Abstract
The emergence of syncytium-inducing (SI) variants of human immunodeficiency virus type 1 (HIV-1) in infected individuals is an indicator of poor prognosis and is often correlated with faster CD4+ cell depletion and rapid disease progression. Interleukin-4 (IL-4) is a pleiotropic cytokine with various immune-modulating functions including induction of immunoglobulin E (IgE) production in B cells, down-regulation of CCR5 (a coreceptor for HIV-1 non-SI [NSI] strains), and up-regulation of CXCR4 (a coreceptor for HIV-1 SI variants). Here we show that homozygosity of a polymorphism in the IL-4 promoter region, IL-4 −589T, is correlated with increased rates of SI variant acquisition in HIV-1-infected individuals in Japan. This mutation was also shown to be associated with elevated serum IgE levels in HIV-1-infected individuals, especially in those at advanced stages of disease. In contrast, neither a triallele polymorphism in IL-10, another Th2 cytokine, nor a biallele polymorphism in the RANTES promoter affected acquisition of the SI phenotype. This finding suggested that IL-4-589T increases IL-4 production in the human body and thus accelerates the phenotypic switch of HIV-1 from NSI to SI and possibly disease progression of AIDS.
Human immunodeficiency virus type 1 (HIV-1) isolates have been classified as T-cell-line-tropic or syncytium-inducing (SI) viruses and macrophagetropic or non-SI (NSI) viruses according to their biological characteristics (3, 5, 7, 38, 39). NSI viruses are more likely to be transmitted through sexual contact (50) and are present early after infection in nearly all HIV-1-infected individuals (7, 34). In contrast, SI variants are usually present in the latter stages of infection in more than half but not all of HIV-1-infected individuals (34, 35). The emergence of SI variants is presumably a sign of poor prognosis and often correlates with faster CD4+ cell depletion and rapid disease progression (7, 13, 30). However, the host factors which affect the rate of conversion of NSI viruses to SI variants are poorly understood.
Interleukin-4 (IL-4) is a pleiotropic cytokine produced primarily by activated CD4+ T lymphocytes, mast cells, and basophils (28). IL-4 has multiple immune response-modulating functions on a variety of cell types (14). It induces immunoglobulin E (IgE) production in B lymphocytes (9) and serves as an important regulator of IgG isotype switching (44). It also regulates the differentiation of precursor T helper cells into those of the Th2 subset that mediate humoral immunity and modulate antibody production (31). With respect to HIV-1 infection, IL-4 was reported to differentially regulate two major HIV-1 coreceptors, CXCR4 for SI variants and CCR5 for NSI viruses (41, 46, 47). IL-4 down-regulates CCR5 expression and thus inhibits replication of HIV-1 NSI isolates in human T cells and macrophages (37, 41, 47). On the other hand, IL-4 up-regulates the expression of CXCR4 (46). In addition, IL-4 stimulates the expression of HIV-1 through activation of viral transcription (41). The combination of these effects of IL-4 on HIV-1 replication may be involved in the phenotypic switch from NSI to SI as well as disease progression in HIV-1 infection. Thus, IL-4 could be an important factor for viral evolution and AIDS pathogenesis.
Recently, polymorphisms in HIV-1 coreceptors and their natural ligand genes have been shown to modify HIV-1 transmission and disease progression (8, 15, 17–21, 25, 33, 36, 42, 48). Rosenwasser et al. reported a polymorphism with a C-to-T exchange at position −589 upstream from the open reading frame of the IL-4 gene, IL-4 −589T, that is associated with increased promoter activity for IL-4 transcription and elevated levels of serum IgE in asthmatic families (32). To evaluate the possible role of this IL-4 polymorphism in acquisition of SI variants and the modulation of disease progression in HIV-1 infection, we analyzed IL-4 genes of 339 HIV-1-infected individuals in Japan and show here that IL-4 −589T is indeed associated with increased rates of acquisition of HIV-1 SI variants.
MATERIALS AND METHODS
Clinical samples.
Blood from 339 HIV-1-infected and 52 non-HIV-1-infected Japanese individuals was collected. HIV-1-infected patients included 137 hemophiliacs who were infected through contaminated blood products between 1982 and 1985. Among 202 HIV-1-infected nonhemophiliacs, 112 reported homosexual or bisexual intercourse, while 90 (58 males and 32 females) reported heterosexual intercourse, as a risk factor upon a structured interview on sexual activity. All hemophiliacs and those who reported homosexual or bisexual intercourse were males. Peripheral blood mononuclear cells (PBMC) were obtained from the blood by using the Ficoll-Hypaque (Amersham Pharmacia) method. DNA was extracted from the PBMC by a method previously described (17). The CD4+ lymphocyte depletion rates were calculated for 128 HIV-1-infected individuals who had five or more CD4+ lymphocyte counts recorded before any kind of antiretroviral therapy started. Of these individuals, 112 had 10 or more CD4+ lymphocyte counts recorded, and 97 had 20 or more. Informed consent was obtained from all individuals described in this study.
Genotyping of IL-4 gene.
