Abstract
Virus-encoded mRNA capping enzymes are attractive targets for antiviral therapy, but functional studies have been limited by the lack of genetically tractable in vivo systems that focus exclusively on the RNA-processing activities of the viral proteins. Here we have developed such a system by engineering a viral capping enzyme—vaccinia virus D1(1-545)p, an RNA triphosphatase and RNA guanylyltransferase—to function in the budding yeast Saccharomyces cerevisiae in lieu of the endogenous fungal triphosphatase (Cet1p) and guanylyltransferase (Ceg1p). This was accomplished by fusion of D1(1-545)p to the C-terminal guanylyltransferase domain of mammalian capping enzyme, Mce1(211-597)p, which serves as a vehicle to target the viral capping enzyme to the RNA polymerase II elongation complex. An inactivating mutation (K294A) of the mammalian guanylyltransferase active site in the fusion protein had no impact on genetic complementation of cet1Δceg1Δ cells, thus proving that (i) the viral guanylyltransferase was active in vivo and (ii) the mammalian domain can serve purely as a chaperone to direct other proteins to the transcription complex. Alanine scanning had identified five amino acids of vaccinia virus capping enzyme—Glu37, Glu39, Arg77, Glu192, and Glu194—that are essential for γ phosphate cleavage in vitro. Here we show that the introduction of mutation E37A, R77A, or E192A into the fusion protein abrogates RNA triphosphatase function in vivo. The essential residues are located within three motifs that define a family of viral and fungal metal-dependent phosphohydrolases with a distinctive capacity to hydrolyze nucleoside triphosphates to nucleoside diphosphates in the presence of manganese or cobalt. The acidic residues Glu37, Glu39, and Glu192 likely comprise the metal-binding site of vaccinia virus triphosphatase, insofar as their replacement by glutamine abolishes the RNA triphosphatase and ATPase activities.
Eukaryotic viruses have evolved diverse strategies to acquire a 5′ cap structure for their mRNAs (4). RNA viruses that encode RNA-dependent RNA polymerases to synthesize their mRNAs either steal the caps from cellular transcripts, encode their own enzymes that cap and methylate the plus-strand transcripts, or circumvent the capping problem by including cis-acting elements in the plus strand that promote cap-independent translation. The mRNAs of most DNA viruses are synthesized by cellular RNA polymerase II (pol II) and are therefore capped by the cellular capping and methylating enzymes. However, vaccinia virus and other poxviruses, which replicate entirely in the cytoplasm, encode and encapsidate their own DNA-dependent RNA polymerase and mRNA capping apparatus (12, 16). African swine fever virus, which has a cytoplasmic replication phase, also encodes and encapsidates its own RNA polymerase and capping enzyme (39). Baculoviruses, which replicate in the nucleus, use pol II to transcribe early genes and then switch at late times to a virus-encoded transcription system that includes RNA polymerase and capping activities (14, 15, 26). Chlorella virus PBCV-1 encodes a capping enzyme (25) but appears not to encode its own RNA polymerase.
Cap formation by the enzymes of DNA viruses, double-stranded RNA viruses, and eukaryotic cells occurs via three sequential reactions: (i) the 5′ triphosphate end of the nascent pre-mRNA is hydrolyzed to a diphosphate by RNA 5′ triphosphatase, (ii) the diphosphate end is capped with GMP by GTP:RNA guanylyltransferase, and (iii) the GpppN cap is methylated by S-adenosylmethonine:RNA (guanine-N7) methyltransferase (48). This “conventional” pathway of cap synthesis was defined using soluble enzymes purified from vaccinia virus particles (12, 54). The mechanisms and structures of cellular and DNA virus capping enzymes have since been delineated through mutagenesis and crystallography (10, 11, 17, 21, 28, 30, 33, 37, 42, 56, 57, 60–62). Several single-stranded RNA viruses have evolved alternative cap synthetic pathways, which entail unconventional chemistry and are less well understood with respect to enzyme structure and mechanisms (1–3, 47).
The genetic and physical organizations of the known virus-encoded mRNA capping enzymes are significantly different from those of metazoan host cells (48). Hence, the viral cap-forming enzymes are potential targets for antiviral drugs that would interfere with capping of pathogen mRNAs but spare the host capping enzymes. For any given virus that provides its own caps, there are a number of questions that need to be addressed before capping can be validated as a target, such as (i) whether the viral gene encoding the capping enzyme is essential for virus replication and (ii) whether the capping activity of the viral gene product is essential for virus replication. These questions have not been answered fully, even where the biochemistry of viral cap formation is well understood. For example, vaccinia virus capping enzyme is a multifunctional protein with RNA triphosphatase, RNA guanylyltransferase, and RNA (guanine-7-) methyltransferase activities (45, 54). The enzyme is a heterodimer of 95- and 33-kDa subunits encoded by the vaccinia virus D1 and D12 genes, respectively. The vaccinia virus D1 and D12 genes are essential for virus replication, insofar as mutations that elicit temperature-sensitive virus growth phenotypes have been mapped to the two capping enzyme subunits (6, 18). However, the genetic landscape is complicated, because vaccinia virus capping enzyme plays a larger role in viral gene expression; it serves as a transcription termination factor during the synthesis of viral early mRNAs (29, 50) and as an initiation factor during the transcription of intermediate genes (55). Amazingly, the D1 and D12 temperature-sensitive mutant viruses display no gross defect in viral gene expression at the restrictive temperature but are instead defective in resolving concatemeric DNA replication intermediates into the hairpin telomeres of the mature viral genome (6, 18). This mysterious phenotype has no obvious connection to the known mRNA-processing or transcription functions of the D1 and D12 proteins. Similar problems arise in interpreting the essentiality of the baculovirus LEF-4 capping enzyme, which is an intrinsic subunit of baculovirus RNA polymerase, because it is not clear if the conditional phenotype of a lef-4 mutant virus is a consequence of failure to transcribe or failure to cap viral mRNAs (15, 26).
