incorporation in HIV-1 particles. Transfection of provirus DNAs yields virus particles that contain comparable amounts of hStau (Fig. 6A, lanes 1 and 2). Genomic RNA encapsidation in HIV-1 is primarily mediated through the association of the packaging psi RNA domain in the 5′ leader sequence with NC (5, 51, 61). Therefore, we initially tested the HIV-1 molecular clone HxBru, in which the 28Cys and 49Cys of NC were mutated to Ser (28C/49C-S [23]) and found that both hStau incorporation and genomic RNA encapsidation were drastically reduced in these virus preparations (Fig. 6A, lane 3). We proceeded to test several other HIV-1 proviruses with NC mutations and deletions (39) and a psi deletion mutant (HXBΔP1 [45]), most of which generate noninfectious virus particles that are significantly impaired in RNA encapsidation. With the exception of the 36C/39C-S NC mutant, transfection of all NC and psi mutant DNA proviral constructs generated virus particles that contained negligible amounts of hStau. Northern blot analyses of genomic RNA encapsidation revealed that the psi and NC mutant constructs yielded virus with drastically reduced levels of genomic 9-kb RNA. In the 36C/39C-S NC mutant virus preparation (Fig. 6A and C, lane 5), hStau is present at 73% of wild-type levels; at the same time we observed clearly detectable levels of genomic RNA encapsidation, consistent with several earlier observations (18, 34, 35, 53). These data indicate that there is a correlation between the levels of genomic RNA encapsidation and hStau incorporation.
FIG. 6.
hStau incorporation quantitatively correlates with genomic RNA content. WT and encapsidation mutant proviral DNAs were transfected into 293T cells as described in Materials and Methods. Supernatants were harvested, and virus was purified as described above. An equal quantity of virus in each well was resolved on a 12% polyacrylamide gel, transferred to nitrocellulose, and probed with anti-hStau (A) and anti-p17 (B). Antigens were revealed by ECL. (C) Results of Northern analysis of encapsidated HIV-1 genomic RNA using RNA that was isolated from equal quantities of virus. Lane 1, pNL4.3; lane 2, HxBru; lane 3, 28C/49C-S NC; lane 4, 15C/18C-S NC; lane 5, 36C/39C-S NC; lane 6 delta 14K-50T NC; lane 7, HXBΔP1.
hStau association with cell and virus-associated HIV-1 RNA.
On the basis of hStau dsRNA-binding capacity and virion incorporation, we addressed the possibility that hStau could bind HIV-1 RNA in HIV-1-producing cells. 293T cells were transfected with either a hStau-HA expressor, both HxBru and hStau-HA, or both HxBru and dsRBDmut. After 48 h, supernatants were harvested, and virus was purified through a sucrose cushion. Virus and cells were lysed in RIPA buffer followed by immunoprecipitation analysis as described in Materials and Methods. Figure 7A shows that equivalent input amounts of hStau protein (wild type or dsRBDmut) was used in each of the immunoprecipitations. Figure 7B shows that the input HIV-1 RNA is approximately equal, when assessed by slot blot analysis as described in Materials and Methods. An identical blot was prepared and probed for GAPDH RNA (74) to control for loading. Figure 7C shows the results of coimmunoprecipitation and RT-PCR analysis of cell lysates. Specific coimmunoprecipitation of HIV-1 RNA with hStau was observed when immune serum to HA was used (lanes 9 and 10). When an anti-TRBP serum was used in this analysis, TRBP and HIV-1 RNA were also coimmunoprecipitated (lanes 7 and 8) (32). HIV-1 RNA was not coimmunoprecipitated using a mouse anti-c-Myc monoclonal antibody (lanes 1 and 2) or preimmune rabbit serum (lanes 3 and 4) in HxBru-hStau-transfected cells, nor was it immunoprecipitated from cells expressing hStau-HA alone using anti-HA (Fig. 7A, lanes 5 and 6). These data demonstrate that hStau associates with genomic HIV-1 RNA in HIV-1-producing cells. Similar analyses were performed using the dsRBDmut in the cotransfections (Fig. 7C, lanes 11 and 12). We did not detect coimmunoprecipitation of HIV-1 RNA.