DNA fragments corresponding to a 1,208-bp upstream noncoding region of the IL-4 gene (2) were PCR amplified by using primer pair 4-1 (5′-GAATTCAATAAAAAACAA-3′) and 4-1190 (5′-GAACAGAGGGGGAAGCA-3′) (Fig. 1). The amplified region contained 1,107 bp of the immediate 5′ upstream region of the major transcription start site, the 5′ (65-bp) untranslated region, and 36 bp of the 5′ coding region (Fig. 1). PCR was performed in a 50-μl reaction mixture containing 1 μg of DNA (40 cycles of 94°C for 30 s, 49.2°C for 30 s, and 72°C for 1 min). Amplified DNA fragments were purified and sequenced by using primers 4-1, 4-200 (5′-ATCTCAAATTCCTGGGCTCAAGT-3′), 4-693 (5′-GGAAGAAGCCAGGTTA-3′), and 4-814 (5′-GGCTGCTGCTGGCTTTTT-3′). Sequencing reactions were performed according to the dideoxy-chain termination method by using an ABI Prism 377 automated DNA sequencer (Applied Biosystems). For genotyping of IL-4 C-589T polymorphism, we performed PCR-restriction fragment length polymorphism analysis according to the protocol described by Noguchi et al. (27), with a modification. Briefly, the region spanning IL-4 −589 was amplified by PCR with primer pair 562m (5′-TAAACTTGGGAGAACATGGT-3′) and 756m (5′-TGGGGAAAGATAGAGTAATA-3′). The underlined base indicates a mismatched at position −592 which was used to introduce the AvaII restriction site when the position −589 was C. PCR was performed for 40 cycles at 94°C for 30 s, 48°C for 30 s, and 72°C for 30 s. Digestion of the 195-bp amplified products with AvaII yielded 177- and 18-bp fragments when position −589 was C.
FIG. 1.
Nucleotide sequence of the IL-4 promoter region. Numbers indicate nucleotide positions relative to the translation start site (marked with a filled triangle). A major transcription start site is indicated by an open triangle. The numbers −1098, −589, −144, and −33 indicate the four polymorphic positions. A C-to-T polymorphism at position −589 was previously reported to be present at −590 (32). A possible consensus site for TATA is underlined. Primer positions used for PCR amplification are underlined with arrows indicating the direction of the primers. Asterisks, #, and a filled arrowhead below the sequence indicate insertions, base changes, and a deletion of two nucleotides, respectively, which were absent in a previous report (33). Each of our sequences possesses these nucleotides. At present, it is unclear whether these differences represent sequence polymorphisms between white and Japanese individuals.
Genotyping of the IL-10 gene.
The segment of the promoter region of the IL-10 gene from −1120 to −533 was amplified as described by Mok et al. (24), using primer pair 10-up (5′-ATCCAAGACAACACTACTAA-3′) and 10-down (5′-TAAATATCCTCAAAGTTCC-3′). The PCR products were purified and sequenced by using the 10-down primer.
Genotyping of the CCR5 promoter region.
We determined the sequence of 688 nucleotides in the upstream noncoding region of the CCR5 gene spanning the entire second and third exons and part of the first and second introns as described previously (15).
HIV-1 isolation, PCR, and sequencing.
Approximately 106 PBMC obtained from HIV-1-infected individuals were cocultivated with the same number of PBMC obtained from healthy donor which were stimulated with phytohemagglutinin P (3 μg/ml) for 3 days. HIV-1 replication was monitored by quantifying the levels of p24 antigens in the culture supernatant (Intracel). Total RNA was extracted from 50 μl of cell-free virus as described previously (35). HIV-1 RNA was reverse transcribed and amplified with nested PCR primers specific for a region spanning the gp120 V1 to V3 of the env gene. The primer used for reverse transcription was E1 (5′-GGTAGAACAGATGCATGAGGAT-3′), and those used for the first round of PCR were E1 and MK601 (5′-TTCTCCAATTGTCCCTCATATCGCCTCCTC-3′). Inner primers used for the second PCR were E2 (5′-ATCAGTTTATGGGATCAAAGAAT-3′) and YT001 (5′-ACAATTTCTGGGTCCCCTCCTGAGGA-3′). The nested PCR products were purified and sequenced using the primer MK650 (5′-AATGTCAGCACAGTACAATGTACAC-3′).
MT2 assay.
One hundred microliters of isolated virus containing 3 ng of p24 was inoculated into 106 MT2 cells. The culture supernatant was harvested 7 days after infection and assayed for p24 levels by enzyme-linked immunosorbent assay (ELISA; Intracel).
Determination of total serum IgE, IgG, IgA, and IgM levels.
Total serum IgE, IgG, IgA, and IgM levels were determined by using commercially available ELISA kit (Dynabott).
Statistical analysis.
The unpaired t test and χ2 test were used.
RESULTS
Sequence variation in the IL-4 promoter.
We determined sequences of 1,170 nucleotides in the upstream noncoding regions of IL-4 genes obtained from 12 non-HIV-1-infected and 36 HIV-1-infected individuals in Japan. Our results confirmed the presence of a previously described polymorphism with a C-to-T change at position −589 upstream from the open reading frame of the IL-4 gene. We also identified three new polymorphisms, T to G at −1098, C to T at −145, and C to T at −33 (Fig. 1). As shown in Table 1, we observed seven genotypes of the IL-4 promoter. Therefore, at least four haplotypes (I, II, III, and IV [Table 1]) are present in Japan. Among them, haplotypes III and IV contained −589T. The C-to-T change at position −589 was completely associated with the C-to-T change at position −33. Haplotypes II and IV contained minor polymorphisms at positions −1098 and −145, respectively.
TABLE 1.
IL-4 genotypes and haplotypes in Japan
| Genotype or haplotype | Genotype/haplotype
|
No. (frequency, %) | |||
|---|---|---|---|---|---|
| −1098a | −589 | −144 | −33 | ||
| Genotype | |||||
| 1 | T/T | C/C | C/C | C/C | 2 (4.2) |
| 2 | T/G | C/C | C/C | C/C | 3 (6.3) |
| 3 | T/G | C/T | C/C | C/T | 2 (4.2) |
| 4 | T/T | C/T | C/C | C/T | 21 (43.8) |
| 5 | T/T | T/T | C/C | T/T | 18 (37.5) |
| 6 | T/T | T/T | C/T | T/T | 1 (2.1) |
| 7 | T/T | T/T | T/T | T/T | 1 (2.1) |
| Total | 48 (100) | ||||
| Haplotype | |||||
| I | T | C | C | C | 29 (30.2) |
| II | G | C | C | C | 5 (5.2) |
| III | T | T | C | T | 59 (61.5) |
| IV | T | T | T | T | 3 (3.1) |
| Total | 96 (100) | ||||
Nucleotide position relative to the translation start site of the IL-4 gene. Major transcription start site is at −65.