Genetic, and ultimately pharmacologic, analysis of viral capping enzymes would be facilitated by the development of in vivo assays in which the functional readout is clearly and exclusively dependent on the capacity of the viral gene product to catalyze cap synthesis. Here we explore the possibility of using the budding yeast Saccharomyces cerevisiae as a genetic model for the study of viral capping enzymes. S. cerevisiae encodes a three-component capping system consisting of separate triphosphatase (Cet1p), guanylyltransferase (Ceg1p), and methyltransferase (Abd1p) gene products (22, 31, 43, 53). All three genes are essential for yeast cell growth. Mutational analyses of Cet1p, Ceg1p, and Abd1p have resulted in the delineation of minimal catalytic domains for each protein and the identification of catalytically important amino acid side chains that comprise the triphosphatase, guanylyltransferase, and methyltransferase active sites (21, 27, 30, 37, 56, 57). By correlating mutational effects on catalysis in vitro with effects on function in vivo, we have shown that the triphosphatase, guanylyltransferase, and methyltransferase activities are essential for yeast cell growth. The feasibility of using yeast growth as a readout of the function of capping enzymes from heterologous sources is underscored by the demonstration that the entire three-component yeast capping apparatus can be replaced in vivo by the two-component mammalian capping system, consisting of a bifunctional triphosphatase-guanylyltransferase protein (Mce1p) and a separate cap methyltransferase (Hcm1p) (41). This result is remarkable because the structure and catalytic mechanism of the mammalian RNA triphosphatase are completely different from those of the yeast RNA triphosphatase (48).
The salient question here is whether a viral capping enzyme can function in place of one or more of the yeast capping enzymes. To address this issue, we tested the ability of the N-terminal triphosphatase-guanylyltransferase domain of the vaccinia virus D1 polypeptide [vD1(1-545)p] to complement the growth of yeast cet1Δ or ceg1Δ cells. We report that the vaccinia virus enzyme is active in vivo, provided that it is targeted to the pol II transcription complex by fusion to a cellular protein that binds to pol II. Mutations of vD1(1-545)p that abrogate triphosphatase activity in vitro abolish complementation of cet1Δ. Additional mutational analysis of vD1(1-545)p defines the triphosphatase active site and suggests a mechanism of metal-dependent catalysis common to DNA viral and fungal capping enzymes. Isogenic strains bearing vaccinia virus and human capping enzymes can be used to screen for cytotoxic compounds that specifically inhibit the poxvirus capping apparatus.
MATERIALS AND METHODS
Yeast expression plasmids for vaccinia virus capping enzyme.
The vD1(1-545) gene encoding the N-terminal triphosphatase-guanylyltransferase domain of vaccinia virus capping enzyme (35, 61) was excised from pET-D1(1-545) with NdeI and HindIII and inserted between the NdeI and HindIII sites of the customized yeast expression vector pYX1 (CEN TRP1), a derivative of pYX132 (Novagen) that contains six tandem histidine codons and a unique NdeI site between the NcoI and BamHI sites. The resulting yeast plasmid, pYX1-vD1(1-545), encodes vaccinia virus D1(1-545)p fused in-frame with an amino-terminal 12-amino-acid leader peptide (MGSHHHHHHSGH). An AatII-NheI fragment containing the vD1(1-545) gene was excised from pYX1-vD1(1-545) and inserted into the yeast multicopy expression plasmid pYX232 (2μm TRP1) to generate p232-vD1(1-545). Expression of the vD1(1-545) gene is under the control of the yeast TPI1 promoter.
Plasmids encoding a chimeric vaccinia virus-mammalian capping enzyme with an N-terminal vD1(1-545)p segment fused to the C-terminal guanylyltransferase domain of the mouse capping enzyme Mce1(211-597)p were constructed as follows. The vD1(1-545) gene was PCR amplified from a pET-D1(1-545) template using an antisense primer that changed the vD1(1-545) stop codon to a His codon and introduced an NdeI restriction site at a position corresponding to the C-terminus. The PCR product was digested with NdeI and then inserted into the NdeI site of pYX1-MCE1(211-597) (CEN TRP1) to yield the fusion gene plasmid pYX1-vD1(1-545)-MCE1(211-597). An AatII-NheI fragment containing the fusion gene was excised from pYX1-vD1(1-545)-MCE1(211-597) and inserted into pYX232 (2μm TRP1) to generate p232-vD1(1-545)-MCE1(211-597). Expression of the vD1(1-545)-MCE1(211-597) gene is under the control of the yeast TPI1 promoter.
CEN TRP1 and 2μm TRP1 plasmids encoding mutated versions of vD1(1-545)-Mce1(211-597)p with alanine substitutions at residue Glu37, Arg77, or Glu192 were constructed as described above, except that PCR amplification of the vaccinia virus component was performed using pET-D1(1-545)-E37A, pET-D1(1-545)-R77A, or pET-D1(1-545)-E192A as a template (60).