FIG. 7.
hStau binds HIV-1 RNA. (A) 293T cells were transfected with HxBru, hStau-HA expressor, HxBru, and hStau-HA or dsRBDmut. At 48 h, cells were lysed in RIPA buffer, and an aliquot containing an equal amount of protein was loaded onto a protein gel and used in Western blot analysis using anti-HA to identify hStau and dsRBDmut proteins. (B) Cytosolic RNA was isolated from cells described above, and an equal amount of RNA was loaded onto duplicate filters in a slot blot and probed for HIV-1 or GAPDH RNA. (C) RT-PCR analysis of immunoprecipitates of transfected 293T cells. 293T cells were transfected with hStau-HA alone (lanes 5 and 6), HxBru and hStau-HA expressor (lanes 1 to 4 and 7 to 10), or HxBru and dsRBDmut (lanes 11 and 12) as described above. At 48 h posttransfection hStau was immunoprecipitated from cell lysates (lanes 9 and 10) as described in Materials and Methods, using an anti-HA antiserum. dsRBDmut was immunoprecipitated under the same conditions (lanes 11 and 12). Anti-TRBP was used as a positive control for HIV-1 RNA coimmunoprecipitation (lanes 7 and 8), and negative controls included using an anti-c-Myc (lanes 1 and 2) or a preimmune rabbit serum (lanes 3 and 4) in the immunoprecipitations. Transfection of hStau-HA DNA alone (lanes 5 and 6) followed by anti-HA immunoprecipitation or the exclusion of RT (lane 13) from the RT reaction served as the specific negative controls. (D) Purified virus produced from HxBru-hStau- and HxBru-dsRBDmut-expressing cells was used in immunoprecipitation analyses. Anti-c-Myc (lanes 1 and 2) and an anti-TRBP (lanes 3 and 4) were used in the immunoprecipitation as nonspecific controls, while an anti-HA specifically coimmunoprecipitated HIV-1 RNA from purified viral lysates from HxBru-hStau-HA-expressing cells (lanes 5 and 6) but not from virus generated from HxBru-dsRBDmut-expressing cells (lanes 7 and 8). Lane 9 shows the RT-PCR result when RT was excluded from the RT reaction. Numbers on the left are sizes of DNA molecular weight standards (in base pairs). Odd-numbered lanes, primer pair a; even-numbered lanes: primer pair b (see Materials and Methods).
When a similar analysis was performed with purified virus from HxBru-hStau-transfected cells (Fig. 7D), we observed a similar coimmunoprecipitation of HIV-1 RNA with hStau using an anti-HA antiserum (Fig. 7B, lanes 5 and 6) but not with the use of the mouse anti-c-Myc monoclonal antibody (lanes 1 and 2) or the anti-TRBP serum (lanes 3 and 4), suggesting that hStau remains associated to the HIV-1 RNA genome during viral assembly and maturation and is associated with the HIV-1 core. As expected, HIV-1 RNA was not detected in the immunoprecipitation of dsRBDmut from virus from HxBru-dsRBDmut-transfected cells (Fig. 7D, lanes 7 and 8).
hStau overexpression in 293T cells.
In our attempts to understand the role of hStau in HIV-1 replication, we performed overexpression studies. 293T cells were transfected with pNL4.3 proviral DNA (plus empty vector) or both pNL4.3 and the hStau-HA expressor. When hStau content in purified virus preparations was assessed, incorporation of hStau into HIV-1 was found to be enhanced when hStau was overexpressed (Fig. 8A). When we relate the signals for hStau to those of p17, hStau incorporation is increased over 2- to 3-fold (2.5-fold ± 0.4 [standard error of the mean {SEM}]). We next examined HIV-1 genomic RNA encapsidation levels by Northern analyses from equal quantities of virus (normalized by p24 ELISA) from the corresponding virus preparations. Genomic RNA content increased severalfold in virus generated from hStau-overexpressing cells (Fig. 8A and B). To confirm these results, similar experiments were performed using Roche's Amplicor RT-PCR kit (27) to quantitate the number of genomic RNA copies per virion. Results from this highly sensitive analysis reflected what we had observed in the Northern analyses: using the same amount of hStau-HA expressor in the transfections, a nearly threefold increase in the number of copies of genomic HIV-1 RNA per virion was observed (Fig. 8C). Using 5 μg of hStau-HA expressor in the transfections slightly enhanced but did not markedly alter the RNA encapsidation levels (Fig. 8C).