Polymorphism in the IL-4 promoter in HIV-1-infected and non-HIV-1-infected individuals.
Since the T allele of the IL-4 promoter at position −589 was reported to be associated with elevated levels of IgE and increased rates of asthma (32), we analyzed additional 303 HIV-1-infected and 40 non-HIV-1-infected subjects by PCR-restriction fragment length polymorphism assay. Table 2 shows the frequency of the IL-4 promoter genotype at position −589 in 339 HIV-1-infected and 52 non-HIV-1-infected individuals in Japan. In non-HIV-1-infected individuals, the frequency of the T allele was 0.69, which is in agreement with the previously reported allele frequency in Japan (27). On the other hand, HIV-1-infected individuals contained more C homozygotes and C/T heterozygotes and fewer T homozygotes than uninfected individuals, showing a slight but significant deviation from Hardy-Weinberg expectation (P = 0.010) and a weak trend toward less T allele frequency (0.64, P = 0.29). This association was specifically seen in individuals who reported sexual intercourse as a risk factor but not in hemophiliac HIV-1-infected individuals. Among individuals who reported sexual intercourse as a risk factor, those who reported heterosexual intercourse showed the most significant deviation from the Hardy-Weinberg expectation (P = 0.0003) and the least frequency of T allele (0.56, P = 0.025). Both men and women in this category showed less T allele frequency than non-HIV-1-infected individuals (0.55 for men and 0.56 for women). This result suggested that the IL-4 −589T polymorphism may be involved in protection against HIV-1 when HIV-1 is transmitted through heterosexual contact.
TABLE 2.
IL-4 genotypes among HIV-1-infected and uninfected individuals
| Clinical status | IL-4 genotype at −589
|
P value (HWEa) | T frequency | P value (t test) | |||
|---|---|---|---|---|---|---|---|
| No. | C/C (%) | C/T (%) | T/T (%) | ||||
| HIV-1 infected | 339 | 13.0 | 46.3 | 40.7 | 0.01 | 0.64 | 0.29 |
| Hemophiliac | 137 | 9.5 | 45.4 | 45.3 | 0.81 | 0.68 | 0.80 |
| Nonhemophiliac | 202 | 15.4 | 47.0 | 37.6 | 0.002 | 0.61 | 0.13 |
| Homosexual/bisexual | 112 | 12.5 | 43.7 | 43.8 | 0.47 | 0.66 | 0.52 |
| Heterosexual | 90 | 18.9 | 51.1 | 30.0 | 0.0003 | 0.56 | 0.025 |
| Male | 58 | 22.4 | 44.8 | 32.8 | 0.001 | 0.55 | 0.035 |
| Female | 32 | 12.5 | 62.5 | 25.0 | 0.03 | 0.56 | 0.093 |
| Uninfected | 52 | 9.6 | 42.3 | 48.1 | 1 | 0.69 | |
HWE, Hardy-Weinberg equilibrium.
IL-4 −589T and disease progression in HIV-1 infection.
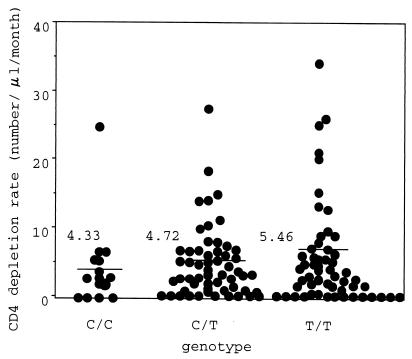
We examined the effect of IL-4 −589T on the rate of disease progression in HIV-1-infected individuals. Among 339 infected patients analyzed, we were able to calculate CD4+ cell depletion rates for 128 (15 C homozygotes, 57 C/T heterozygotes, and 56 T homozygotes) before any kind of antiretroviral therapy started. There was no significant difference in the CD4+ T cell counts at the beginning of the observation period among C homozygotes (mean ± standard deviation was 415 ± 152 cells/μl), heterozygotes (485 ± 212 cells/μl), and T homozygotes (468 ± 191 cells/μl). As shown in Fig. 2, there is a weak but apparent trend toward faster loss in the T homozygotes (5.46 ± 7.30 cells/μl/month) than heterozygotes (4.72 ± 5.18 cells/μl/month) or C homozygotes (4.33 ± 6.18 cells/μl/month), although the difference was not statistically significant. In a cohort of 77 HIV-1-infected hemophiliacs who have been followed up for at least 9 years after the primary infection, T homozygotes also showed a weak trend toward faster CD4+ cell depletion than others, but the difference was again not statistically significant (data not shown).
FIG. 2.
Effect of sequence polymorphism in the IL-4 promoter region on CD4+ lymphocyte depletion rate. The CD4+ lymphocyte depletion rate of each HIV-1-infected individual is represented by a circle; a horizontal bar represents the mean CD4+ lymphocyte depletion rate of each genotype group.
IL-4 −589T is associated with acquisition of the SI phenotype.