Plasmids encoding a mutated version of vD1(1-545)-MCE1(211-597)p in which the active-site lysine nucleophile of the mouse guanylyltransferase domain (Lys294) was replaced by alanine were constructed as follows. The K294A mutation was introduced into the MCE1(211-597) gene by PCR amplification with mutagenic primers using the two-stage overlap extension method. pYX1-MCE1(211-597) served as the template for the first-round PCR. The product of the second-stage PCR was digested with StuI and NsiI and then inserted into StuI- and NsiI-digested pYX1-vD1(1-545)-MCE1(211-597) in lieu of the wild-type gene fragment. An AatII-NheI fragment containing the fusion gene was excised from pYX1-vD1(1-545)-MCE1(211-597)-K294A and inserted into pYX232 (2μm TRP1) to generate p232-vD1(1-545)-MCE1(211-597)-K294A.
The presence of the desired mutations in the yeast plasmids was confirmed in every case by DNA sequencing; the inserted fragments were sequenced completely to exclude the acquisition of unwanted mutations during the amplification and cloning procedure.
Yeast strains.
Strain YBS20 (MATa trp1 his3 ura3 leu2 ade2 can1 cet1::LEU2 p360-CET1) is deleted at the chromosomal CET1 locus encoding yeast RNA triphosphatase. Growth of YBS20 depends on maintenance of plasmid p360-CET1 (CEN URA3 CET1). Strain YBS2 (MATa ura3 trp1 lys2 leu2 ceg1::hisG pGYCE-360) is deleted at the chromosomal CEG1 locus encoding yeast RNA guanylyltransferase. Growth of YBS2 depends on maintenance of plasmid pGYCE-360 (CEN URA3 CEG1). Strain YBS50 (MATa leu2 ade2 trp1 his3 ura3 can1 ceg1::hisG cet1::LEU2 p360-CET1/CEG1) is deleted at the chromosomal CET1 and CEG1 loci (41). Growth of YBS50 is contingent on the maintenance of plasmid p360-CET1/CEG1 (CEN URA3 CET1 CEG1).
T7-based plasmids for expression of vD1(1-545)p in bacteria.
vD1(1-545) mutations E37D, E37Q, E39D, E39Q, E192D, E192Q, E194D, and E194Q were programmed by synthetic oligonucleotides, using the two-stage PCR-based overlap extension strategy. NdeI-BglII restriction fragments of the PCR-amplified mutated vD1(1-545) DNAs were inserted into the T7-based expression plasmid pET16b that had been digested with NdeI and BamHI. The resulting plasmids contained the mutated vD1(1-545) coding sequence fused in frame with a 63-bp 5′ leader sequence that encodes 10 consecutive histidine residues. The presence of the desired mutations was confirmed in each case by sequencing the entire 1.7-kbp insert; the occurrence of PCR-generated mutations outside the targeted region was thereby excluded.
Expression and purification of recombinant vD1(1-545)p.
The recombinant His-tagged vaccinia virus proteins were purified by nickel-agarose chromatography of soluble extracts of 1-liter cultures of Escherichia coli BL21(DE3) bearing wild-type or mutated pET-His-D1(1-545) plasmids as described previously (61). The 200 mM imidazole eluate fractions of each preparation were dialyzed and purified further by phosphocellulose chromatography (61). The polypeptide compositions of the column fractions were monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Protein concentrations were determined using the Bio-Rad dye reagent with bovine serum albumin as a standard.
RESULTS
Engineering vaccinia virus capping enzyme to function in yeast.
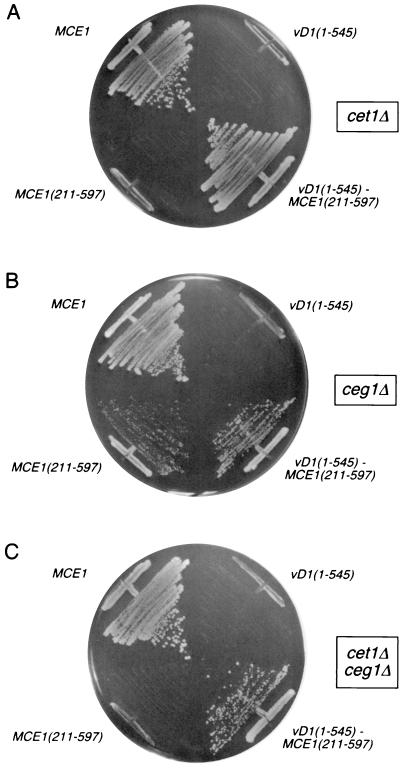
vD1(1-545)p is an autonomous trypsin-resistant domain with RNA triphosphatase and RNA guanylyltransferase activities equivalent to those of the native vaccinia virus capping enzyme (35, 36, 44, 61). The triphosphatase and guanylyltransferase active sites are distinct, but the structural elements that comprise the active sites are partially interdigitated within the primary structure, such that vD1(1-545)p cannot be separated into catalytically active subdomains. To express vaccinia virus triphosphatase-guanylyltransferase in yeast, we cloned the vD1(1-545) gene into a yeast 2μm plasmid and placed it under the transcriptional control of the strong constitutive TPI1 promoter. The capacity of vD1(1-545)p to replace the yeast RNA triphosphatase Cet1p was tested by plasmid shuffle in yeast cet1Δ cells that contain CET1 on a URA3 plasmid. The cet1Δ strain is unable to form colonies on medium containing 5-fluoro-orotic acid (5-FOA), a drug that selects against the URA3 plasmid, unless it is transformed with a second plasmid bearing CET1 or a functional homologue from another source. For example, transformation with a TRP1 plasmid bearing the MCE1 gene, which encodes the 597-amino-acid mammalian triphosphatase-guanylyltransferase, allowed growth of cet1Δ cells on 5-FOA, whereas a plasmid expressing Mce1(211-597)p, the C-terminal guanylyltransferase domain of mammalian capping enzyme, did not (Fig. 1A). The key finding was that expression of vD1(1-545)p did not complement the cet1Δ mutation (Fig. 1A).