The titer of virus from hStau-overexpressing cells was tested, and the results from these analyses are shown in Fig. 8D. Equal amounts of virus generated in cells expressing pNL4.3 or pNL4.3 and hStau-HA were used to infect BF-24 monocytic indicator cells (these cells contain an integrated CAT gene downstream of the HIV-1 long terminal repeat). At 48 h posttransfection, virus was harvested and purified by ultracentrifugation. We observed a marked impairment in the infectivity of virus derived from hStau-overexpressing cells (Fig. 8D). Data from three independent experiments showed a 6.7-fold (±1.4 [SEM]) reduction in infectivity in virus harvested from hStau-overexpressing cells, using 5 μg in the transfections (Fig. 8D). A dose-dependent effect on viral infectivity was observed, but this too was maximal at the concentration used in the experiments presented in Fig. 8D (5 μg). Evaluation of the infectivity of virus from hStau-overexpressing cells using the MAGI assay concurred with the results obtained in BF-24 assays. These consistently revealed a fourfold (±0.3) reduction in infectivity using 5 μg of hStau-HA in the transfections (data not shown).
DISCUSSION
hStau incorporation in HIV-1 likely reflects an important biological function for this host cell dsRNA-binding protein in HIV-1 replication. hStau is incorporated in HIV-1, as shown in two gradient analyses and a subtilisin assay (Fig. 1 and 2). hStau is not passively incorporated into HIV-1 due to its dsRNA-binding capacity since PKR and TRBP, which possess similar dsRNA-binding motifs (12) and have comparable cellular distributions, are not detectable in HIV-1 preparations (Fig. 3B). Both TRBP and hStau bind genomic HIV-1 RNA (Fig. 7C) but only hStau is incorporated (Fig. 3B), suggesting a separable and functional difference. And finally, hStau is not detectable in Gag VLPs (Fig. 5), and virion-incorporated levels of hStau correlate with the genomic RNA content in virions (Fig. 6), both evidence that suggests the dependence on genomic RNA encapsidation, and not p55Gag binding, for example, for incorporation. Whereas several other host cell RNA-binding proteins are capable of binding HIV-1 RNA, among which are hnRNP A1 (9), CRM1 (10), and poly(A)-binding protein (1), there is no evidence to show that these proteins are virion incorporated, indicating that HIV-1 RNA binding does not necessarily lead to incorporation.
We have ruled out the involvement of several viral proteins in hStau incorporation, including Env, Vpr, Pol (RT, IN, and protease), Vif, Vpu, and Nef, with the use of mutant proviral clones. Preliminary two-hybrid and coimmunoprecipitation data show that hStau does not interact strongly with p55Gag. Furthermore, hStau is not incorporated in VLPs (which do not encapsidate RNA [Fig. 5]). These data collectively indicate that an RNA genome interaction(s) is likely to be the main mechanism for incorporation. We cannot, however, rule out contributions from other factors that may come into play during HIV-1 replication.
In this study we have attempted to define the determinant by which hStau is incorporated in HIV-1 particles: it requires an intact functional dsRBD (dsRBD3 [Fig. 4]). A single point mutation (F135AhStau) abolishes RNA binding (Luo and DesGroseillers, submitted) and drastically reduces hStau HIV-1 incorporation. We have no information about the HIV-1 RNA-binding sites of hStau except that it binds the TAR RNA in vitro (Wickham and DesGroseillers, unpublished), though structured RNA domains characteristic of retrovirus leader sequences (37) could potentially serve as binding sites. It is possible that the F135A mutation in the dsRNA-binding mutant protein affects other types of interactions, in addition to RNA binding. In TRBP, for instance, both TRBP dimerization and TRBP-PKR interaction are mediated through dsRBD (Anne Gatignol, personal communication; 16). Very little is known about interacting partners of hStau except that it can bind the influenza virus NS1 protein (25) and can also exist as a homodimer (Luo and DesGroseillers, submitted). Further analyses will be required to determine whether these interactions are mediated through dsRBD3.