IL-4 is known to up-regulate CXCR4 expression (46) while causing a dramatic reduction of CCR5 expression (41, 47). CXCR4-tropic HIV-1 strains grow in IL-4-treated CD4+ T cells much more rapidly and to higher titers than CCR5-tropic HIV-1 strains do (37). These findings prompted us to examine the effect of the IL-4 polymorphism on rates of SI virus acquisition in HIV-1-infected individuals. We analyzed primary virus isolates obtained from PBMC of 115 HIV-1-infected individuals (12 C homozygotes, 55 C/T heterozygotes, and 48 T homozygotes). Those PBMC were collected between 1994 and 1998. Among 115 patients, 62 had never been treated with antiretroviral drugs, and 16 were treated with mononucleoside analogue, 23 were treated with dual-nucleoside analogue combination, and 14 were received combination therapy containing protease inhibitor at the time of virus isolation. Viruses were repeatedly isolated from 15 individuals, but viruses isolated at the latest time points were used for further analysis. HIV-1 genomic regions corresponding to the gp120 V1 to V3 loops were amplified by reverse transcription-PCR, and nucleotide sequences of the amplified fragments were determined. An HIV-1 isolate with a basic amino acid residue at position 11 and/or position 25 in the V3 loop was regarded as an SI virus. In 12 HIV-1 isolates which exhibited V3 loops with elevated positive charges (>+5) despite a lack of basic amino acid residues at position 11 or 25, we examined their ability to grow in an MT2 T-cell line. A virus isolate which could grow in MT2 cells was regarded as an SI virus. Among 115 viruses isolated from 115 infected individuals, 41 appeared to be SI viruses, while the remaining 74 were classified as NSI viruses.
SI viruses were isolated from 50.0% of T homozygotes (24 of 48), 23.6% of C/T heterozygotes (13 of 55), and 33.3% of C homozygotes (4 of 12), showing a marked increase in SI virus frequency in T homozygotes (Table 3, P = 0.0091). The average CD4+ cell counts at the time of virus isolation were 236 ± 207 cells/μl for C homozygotes and C/T heterozygotes (subjects C) and 204 ± 203 cells/μl for T homozygotes (subjects T), indicating no significant difference in the average CD4+ cell counts between subjects C and T (P = 0.45). SI viruses were more frequently isolated from subjects T than subjects C either in treatment-naive patients (Table 3, P = 0.13) or in treated patients (Table 3, P = 0.029). The average CD4+ cell counts at the time of virus isolation were 262.6 ± 235.6 cells/μl for treatment-naive patients and 184.0 ± 155.5 cells/μl for treated patients, indicating that treatment-naive patients were at a less advanced stage of disease than treated patients. It is possible that the loss of statistical significance in differences between subjects T and C in treatment-naive patients was due to low frequency of advanced-stage patients, who had most likely acquired SI variants. Furthermore, SI viruses were more frequently isolated from subjects T than from subjects C even when we analyzed cases whose CD4+ cell counts were below 200 cells/μl (Table 3, P = 0.025). In those cases, a weak trend toward faster loss of CD4+ cell was again observed, but the difference was not statistically significant (8.40 ± 6.09 cells/μl/month for subjects C and 9.85 ± 7.79 cells/μl/month for subjects T, P = 0.61). Fifty-six percent of subjects T (14 of 25) and 54% of subjects C (14 of 26) in patients whose CD4+ cell counts were below 200 cells/μl received antiretroviral drugs, indicating that subjects T and subjects C with low CD4 cell counts were almost equally treated. Again, treatment did not affect the rates of SI virus acquisition in patients whose CD4+ cell counts were below 200 cells/μl, since SI viruses were more frequently isolated from subjects T than from subjects C in both treatment-naive and treated groups (data not shown). These results excluded the possibility that subjects T analyzed here were patients at a more advanced stage, who had most likely acquired SI viruses, than subjects C. The different rates of SI virus acquisition was also apparent between subjects T and C in a cohort of 56 HIV-1-infected hemophiliacs who were followed up for at least 9 years after the primary infection (Table 3, P = 0.013). All of the above results clearly demonstrated that the IL-4 −589T allele is associated with increased rates of SI virus acquisition.
TABLE 3.
IL-4 promoter genotype distribution in HIV-1-infected individuals with and without SI variants
| Category | Virus phenotype (no.) | No. (frequency, %) with IL-4 promoter genotype at position −589
|
P value | |
|---|---|---|---|---|
| C/C, C/T | T/T | |||
| Total | SI (41) | 17 (25.4) | 24 (50.0) | 0.0091 |
| NSI (74) | 50 (74.6) | 24 (50.0) | ||
| Total (115) | 67 (100) | 48 (100) | ||
| Treatment naive | SI (22) | 11 (28.2) | 11 (47.8) | 0.13 |
| NSI (40) | 28 (71.8) | 12 (52.2) | ||
| Total (62) | 39 (100) | 23 (100) | ||
| Treated | SI (19) | 6 (21.4) | 13 (52.0) | 0.029 |
| NSI (34) | 22 (78.6) | 12 (48.0) | ||
| Total (53) | 28 (100) | 25 (100) | ||
| Low CD4+ counts (<200/μl) | SI (26) | 9 (34.6) | 17 (68.0) | 0.025 |
| NSI (25) | 17 (65.4) | 8 (32.0) | ||
| Total (51) | 26 (100) | 25 (100) | ||
| Hemophiliacs | SI (23) | 7 (24.1) | 16 (59.3) | 0.013 |
| NSI (33) | 22 (75.9) | 11 (42.3) | ||
| Total (56) | 29 (100) | 27 (100) | ||
| Subtype B | SI (33) | 11 (21.1) | 22 (51.2) | 0.0039 |
| NSI (62) | 41 (78.9) | 21 (48.8) | ||
| Total (95) | 52 (100) | 43 (100) | ||
When we excluded patients infected with HIV-1 subtypes other than clade B, the difference in the rates of SI virus acquisition between subjects C and T became more prominent (Table 3, P = 0.0039). This finding suggests that HIV-1 subtypes other than clade B may evolve differently in infected individuals than subtype B. This view may be relevant to recent findings suggesting that different subtypes of HIV-1 show wide variations in their rates of phenotypic shift from NSI to SI virus (29).