FIG. 1.
Complementation of yeast cet1Δ and ceg1Δ mutations by expression of a vD1(1-545)-Mce1(211-597)p chimeric capping enzyme. Yeast strains YBS20 (cet1Δ), YBS2 (ceg1Δ), and YBS50 (cet1Δ ceg1Δ) were transformed with 2μm TRP1 plasmids containing either vD1(1-545) or vD1(1-545)-MCE1(211-597). Control transformations were performed with CEN TRP1 plasmids containing MCE1 or MCE1(211-597). Single Trp1+ transformants were patched to agar plates lacking tryptophan and then streaked on agar medium containing 5-FOA (0.75 mg/ml). The plates were photographed after incubation for 4 days at 30°C.
Similar tests of the ability of vD1(1-545)p to substitute for yeast guanylyltransferase Ceg1p were performed by plasmid shuffle into a ceg1Δ strain bearing a URA3 CEG1 plasmid (Fig. 1B). Cell expressing the full-length mammalian Mce1p grew on 5-FOA. So did cells expressing the C-terminal guanylyltransferase domain Mce1(211-5497)p, but as noted previously (24), MCE1(211-597) cells formed smaller colonies than MCE1 cells (Fig. 1B). We found that expression of vD1(1-545)p did not complement the ceg1Δ mutation (Fig. 1B). Thus, the vaccinia virus capping enzyme, by itself, was unable to function in yeast, either as a triphosphatase or as a guanylyltransferase.
A plausible explanation for why vD1(1-545)p could not replace Cet1p or Ceg1p is that the vaccinia virus capping enzyme failed to localize to the intranuclear sites of pre-mRNA synthesis. We and others have shown that specific capping of nascent pre-mRNAs is achieved via the binding of the guanylyltransferase component of the cellular capping apparatus to the phosphorylated carboxyl-terminal domain (CTD) of elongating pol II (8, 9, 23, 24, 34, 46, 62). The CTD, consisting of tandem repeats of a heptapeptide of the consensus sequence YSPTSPS, is extensively phosphorylated in the context of the transcription elongation complex. The guanylyltransferase domain Mce1(211-597)p of mammalian capping enzyme binds specifically to the phosphorylated CTD but not to unmodified CTD. The triphosphatase domain of mammalian capping enzyme Mce1(1-210)p does not bind the CTD but is normally brought along via its linkage in cis to the guanylyltransferase. In yeast, the guanylyltransferase Ceg1p binds to CTD-PO4, whereas the triphosphatase Cet1p does not. Formation of a Cet1p-Ceg1p complex in trans allows the yeast guanylyltransferase to chaperone the triphosphatase to the transcription complex (20, 27). The reason why yeast MCE1(211-597) cells grow slowly (Fig. 1B) is that mouse guanylyltransferase has low affinity for the yeast triphosphatase (20).
Vaccinia virus capping enzyme forms a binary complex in solution with vaccinia virus RNA polymerase (16). This interaction facilitates the capping of nascent mRNA chains as soon as their 5′ ends are extruded from the RNA binding pocket on the elongating polymerase (16). The poor efficiency of capping of RNAs transcribed by T7 RNA polymerase in vaccinia virus-infected cells is likely a manifestation of the vaccinia virus capping enzyme-vaccinia polymerase interaction (13). Although several of the subunits of vaccinia virus RNA polymerase are structurally homologous to those of pol II, the viral enzyme has no equivalent of the CTD. In fact, the vaccinia virus capping enzyme is unable to bind to CTD-PO4 in vitro under conditions permissive for CTD-PO4 binding by cellular capping enzymes (34).
In light of these observations, we envisioned a scenario whereby vaccinia virus capping enzyme might be made to function in yeast if it could be correctly targeted to the pol II elongation complex. To accomplish this targeting, we engineered a chimeric capping enzyme gene, vD1(1-545)-MCE1(211-597), encoding a product in which the vaccinia virus triphosphatase-guanylyltransferase is fused to the C-terminal guanylyltransferase domain of the mammalian capping enzyme. We showed recently that mammalian guanylyltransferase can act as chaperone in cis for a catalytic domain of yeast triphosphatase Cet1p that lacks the ability to bind to Ceg1p (27). We also found that mammalian guanylyltransferase can target Cth1p, an S. cerevisiae RNA triphosphatase not normally involved in capping, and thereby convert it into a cap-forming enzyme in vivo (37). These results suggested that the mammalian guanylyltransferase can be used as a vehicle to deliver heterologous proteins to the pol II transcription elongation complex in vivo.