While we do not completely understand hStau's function in HIV-1 replication, we demonstrate that hStau overexpression can enhance severalfold the abundance of genomic HIV-1 RNA in virions. Both Northern analysis and a very sensitive RT-PCR method reveal an almost threefold increase in the number of encapsidated RNA molecules per virion (Fig. 8). The effect approximated a dose-response curve, although genomic RNA content appeared to plateau when 5 μg of hStau-HA expressor was included in the transfection (Fig. 8C). Furthermore, in proviruses harboring NC mutations, a loss of selectivity of genomic RNA results in enhanced levels of subgenomic and cellular RNA packaging. hStau function, which is likely to be dependent on RNA binding, appears to be somehow involved in NC function such that in NC mutants that prevent selective genomic RNA encapsidation, hStau incorporation is also abrogated (Fig. 6). While NC is the principal determinant for selective genomic RNA encapsidation via binding to the psi packaging signal (18, 20), how and where hStau acts remain to be established. However, hStau's capacity to drive RNA encapsidation points to a role of this protein at the level of the selection of genomic RNA molecules destined for encapsidation.
Earlier work from several investigators has suggested that there are constraints to the length of the encapsidated genomic RNA in retroviruses (33, 38). While this appears to be obvious, especially since there must be a packaging limit of the retroviral core, more recent data indicate that the understanding of this assembly step is still evolving. For instance, there is evidence to suggest that two RNA dimers (i.e., four copies) can be coencapsidated in RSV (64), although the RNA molecules tested were shorter than the genomic RNA. More recently, however, an RNA of ≈20 kb, over two times the wild-type size (68), was shown to be packaged in MLV particles, resulting in impaired infectivity. These data suggested that replication defects associated with lengthened genomes are evident at several replication steps and may be less pronounced at the stage of RNA packaging. The infectivity of virus from hStau-overexpressing cells is also significantly impaired. However, in our study it is the number of copies per virion, not the length, that is increased (Fig. 8). Our data are consistent with the possibility that it is not only the quantity of RNA packaged that is primarily responsible for the replication defect, but defects at multiple steps of replication including encapsidation, reverse transcription, and integration can collectively contribute to impaired infectivity (68). It remains to be determined how the increase in genomic RNA encapsidation mediated by hStau can affect virion infectivity in the HIV-1 replication cycle.
Although several cellular RNA-binding proteins such as RNA helicase A, hnRNP A1, eukaryotic initiation factor 5A, and poly(A)-binding protein are reported to influence HIV-1 gene expression at several levels including RNA splicing, nucleocytoplasmic transport, RNA metabolism, and translation (1, 8, 46, 55), recent evidence suggests that mammalian Stau, like its Drosophila counterpart, is involved in RNA transport (43). In HIV-1 replication, hStau appears to be involved in pre- and postassembly since it is found associated with HIV-1 RNA in both cytosolic and purified viral lysates (Fig. 7C and D). Moreover, both hStau HIV-1 RNA binding in virus-producing cells and its RNA binding and transport function in Drosophila raise the intriguing possibility that hStau is involved in the transport of HIV-1 genomic RNA. Its presence in several retroviruses (Fig. 3A) also suggests that it is implicated in a mechanism that is common to all of these viruses.
We have calculated that only a few molecules (≈2 to 5) of hStau are incorporated per HIV-1 virion (A. J. Mouland and E. A. Cohen, unpublished data). We are able to enhance this amount by overexpressing hStau in cells (Fig. 8). In antisense studies not presented here, we were able to downmodulate hStau viral incorporation by 40 to 50% (Mouland and Cohen, unpublished). In both of these conditions, the result is the production of virus with impaired infectivity, suggesting that an appropriate amount of incorporated hStau is a requirement to generate infectious viral particles. Similar conclusions were made for HIV-1-incorporated levels of cyclophilin A (75) and for several cellular adhesion molecules in the HIV-1 envelope (59). On the other hand, the amounts of several cellular proteins embedded within the HIV-1 envelope appear to be variable (3). More work will be required to study hStau's mechanism of action; however, our data show that hStau's HIV-1 incorporation levels appear to influence the production of infectious viral particles.
The studies presented here are not the first to implicate hStau in virus-host interactions. For instance, the nonstructural protein NS1, a protein involved in blocking nucleocytoplasmic transport of cellular polyadenylated RNAs during influenza virus infection, can bind hStau as shown in two-hybrid analysis and in in vitro binding assays (25). It is believed that both protein and RNA binding are involved since hStau and NS1 physically interact and both bind dsRNA. Likewise, RNA and protein interactions have recently been shown to be critical for dStau's function (67). Based on the data presented here, hStau's function during HIV-1 expression appears to principally rely on an RNA-binding event. Inasmuch as the Gag-Gag dimerization depends on RNA (13) and elongation factor 1α incorporation of HIV-1 is dependent on both protein and RNA interactions (14), it is likely that both types of interactions will be exposed as important elements in hStau's function during HIV-1 replication.