In contrast, triallelic polymorphisms in the IL-10 promoter (24), another Th2 cytokine, and biallelic polymorphisms in the RANTES promoter, which affects RANTES expression (17), did not affect acquisition of the SI variant (Table 4). However, CCR5-927T (15), which is associated with a delay in HIV-1 disease progression and is almost completely linked to CCR2- 64I (24), showed an effect similar to that of IL-4 −589T on SI virus acquisition. As shown in Table 5, homozygosity and heterozygosity of this allele are also associated with increased rates of SI virus acquisition. This result is consistent with the recent finding that CCR2-64I is associated with increased prevalence of SI variants in an Amsterdam cohort of HIV-1-infected Caucasians (43). Our data showed further that individuals wild-type for CCR5-927 and wild-type or heterozygous for IL-4 −589 showed a strikingly low frequency of SI virus acquisition (Table 5).
TABLE 4.
Genotype distribution of IL-10 and RANTES promoter in HIV-1-infected individuals with and without SI variants
| Virus phenotype (no.) | No. (frequency, %) with:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IL-10 promoter genotype at position −1087
|
P value | IL-10 promoter genotype at positions −824 and −597
|
P value | RANTES promoter genotype at position −28
|
P value | ||||
| A/G | A/A | CC/CC, TA/CC | TA/TA | C/C | C/G, G/G | ||||
| SI (29) | 2 (33.3) | 27 (32.9) | 0.98 | ||||||
| NSI (59) | 4 (66.7) | 55 (67.1) | |||||||
| Total (88) | 6 (100) | 76 (100) | |||||||
| SI (31) | 18 (34.6) | 13 (31.0) | 0.71 | ||||||
| NSI (63) | 34 (65.4) | 29 (69.0) | |||||||
| Total (94) | 52 (100) | 42 (100) | |||||||
| SI (30) | 22 (29.7) | 8 (33.3) | |||||||
| NSI (68) | 52 (70.3) | 16 (66.7) | |||||||
| Total (98) | 74 (100) | 24 (100) | 0.74 | ||||||
TABLE 5.
Genotype distribution of CCR5 and IL-4 promoter in HIV-1-infected individuals with and without SI variants
| Virus phenotype (no.) | No. (frequency, %)
|
|||||
|---|---|---|---|---|---|---|
| CCR5 promoter genotype at position 927
|
P value | CCR5 and IL-4 genotype (CCR5 927)*(IL-4 −589)
|
P value | |||
| C/C | C/T, T/T | (C/C)*(C/C, C/T) | (C/T, T/T)*(C/C, C/T) (C/T, T/T)*(T/T) (C/C)*(T/T) | |||
| SI (40) | 20 (29.0) | 20 (44.4) | 0.098 | 5 (13.5) | 35 (45.5) | 0.0019 |
| NSI (74) | 49 (71.0) | 25 (55.6) | 32 (86.5) | 42 (54.5) | ||
| Total (114) | 69 (100) | 45 (100) | 37 (100) | 77 (100) | ||
Effects of SI virus acquisition on HIV-1 disease progression.
The emergence of SI variants is a sign of poor prognosis (7, 13, 30). We confirmed the association of SI virus acquisition with faster rates of disease progression, since the CD4+ cell depletion rate of 20 HIV-1-infected individuals with SI viruses (mean ± standard deviation was 11.8 ± 9.49 cells/μl/month) was significantly higher than that of 54 HIV-1-infected individuals without SI viruses (4.23 ± 4.76 cells/μl/month, P = 0.000019). Analysis of the CD4+ cell depletion rates of cases with SI variants suggested that the nine subjects T with SI variants showed a tendency to progress more rapidly (14.73 ± 10.94/μl/month) than 11 subjects C with SI variants (9.50 ± 7.85/μl/month), although this tendency was not statistically significant (P = 0.23).
IL-4 promoter polymorphism is associated with higher level of serum IgE in HIV-1-infected individuals.
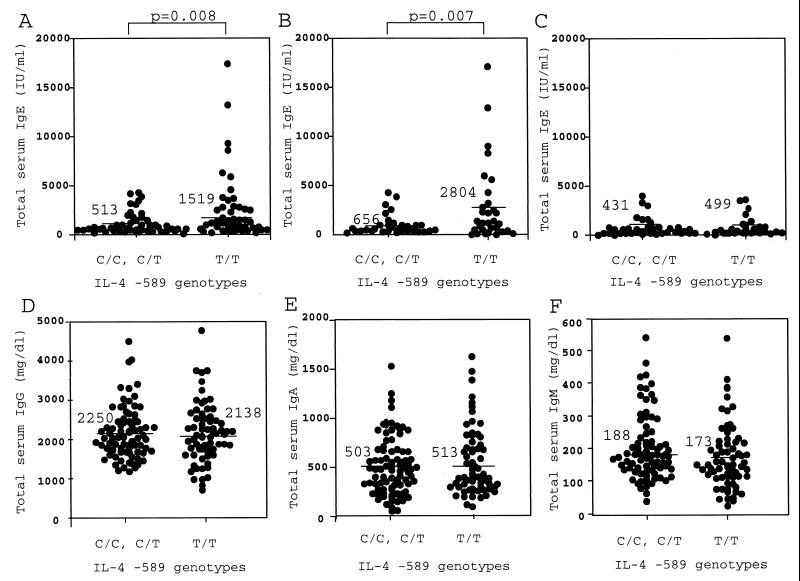
Rosenwasser et al. reported that IL-4 −589T is associated with elevated levels of total serum IgE in asthmatic families (32), while Noguchi et al. reported that there is no such association of total serum IgE with this polymorphism in Japanese asthmatic and control families (27). To evaluate the association of the IL-4 polymorphism with IgE production in HIV-1-infected individuals, total serum IgE levels were determined in 159 HIV-1-infected individuals. As shown in Fig. 3A, a significant difference was observed in serum IgE levels between subjects C (513 ± 894 IU/ml) and subjects T (1,519 ± 2,976 IU/ml, P = 0.008). This difference was specifically seen in cases whose CD4+ cell counts were below 100 cells/μl (Fig. 3B, 2,804 ± 4,045 for subjects T and 656 ± 1,017 for subjects C, P = 0.007) but not in cases whose CD4+ cell counts were above 100 cells/μl (Fig. 3C, 499 ± 864 for subjects T and 431 ± 812 for subjects C). In contrast, there was no significant difference in the level of total IgG (Fig. 3D), IgA (Fig. 3E), IgM (Fig. 3F), CD4+ cell counts (not shown), and eosinophil counts (not shown) between subjects C and T.