The instructive finding was that either cet1Δ or ceg1Δ cells transformed with the vD1(1-545)-MCE1(211-597) fusion gene on a 2μm TRP1 plasmid readily gave rise to 5-FOA-resistant colonies (Fig. 1A and B). Indeed, vD1(1-545)-MCE1(211-597) complemented the growth of a yeast cet1Δ ceg1Δ strain in which the yeast triphosphatase and guanylyltransferase were both deleted (Fig. 1C). vD1(1-545)-MCE1(211-597) cells grew well on rich medium at either 30 or 37°C (the latter being the natural growth temperature for vaccinia virus in the mammalian host). The colony size of vD1(1-545)-MCE1(211-597) cells on rich medium was similar to that of MCE1 cells (not shown). We conclude that vaccinia virus capping enzyme is functional in yeast when it is targeted appropriately to the yeast transcription complex.
Vaccinia virus guanylyltransferase is active in yeast.
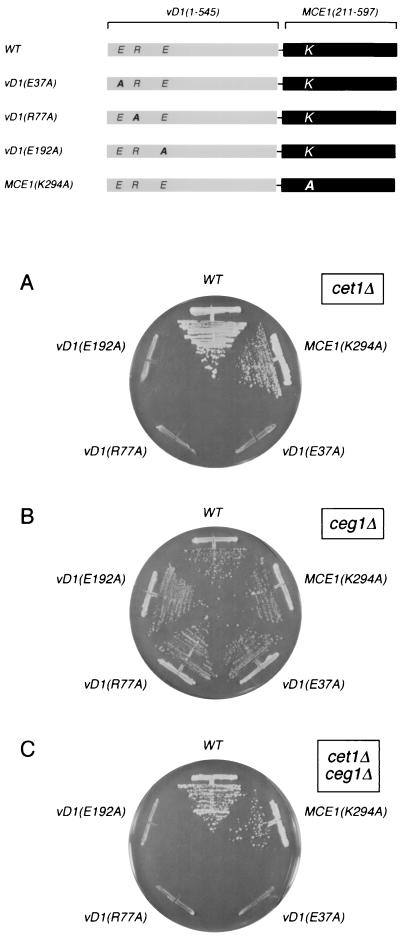
vD1(1-549)-MCE1(211-597) cells contain a single source of RNA triphosphatase activity derived from the vaccinia virus component of the fusion protein, but they contain two potential guanylyltransferase activities: one from the N-terminal vaccinia virus domain and one from the C-terminal mammalian domain. In order to determine whether the vaccinia virus guanylyltransferase could sustain cell growth when it is the only guanylyltransferase present, we introduced an inactivating missense mutation into the mouse guanylyltransferase domain. RNA guanylyltransferases are structurally and mechanistically conserved among fungi, mammals, and DNA viruses. The signature features of the guanylyltransferases are a two-step ping-pong reaction mechanism of nucleotidyl transfer through a covalent enzyme-(lysyl-N)-GMP intermediate (49). In the first step, a lysine nucleophile on the enzyme attacks the α phosphorus of GTP to form the covalent intermediate and expel pyrophosphate. In the second step, a nonbridging β phosphate oxygen of diphosphate-terminated RNA attacks the covalently bound guanylate to form the GpppN cap. A set of six conserved peptide motifs comprises the active site (17, 48, 56). The lysine nucleophile that becomes covalently attached to GMP is located within motif I (KxDG). The active-site lysine of Mce1p is Lys294. Substitution of alanine for Lys294 abrogates the guanylyltransferase activity and precludes complementation of ceg1Δ cells by mammalian capping enzyme (24, 59, 62).
A mutant chimeric capping enzyme gene, vD1(1-545)-MCE1(211-597)-K294A, was cloned into the 2μm TRP1 vector and tested for activity in yeast by plasmid shuffle. Inactivation of the mammalian guanylyltransferase had no effect on the ability of the fusion protein to support the growth of cet1Δ cells (Fig. 2A), ceg1Δ cells (Fig. 2B), or doubly deleted cet1Δ ceg1Δ cells (Fig. 2C). We conclude that (i) vaccinia virus capping enzyme is functional in yeast as the sole source of RNA triphosphatase and guanylyltransferase activities and (ii) the mutant mammalian domain can serve as a vehicle for targeting of the viral capping enzymes without playing a catalytic role in cap synthesis.
FIG. 2.
Mutational analysis of the vD1(1-545)-MCE1(211-597) fusion defines residues in vaccinia virus capping enzyme required for cap formation in vivo in yeast. The polypeptides encoded by the “wild-type” (WT) fusion gene vD1(1-545)-MCE1(211-597) and mutant alleles vD1(E37A), vD1(R77K), vD1(E192A), and MCE1(K294A) are depicted as horizontal bars with the N termini at the left and the C termini at the right. The positions of residues mutated in the vaccinia virus RNA triphosphatase subdomain (E37, R77, and E192) and the mouse guanylyltransferase domain (K294) are indicated. Complementation of the yeast cet1Δ, ceg1Δ, and cet1Δ ceg1Δ strains by the fusion genes on 2μm TRP1 plasmids was tested by plasmid shuffle as described for Fig. 1. Trp1+ isolates were streaked on agar medium containing 5-FOA. The plates were photographed after incubation for 4 days at 30°C.
In vivo effects of mutations in the putative active site of vaccinia virus RNA triphosphatase.