Future studies to investigate the detailed mechanism of action of hStau will provide important clues to the understanding of the involvement of host cell proteins in the late steps of the retroviral replication cycle.
ACKNOWLEDGMENTS
We thank Sundararajan Venkatesan, Anne Gatignol, and Antonis Koromilas for antisera; Guy Lemay, Lawrence Kleiman, Anne Gatignol, Eliane Meurs, and George Pavlakis for plasmid constructs; Mark Wainberg, Bernard Massie, Ali Ahmad, and José Menezes for providing virus stocks; David Ott for purified microvesicles and advice on subtilisin assays; Julie Deschambeault for performing MLV RT assays; Alice Telesnitsky for sharing data prior to publication; and Dominique Bergeron and Xiao-Jian Yao for comments. Anti-IN serum, HIV-2 (ROD), and pNL4.3 proviral DNA were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID.
A.J.M. was a recipient of a National Health Research Development Program (NHRDP)/Medical Research Council (MRC) of Canada postdoctoral fellowship for most of this work and is a recipient of a scholarship from the Fonds de recherche en santé du Québec. E.A.C. is a recipient of a Scientist Award from the MRC. This work was supported by grants from the Canadian Foundation for AIDS Research (A.J.M.), the NHRDP (L.D.), the Natural Sciences and Engineering Research Council of Canada (L.D.), the Fonds pour la formation des chercheurs et l'aide à la recherche (E.A.C.), and the MRC of Canada (E.A.C.).
REFERENCES
- 1.Afonina E, Neumann M, Pavlakis G N. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem. 1997;272:2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 2.Arn E A, Macdonald P M. Motors driving mRNA localization: new insights from in vivo imaging. Cell. 1998;95:151–154. doi: 10.1016/s0092-8674(00)81745-6. [DOI] [PubMed] [Google Scholar]
- 3.Bastiani L, Laal S, Kim M, Zolla-Pazner S. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J Virol. 1997;71:3444–3450. doi: 10.1128/jvi.71.5.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adapter complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bess J W, Jr, Gorelick R J, Bosche W J, Henderson L E, Arthur L O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 8.Bevec D, Jaksche H, Oft M, Wohl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, Lottspeich F, Hauber J. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 9.Black A C, Luo J, Chun S, Bakker A, Fraser J K, Rosenblatt J D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes. 1996;12:275–285. doi: 10.1007/BF00284648. [DOI] [PubMed] [Google Scholar]
- 10.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broadus J, Fuerstenberg S, Doe C Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 12.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 13.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimarelli A, Luban J. Translation elongation factor 1α interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 18.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 20.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 21.Deschambeault J, Lalonde J P, Cervantes-Acosta G, Lodge R, Cohen E A, Lemay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorfman T, Luban J, Goff S P, Haseltine W A, Göttlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and retrovirus assembly. Virology. 1983;130:415–426. doi: 10.1016/0042-6822(83)90096-x. [DOI] [PubMed] [Google Scholar]
- 25.Falcon A M, Fortes P, Marion R M, Beloso A, Ortin J. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 1999;27:2241–2247. doi: 10.1093/nar/27.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrandon D, Elphick L, Nusslein-Volhard C, St. Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 27.Fischer M, Huber W, Kallivroussis A, Ott P, Opravil M, Luthy R, Weber R, Cone R W. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37:1260–1264. doi: 10.1128/jcm.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fortin J F, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol. 1997;71:3588–3596. doi: 10.1128/jvi.71.5.3588-3596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortin J F, Cantin R, Tremblay M J. T cells expressing activated LFA-1 are more susceptible to infection with human immunodeficiency virus type 1 particles bearing host-encoded ICAM-1. J Virol. 1998;72:2105–2112. doi: 10.1128/jvi.72.3.2105-2112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 31.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatignol A, Buckler C, Jeang K T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsl kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelinas C, Temin H M. Nondefective spleen necrosis virus-derived vectors define the upper size limit for packaging reticuloendotheliosis viruses. Proc Natl Acad Sci USA. 1986;83:9211–9215. doi: 10.1073/pnas.83.23.9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grandgenett D P, Goodarzi G. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci. 1994;3:888–897. doi: 10.1002/pro.5560030604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison G P, Hunter E, Lever A M. Secondary structure model of the Mason-Pfizer monkey virus 5′ leader sequence: identification of a structural motif common to a variety of retroviruses. J Virol. 