FIG. 3.
Serum immunoglobulin levels in infected individuals. The serum immunoglobulin level of each HIV-1-infected individual is represented by a circle; a horizontal bar represents the mean titer. Statistical significance for each difference is indicated. (A) Levels of serum IgE in 159 HIV-1-infected individuals; (B) levels of serum IgE in 65 HIV-1-infected individuals whose CD4+ counts were less than 100; (C) levels of serum IgE in 91 HIV-1-infected individuals whose CD4+ counts were more than 100; (D) levels of serum IgG levels in 159 HIV-1-infected individuals; (E) levels of serum IgA levels in 159 HIV-1-infected individuals; (F) levels of serum IgM levels in 159 HIV-1-infected individuals.
DISCUSSION
Although the loss of T lymphocytes with helper functions is a central feature of HIV-1-induced immune deficiency, other signs of immune dysfunction, such as early loss of cellular responses to recall antigens (4, 29), polyclonal B-cell activation (1), and hypergammaglobulinemia including hyper-IgE syndrome (49), are present in HIV-1 infected individuals. A switch to Th2 immune responses has been proposed to affect a viral phenotypic shift from NSI to SI (37, 41, 47) and AIDS pathogenesis (6), although conflicting results were also reported (10). In this report, we showed that a single point mutation in the IL-4 promoter, −589T, which is completely linked to the newly identified polymorphism −33T, is associated with the acquisition of SI variants and elevated levels of total IgE in HIV-1-infected individuals. The IL-4 −589T mutation was previously shown to increase promoter activity of IL-4 in luciferase reporter gene constructs (32), suggesting that this mutation increases IL-4 expression in the human body. This view is relevant to the recent finding that HIV-1-infected individuals with SI variants showed higher concentrations of serum IL-4 than those who remained negative for SI variants (40).
IL-4 down-regulates CCR5, a major coreceptor of HIV-1 NSI strains which are predominantly transmitted via sexual contact (41, 47). Therefore, we speculated that IL-4 −589T has a protective effect against HIV-1 infection. Indeed, we observed a significantly lower frequency of IL-4 −589T in both males and females who reported heterosexual contact as a risk factor. However, a lower frequency of IL-4 −589T was not observed in hemophiliacs or in individuals who reported homosexual and bisexual contact. It is possible that the protective effect of IL-4 −589T against HIV-1 transmission via CCR5 down-modulation may not be effective enough to prevent viral transmission through homosexual contact or direct injection of HIV-1 into the blood. Consistent with this notion, heterozygosity of CCR5Δ32 was reported to confer partial resistance to HIV-1 transmission through heterosexual contact but not through homosexual contact (12). An alternative explanation for lower frequency of the −589T allele in HIV-1-infected individuals would be decreased rate of survival of those with T allele. Although this cannot be ruled out definitively in our cross-sectional study, it is unlikely since we observed only a weak tendency toward rapid disease progression of T homozygotes.
Since IL-4 up-regulates expression of HIV-1 through activation of viral transcription (41), it was tempting to speculate that IL-4 −589T accelerates disease progression in HIV-1 infection. It is also possible that IL-4 −589T accelerates disease progression through suppression of cellular immunity, which plays an important role in controlling HIV-1 in infected individuals. Although the average CD4+ cell depletion rates of subjects T were slightly higher than those of subjects C (Fig. 2), the difference was not statistically significant. It is possible that the disease-accelerating effect of IL-4 −589T may be offset by the protective effect of IL-4 −589T via CCR5 down-modulation. Further analysis in well-organized cohorts is required since the data obtained from our cross-sectional analysis are not adequate to discriminate subtle differences statistically.
SI variants observed in approximately half of all HIV-1-infected individuals signify poor prognosis and often correlate with faster CD4+ cell depletion and rapid disease progression (7, 13, 30). The average CD4+ cell depletion rate of subjects T with SI variants was higher than that of subjects C with SI variants, although the difference was not statistically significant possibly due to the limited numbers of subjects available. It is possible that the protective effect of IL-4 −589T against HIV-1 disease progression via CCR5 down-modulation is dominant in the absence of SI variants, but once an SI variant emerges, IL-4 −589T no longer exerts a protective effect and rather accelerates HIV-1 disease progression. This complex effect of IL-4 −589T on HIV-1 disease progression may explain our failure to detect a significant impact of this polymorphism on disease progression (Fig. 2), despite its clear effect on the rate of SI variant acquisition. At present, it is difficult to determine whether this polymorphism accelerates disease progression after emergence of SI virus. Further analysis of a well-defined cohort in relation to the presence or absence of SI variants is also required to clarify this point.