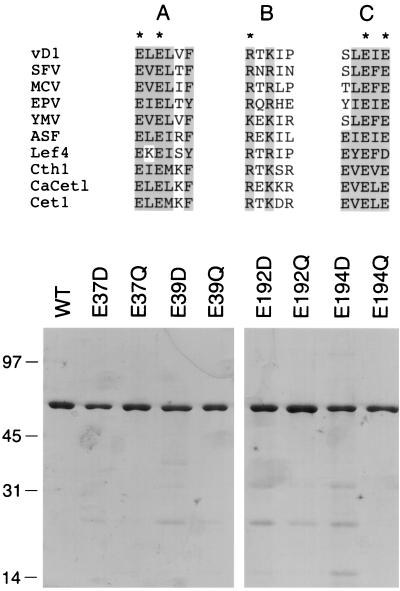
Two distinct classes of eukaryotic RNA triphosphatases have been described. The RNA triphosphatases of mammals and other metazoan species belong to a superfamily of phosphatases that act via formation and hydrolysis of a covalent enzyme-(cysteinyl-S-)-phosphate intermediate (24, 52, 58). The metazoan RNA triphosphatase reaction requires no metal cofactor. In fact, metazoan RNA triphosphatases are inhibited by divalent cations. In contrast, the RNA triphosphatases of DNA viruses (poxviruses and baculoviruses), the budding yeast S. cerevisiae, and the pathogenic fungus Candida albicans are strictly dependent on a divalent cation cofactor (14, 21, 26, 38, 51). The viral-fungal triphosphatase family is defined by three conserved collinear motifs (A, B, and C) that include clusters of acidic and basic amino acids that are essential for catalytic activity (21, 37) (Fig. 3). Five essential residues within these motifs were initially uncovered by alanine scanning mutagenesis of the vaccinia virus RNA triphosphatase (60, 61). Single alanine substitutions for residues Glu37, Glu39, Arg77, Glu192, and Glu194 of vD1(1-545)p reduced triphosphatase activity by two to three orders of magnitude without affecting the guanylyltransferase activity of the mutant proteins (60).
FIG. 3.
Purification of wild-type and mutated versions of vaccinia virus D1(1-545)p. Conserved motifs A, B, and C of the RNA triphosphatases of vaccinia virus (vD1), Shope fibroma virus (SFV), molluscum contagiosum virus (MCV), Melanoplus sanguinipes entomopoxvirus (EPV), Yaba monkey tumor virus (YMV), African swine fever virus (ASF), Autographa californica baculovirus (Lef4), S. cerevisiae (Cth1 and Cet1), and C. albicans (CaCet1) are aligned in the top panel. Vaccinia virus D1 residues that are conserved in the other proteins are shaded. The five amino acids in vD1(1-545)p that were found by alanine scanning to be essential for triphosphatase activity are indicated by asterisks. Protein purification is shown in the bottom panel. Aliquots (0.5 μg) of the phosphocellulose preparations of recombinant wild-type (WT) vD1(1-545) and the indicated mutant proteins were electrophoresed through a 10% polyacrylamide gel containing 0.1% SDS. Polypeptides were visualized by staining with Coomassie Blue dye. The positions and sizes (in kilodaltons) of marker proteins are indicated on the left.
Three of the triphosphatase-inactivating mutations in motifs A, B, and C (E37A, R77A, and E192A) were introduced into the chimeric capping enzyme vD1(1-545)-Mce1(211-597)p, and their in vivo effects were tested by plasmid shuffle (Fig. 2). The E37A, R77A, and E192A alleles were incapable of complementing the growth of cet1Δ cells (Fig. 2A) but retained their activity in complementing the growth of ceg1Δ cells (Fig. 2B). These results establish a correlation between the catalytic activity of vaccinia virus RNA triphosphatase in vitro and its function in vivo. The findings support the hypothesis that motifs A, B, and C comprise the triphosphatase active site.
Conservative mutations of the active site of vaccinia virus RNA triphosphatase.
We hypothesized previously that the glutamates in vaccinia virus motifs A and C facilitate catalysis by coordinating the essential divalent cation(s) (60, 61). The recently reported crystal structure of the S. cerevisiae RNA triphosphatase revealed that the glutamates in motif A and motif C do indeed comprise the metal-binding site of Cet1p (28). Mutational analysis showed that the glutamates are essential for Cet1p triphosphatase activity, and other studies indicate that their equivalents in motifs A and C of the C. albicans RNA triphosphatase CaCet1p (38) and baculovirus LEF4 (26) are also essential for catalysis.
The expectation is that if an acidic side chain is critical for metal binding, then replacement of a metal-binding glutamate by glutamine would have a significant effect on triphosphatase activity. Therefore, we tested the effects of conservative substitutions in residues Glu37, Glu39, Glu192, and Glu194 on vD1(1-545)p RNA triphosphatase activity in vitro. Each position was changed to glutamine and aspartic acid. The mutant proteins were expressed in E. coli as His-tagged fusions and purified from soluble bacterial extracts by Ni-agarose and phosphocellulose column chromatography. SDS-polyacrylamide gel electrophoresis analysis of the phosphocellulose preparations revealed a predominant 60-kDa polypeptide corresponding to vD1(1-545)p (Fig. 3). The guanylyltransferase activity of each of the phosphocellulose preparations was demonstrated by label transfer from [α-32P]GTP to the enzyme to form a 60-kDa covalent enzyme-GMP complex (not shown). The fact that the mutant enzymes retained guanylyltransferase activity indicated that the conservative mutations did not affect the global folding of the vaccinia virus protein.