1995;69:2175–2186. doi: 10.1128/jvi.69.4.2175-2186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman S A, Coffin J M. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science. 1987;236:845–848. doi: 10.1126/science.3033828. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, Khorchid A, Wang J, Parniak M A, Darlix J L, Wainberg M A, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into human immunodeficiency virus type 1. J Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiebler M A, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion R M, Ortin J, Dotti C G. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim-Ha J, Kerr K, Macdonald P M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 42.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhrmann M, Luo M, Kaether C, DesGroseillers L, Dotti C G, Kiebler M A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavallée C, Yao X J, Ladha A, Göttlinger H, Haseltine W A, Cohen E A. Requirement of the Pr55gag precursor for incorporation of the Vpr product into human immunodeficiency virus type 1 viral particles. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lever A, Göttlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Tang H, Mullen T M, Westberg C, Reddy T R, Rose D W, Wong-Staal F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li P, Yang X, Wasser M, Cai Y, Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- 48.Liu B, Dai R, Tian C J, Dawson L, Gorelick R, Yu X F. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marion R M, Fortes P, Beloso A, Dotti C, Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miele G, Mouland A, Harrison G P, Cohen E, Lever A M. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizuno A, Ido E, Goto T, Kuwata T, Nakai M, Hayami M. Mutational analysis of two zinc finger motifs in HIV type 1 nucleocapsid proteins: effects on proteolytic processing of Gag precursors and particle formation. AIDS Res Hum Retroviruses. 1996;12:793–800. doi: 10.1089/aid.1996.12.793. [DOI] [PubMed] [Google Scholar]
- 54.Mouland A J, Bevan S, White J H, Hendy G N. Human chromogranin A gene. Molecular cloning, structural analysis, and neuroendocrine cell-specific expression. J Biol Chem. 1994;269:6918–6926. [PubMed] [Google Scholar]
- 55.Najera I, Krieg M, Karn J. Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J Mol Biol. 1999;285:1951–1964. doi: 10.1006/jmbi.1998.2473. [DOI] [PubMed] [Google Scholar]
- 56.Ott D E, Coren L V, Johnson D G, Sowder R C, II, Arthur L O, Henderson L E. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res Hum Retroviruses. 1995;11:1003–1006. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- 57.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paquette J S, Fortin J F, Blanchard L, Tremblay M J. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J Virol. 1998;72:9329–9336. doi: 10.1128/jvi.72.11.9329-9336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perotti M E, Tan X, Phillips D M. Directional budding of human immunodeficiency virus from monocytes. J Virol. 1996;70:5916–5921. doi: 10.1128/jvi.70.9.5916-5921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poon D T, Li G, Aldovini A. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 63.Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 64.Sakalian M, Wills J W, Vogt V M. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuldt A J, Adams J H, Davidson C M, Micklem D R, Haseloff J, Johnston D S, Brand A H. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin N-H, Hartigan-O'Connor D, Pfeiffer J K, Telesnitsky A. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J Virol. 2000;74:2694–2702. doi: 10.1128/jvi.74.6.2694-2702.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 70.Tang Y, Winkler U, Freed E O, Torrey T A, Kim W, Li H, Goff S P, Morse H C. Cellular motor protein KIF-4 associates with retroviral Gag. J Virol. 1999;73:10508–10513. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terwilliger E F, Cohen E A, Lu Y C, Sodroski J G, Haseltine W A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci USA. 1989;86:5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickham L, Duchaine T, Luo M, Nabi I R, DesGroseillers L. Mammalian staufen is a double-stranded-RNA-and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yakunin A F, Hallenbeck P C. A luminol/iodophenol chemiluminescent detection system for western immunoblots. Anal Biochem. 1998;258:146–149. doi: 10.1006/abio.1998.2571. [DOI] [PubMed] [Google Scholar]
- 74.Yao X J, Mouland A J, Subbramanian R A, Forget J, Rougeau N, Bergeron D, Cohen E A. Vpr stimulates viral expression and induces cell killing in human immunodeficiency virus type 1-infected dividing Jurkat T cells. J Virol. 1998;72:4686–4693. doi: 10.1128/jvi.72.6.4686-4693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin L, Braaten D, Luban J. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J Virol. 1998;72:6430–6436. doi: 10.1128/jvi.72.8.6430-6436.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]