In contrast to our results, Noguchi et al. (27) and Walley and Cookson (45) reported that there is no statistically significant association between elevated levels of total serum IgE and IL-4 −589T. As described above, the difference in total serum IgE levels between subjects C and T was seen only in cases where CD4+ cell counts dropped below 100/μl. Therefore, it is possible that elevated serum levels of IgE observed in the present study may be due to the combined effect of IL-4 −589T and the dominance of Th2 type CD4+ T lymphocytes caused by prolonged destruction of CCR5-expressing Th1 type CD4+ T lymphocytes by HIV-1 NSI virus.
In conclusion, we have demonstrated that a polymorphism in the IL-4 promoter is associated with an increased rate of HIV-1 SI virus acquisition and elevated levels of serum IgE. The signal of IL-4 is conferred to effector cells through binding to the α chain of the IL-4 receptor (IL-4Rα). Recently, the polymorphisms in the IL-4Rα gene were reported to influence the signal transduction pathway of IL-4 (11, 16, 22, 23). Thus, it is important to investigate the effects of those polymorphisms in the IL-4Rα gene on the rate of SI virus acquisition and disease progression in HIV-1 infection.
ACKNOWLEDGMENTS
We thank David Chao for critical discussions.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture, the Ministry of Health and Welfare, the Science and Technology Agency of Japanese Government, and the Organization for Pharmaceutical Safety and Research (OPSR).
REFERENCES
- 1.Amadori A, Chieco-Bianchi L. B-cell activation and HIV-1 infection: deeds and misdeeds. Immunol Today. 1990;11:374–379. doi: 10.1016/0167-5699(90)90144-x. [DOI] [PubMed] [Google Scholar]
- 2.Arai N, Nomura D, Villaret D, Malefijt D, Seiki M, Yoshida M, Minoshima S, Fukuyama R, Maekawa M, Kudoh J, Shimizu N, Yokota K, Abe E, Yokota T, Takebe Y, Arai K. Complete nucleotide sequence of the chromosomal gene for human IL-4 and its expression. J Immunol. 1989;142:274–282. [PubMed] [Google Scholar]
- 3.Asjo B, Albert J, Karlsson A, Morfeldt-Manson L, Biberfeld G, Lidman K, Fenyo E M. Replicative properties of human immunodeficiency viruses from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 4.Blazquez M V, Madueno J A, Gonzalez R, Jurado R, Bachochin W W, Pena J, Munoz E. Selective decrease of CD26 expression in T cells from HIV-1 infected individuals. J Immunol. 1992;149:3073–3077. [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 6.Clerici M, Shearer G M. A TH1→TH2 switch is a critical step in the etiology of HIV infection. Immunol Today. 1993;14:107–111. doi: 10.1016/0167-5699(93)90208-3. [DOI] [PubMed] [Google Scholar]
- 7.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.Del Prete G, Maggi E, Parronchi P, Chetrien I, Tiri A, Macchia D, Ricci M, Banchereau J, de Vries J, Romagnani S. IL-4 is essential factor for the IgE synthesis induced in vitro by human T cell clones and their supernatants. J Immunol. 1988;140:4193–4198. [PubMed] [Google Scholar]
- 10.Graziosi C, Pantaleo G, Gantt K R, Fortin J-P, Demarest J F, Cohen O J, Sekaly R P, Fauci A S. Lack of evidence for the dichotomy of TH1 and TH2 predominance In HIV-1-infected individuals. Science. 1994;265:248–252. doi: 10.1126/science.8023143. [DOI] [PubMed] [Google Scholar]
- 11.Hershey G K K, Friedrich M F, Esswein L A, Thomas M L, Chatila T A. The association of atopy with a gain-of-function mutation in the α subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–1725. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman T L, MacGregor R R, Burger H, Mick R, Doms R W, Collman R G. CCR5 genotypes in sexually active couples discordant for human immunodeficiency virus type 1 infection status. J Infect Dis. 1997;176:1093–1096. doi: 10.1086/516519. [DOI] [PubMed] [Google Scholar]
- 13.Ida S, Gatanaga H, Shioda T, Nagai Y, Kobayashi N, Shimada K, Kimura S, Iwamoto A, Oka S. HIV type 1 V3 variation dynamics in vivo: long-term persistence of non-syncytium-inducing genotypes and transient presence of syncytium-inducing genotypes during the course of progressive AIDS. AIDS Res Hum Retroviruses. 1997;13:1597–1609. doi: 10.1089/aid.1997.13.1597. [DOI] [PubMed] [Google Scholar]
- 14.Keegan A D, Nelms K, Wang L-M, Pierce J H, Paul W E. Interleukin 4 receptor: signaling mechanisms. Immunol Today. 1994;15:423–432. doi: 10.1016/0167-5699(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 15.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 16.Kruse S, Japha T, Tedner M, Sparholt S H, Forster J, Kuehr J, Deichmann K A. The polymorphisms S503P and Q576R in the interleukin-4 receptor α gene are associated with atopy and influence the signal transduction. Immunology. 1999;96:365–371. doi: 10.1046/j.1365-2567.1999.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Chao D, Nakayama E E, Taguchi H, Goto M, Xin X, Takamatsu J, Saito H, Ishikawa Y, Akaza T, Juji T, Takebe Y, Ohishi T, Fukutake K, Maruyama Y, Yashiki S, Sonoda S, Nakamura T, Nagai Y, Iwamoto A, Shioda T. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci USA. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 19.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O'Brien T R, Hilgartner M W, Vlahov D, O'Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 20.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M the Multicenter AIDS Cohort Study (MACS) CCR5 promoter polymorphism and HIV-1 disease progression. Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 21.Michael N L, Louie L G, Rohrbaugh A L, Schultz K A, Dayhoff D E, Wang C E, Sheppard H W. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat Med. 1997;3:1160–1162. doi: 10.1038/nm1097-1160. [DOI] [PubMed] [Google Scholar]