The RNA triphosphatase activities of the phosphocellulose preparations of wild-type and mutant vD1(1-545)p were assayed by the release of 32Pi from γ-32P-labeled poly(A) during a 5-min incubation in the presence of magnesium chloride. The specific activities of the vD1(1-545)p mutants were calculated from the slopes of the titration curves in the linear range of enzyme dependence. The activities of the mutants were normalized to that of the wild-type enzyme and are shown in Table 1. The salient findings were that the E37Q, E39Q, and E192Q mutations abolished the RNA triphosphatase activity of vaccinia virus capping enzyme, whereas the E194Q protein displayed nearly wild-type specific activity (Table 1). Introduction of aspartic acid at positions 37 and 39 restored RNA triphosphatase activity to 29 and 10% of the wild-type level, respectively (Table 1). We surmise that the negative charges on the two positions in motif A are critical for catalysis and that shortening the distance from the main chain to the carboxylate has a modest negative effect on activity. Remarkably, the introduction of aspartic acid in lieu of Glu192 in motif C elicited a twofold increase in RNA triphosphatase activity compared to wild-type vD1(1-545)p. Thus, position 192 must be acidic, but the shorter linker arm of aspartate is preferable to the native glutamate. These data support a direct role for Glu37, Glu39, and Glu192 in coordinating the metal, and they imply some steric flexibility in the metal-binding site of vaccinia virus triphosphatase. Note that RNA triphosphatase activity was also enhanced relative to that of the wild type when Glu194 in motif C was replaced by aspartic acid. Given that the E194A mutation abolished activity (60), while the E194Q and E194D changes restored activity (Table 1), we infer that hydrogen bonding rather than electrostatic interactions of this residue are critical for catalysis.
TABLE 1.
Mutational effects on the phosphohydrolase activity of vaccinia virus D1(1-545)pa
| Motif | Mutant | Sp act (% of wild type)b
|
|||
|---|---|---|---|---|---|
| RNA triphosphatase with MgCl2 | ATPase with:
|
||||
| MgCl2 | MnCl2 | CoCl2 | |||
| A | E37D | 29 | 2 | 1 | 2 |
| E37Q | 0.1 | 0.4 | <0.1 | <0.1 | |
| E39D | 10 | 2 | 0.3 | 0.3 | |
| E39Q | 0.1 | 0.2 | 0.1 | <0.1 | |
| C | E192D | 240 | 150 | 34 | 14 |
| E192Q | 1 | 0.5 | <0.1 | 0.2 | |
| E194D | 230 | 45 | 5 | 0.8 | |
| E194Q | 81 | 9 | 0.3 | 1 | |
The phosphocellulose preparations of wild-type and mutant D1(1-545)p were assayed for RNA triphosphatase and ATPase activities as follows. RNA triphosphatase reaction mixtures (10 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol, 2 mM MgCl2, 1 μM γ-32P-labeled poly(A), and serial twofold dilutions of enzyme were incubated for 5 min at 37°C and then quenched with formic acid. ATPase reaction mixtures (10 μl) containing 1 mM [γ-32P]ATP, serial twofold dilutions of enzyme, and either (i) 50 mM Tris-HCl (pH 8.0) and 10 mM MgCl2, (ii) 50 mM Tris-HCl (pH 7.5) and 1 mM MnCl2, or (iii) 50 mM Tris-acetate (pH 6.5) and 2 mM CoCl2 were incubated for 30 min at 37°C. The reaction products were analyzed by polyethyleneimine-cellulose thin-layer chromatography (61), and the release of 32Pi was quantitated by scanning the thin-layer chromatography plate with a phosphorimager.
Specific activities were calculated from the slopes of the titration curves and then normalized to the specific activity of the wild-type enzyme under each reaction condition (defined as 100%). From the wild-type specific activities we calculated turnover numbers of 1.7 s−1 for magnesium-dependent RNA triphosphatase, 16 s−1 for magnesium-dependent ATPase, 10 s−1 for manganese-dependent ATPase, and 49 s−1 for cobalt-dependent ATPase.
Effects of conservative mutations on metal-dependent ATPase activity.
The metal-dependent viral and fungal RNA triphosphatases also have an intrinsic capacity to hydrolyze nucleoside triphosphates to nucleoside diphosphates and inorganic phosphate in the presence of a divalent cation cofactor—either manganese or cobalt in the case of the yeast and baculovirus enzymes and either manganese, cobalt, or magnesium in the case of vaccinia virus capping enzyme (14, 21, 26, 37, 38, 45, 51). The nucleoside triphosphatase activities of wild-type and mutant vD1(1-545)p proteins were assayed by the release of 32Pi from [γ-32P]ATP (1 mM) during a 30-min reaction in the presence of 10 mM magnesium chloride (the optimal magnesium concentration for ATP hydrolysis [data not shown]). The relative specific activities of the vD1(1-545)p motif A and C mutants (normalized to the wild-type activity) are shown in Table 1. The motif A mutational effects on magnesium-dependent ATPase generally mirrored the effects on RNA triphosphatase, insofar as ATPase activity was abolished by introducing glutamine in place of Glu37 and Glu39. However, the extent of restoration of ATPase activity when aspartic acid was introduced at positions 37 and 39 was about an order of magnitude less than it was for RNA triphosphatase (Table 1). Thus, ATP hydrolysis was more stringent in its dependence on glutamates in motif A. In motif C, a glutamine at position 192 eliminated ATPase activity, which was restored to greater-than-wild-type levels when an aspartic acid was present at this position. This was the same mutational effect seen for RNA triphosphatase. At position Glu194, the level of residual activity with glutamine was much lower for ATPase (9%) than for RNA triphosphatase (81%). Similarly, the E194D change, although it restored activity compared to E194Q, had a relatively greater impact on ATPase than on RNA triphosphatase.