- 22.Mitsuyasu H, Izuhara K, Mao X-Q, Gao P-S, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin J M. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998;19:119–120. doi: 10.1038/472. [DOI] [PubMed] [Google Scholar]
- 23.Mitsuyasu H, Yanagihara Y, Mao X-Q, Gao P-S, Arinobu Y, Ihara K, Takabayashi A, Hara T, Enomoto T, Sasaki S, Kawai M, Hamasaki N, Shirakawa T, Hopkin J M, Izuhara K. Cutting edge: dominant effect of Ile50Val variant of the human IL-4 receptor alpha-chain in IgE synthesis. J Immunol. 1999;162:1227–1231. [PubMed] [Google Scholar]
- 24.Mok C C, Lanchbury J S, Chan D W, Lau C S. Interleukin-10 promoter polymorphisms in southern Chinese patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41:1090–1095. doi: 10.1002/1529-0131(199806)41:6<1090::AID-ART16>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O'Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 26.Noesel C J M, Gruters R A, Terpstra F G, Schellekens P T A, Lier R A W, Miedema F. Functional and phenotypic evidence for a selective loss of memory T cells in asymptomatic human immunodeficiency virus-infected men. J Clin Investig. 1990;86:293–299. doi: 10.1172/JCI114698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T, Yanagi H, Matsui A, Hamaguchi H. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy. 1998;28:449–453. doi: 10.1046/j.1365-2222.1998.00256.x. [DOI] [PubMed] [Google Scholar]
- 28.Paul W E. Interleukin-4: a prototypic immunoregulatory cytokine. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 29.Peeters M, Vincent R, Perret J-L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 30.Richman D D, Bozette S A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 31.Romagnani S. Biology of human TH1 and TH2 cells. J Clin Immunol. 1995;15:121–129. doi: 10.1007/BF01543103. [DOI] [PubMed] [Google Scholar]
- 32.Rosenwasser L J, Klemm D J, Dresback J K, Inamura H, Mascali J J, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25(Suppl. 2):74–78. doi: 10.1111/j.1365-2222.1995.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 33.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-F, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Geoges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 34.Schuitemaker H, Koot M, Koostra N A, Dercksen M W, DeGoede T E Y, Van Steenwijk R P, Lange J M A, Schattenkerk J K M E, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan M K, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, Buchbinden S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O'Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka Y, Koyanagi Y, Tanaka R, Kumazawa Y, Nishimura T, Yamamoto N. Productive and lytic infection of human CD4+ type 1 helper T cells with macrophage-tropic human immunodeficiency virus type 1. J Virol. 1997;71:465–470. doi: 10.1128/jvi.71.1.465-470.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tersmette M, Gruters R A, DeWolf F, DeGoede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tersmette M, Lange J M A, DeGoede R E Y, DeWolf F, Schattenkerk J K M E, Schellekens P T A, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 40.Torres Y, Medrano F J, Rey C, Calderon E J, Sanchez-Quijano A, Lissen E, Leal M. Evidence for a role of T-helper type 2 cytokines in the acquisition of human immunodeficiency virus syncytium-inducing phenotype. Eur J Clin Investig. 1998;28:930–936. doi: 10.1046/j.1365-2362.1998.00368.x. [DOI] [PubMed] [Google Scholar]
- 41.Valentin A, Lu W, Rosati M, Schneider R, Albert J, Karlsson A, Pavlakis G N. Dual effect of interleukin-4 on HIV-1 expression: implications for phenotypic switch and disease progression. Proc Natl Acad Sci USA. 1998;95:8886–8891. doi: 10.1073/pnas.95.15.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rij R P, Broersen S, Goudsmit J, Coutinho R A, Schuitemaker H. The role of a stromal cell-derived factor-1 chemokine gene variant in the clinical course of HIV-1 infection. AIDS. 1998;12:F85–F90. [PubMed] [Google Scholar]
- 43.van Rij R P, de R. Husman A-M, Brouwer M, Goudsmit J, Coutinho R A, Schuitemaker H. Role of CCR2 genotype in the clinical course of syncytium-inducing (SI) or non-SI human immunodeficiency virus type 1 infection and in the time to conversion to SI virus variants. J Infect Dis. 1998;178:1806–1811. doi: 10.1086/314522. [DOI] [PubMed] [Google Scholar]
- 44.Vitetta E S, Ohara J, Myers C, Layton J, Kramer P H, Paul W E. Serological, biochemical, and functional identity of B cell-stimulatory factor 1 and B cell differentiation factor for IgG1. J Exp Med. 1985;162:1726–1732. doi: 10.1084/jem.162.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walley A J, Cookson W O. Investigation of an interleukin-4 promoter polymorphism for associations with asthma and atopy. J Med Genet. 1996;33:689–692. doi: 10.1136/jmg.33.8.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Harada A, Matsushita S, Matsumi S, Zhang Y, Shioda T, Nagai Y, Matsushima K. IL-4 and a glucocorticoid up-regulate CXCR4 expression on human CD4+ T lymphocytes and enhance HIV-1 replication. J Leukoc Biol. 1998;64:642–649. doi: 10.1002/jlb.64.5.642. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Roderiquez G, Oravecz T, Norcross M A. Cytokine regulation of human immunodeficiency virus type 1 entry and replication in human monocytes/macrophages through modulation of CCR5 expression. J Virol. 1998;72:7642–7647. doi: 10.1128/jvi.72.9.7642-7647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O'Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O'Brien S J ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 49.Wright D N, Nelson R P, Jr, Ledford D K, Fernandez-Caldas E, Trudeau W L, Lockey R F. Serum IgE and human immunodeficiency virus (HIV) infection. J Allergy Clin Immunol. 1990;85:445–452. doi: 10.1016/0091-6749(90)90154-v. [DOI] [PubMed] [Google Scholar]
- 50.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]