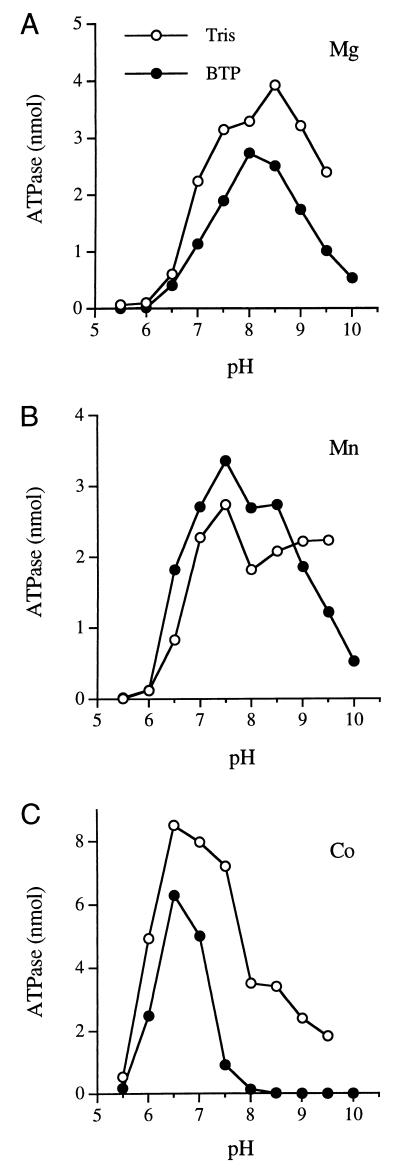
Activation of the ATPase of vaccinia virus capping enzyme by manganese and cobalt has not been well characterized. Optimal conditions for ATP hydrolysis by vD1(1-545)p in the presence of MnCl2 or CoCl2 were established by varying the divalent cation concentration and pH of the reaction mixture. ATPase activity at pH 8 was optimal at 1 mM MnCl2 or 2 mM CoCl2 (data not shown). Remarkably, the pH optimum for ATP hydrolysis was strongly influenced by the choice of divalent cation cofactor. We assayed ATPase in bis-Tris-propane (BTP) buffers at pH 5.5 to 10 in the presence of 10 mM MgCl2, 1 mM MnCl2, or 2 mM CoCl2 (Fig. 4). Magnesium-dependent ATPase was optimal at pH 8 to 8.5 and was virtually nil at pH 6 (Fig. 4A). Manganese-dependent ATPase was optimal at pH 7.5 and minimal at pH 6. In contrast, cobalt-dependent ATPase peaked at pH 6.5 (Fig. 4C). Cobalt supported considerable activity at pH 6 (unlike magnesium and manganese) but was inactive in BTP buffer at pH 8 to 8.5, which is the favored pH range of BTP buffer for magnesium and manganese cofactor activity. We observed similar pH optima when Tris-acetate and Tris-HCl buffers were tested in the range of pH 5.5 to 9.5, except that cobalt-dependent ATPase was less sensitive to alkaline pH conditions in Tris-HCl buffer than in BTP buffer (Fig. 4C). The molecular basis for the distinctive acid shift in the pH dependence of ATP hydrolysis in cobalt is unclear.
FIG. 4.
Effect of pH on metal-dependent ATP hydrolysis by vaccinia virus D1(1-545)p. Reaction mixtures (10 μl) containing 50 mM buffer (either BTP [pH 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, or 10.0] or Tris [Tris-acetate at pH 5.5, 6.0, 6.5, or 7.0 or Tris-HCl at pH 7.5, 8.0, 8.5, 9.0, or 9.5]), 5 ng of vD1(1-545)p, 1 mM [γ-32P]ATP, and either 10 mm MgCl2 (A), 1 mM MnCl2 (B), or 2 mM CoCl2 (C) were incubated for 30 min at 37°C. The reaction products were analyzed by polyethyleneimine-cellulose thin-layer chromatography (61). ATPase activity is plotted as a function of pH.
The effects of motif A and C mutations on the manganese- and cobalt-dependent ATPase activities of vD1(1-545)p at pH 8 and 6.5, respectively, are shown in Table 1. The vaccinia virus enzyme displayed a stringent requirement for glutamate at positions 37 and 39 to support ATP hydrolysis in cobalt and manganese; there was no recovery of function when these positions were occupied by aspartic acid. In motif C, the E194Q and E194D changes elicited more severe defects in manganese- and cobalt-dependent ATPase than they did on activity in magnesium (Table 1). The same was true of the E192D mutation.
DISCUSSION
Viral RNA capping enzymes are attractive targets for antiviral drugs because the properties of the viral enzymes are often different from those of the host cell enzymes. This is especially so for the metal-dependent RNA triphosphatases of DNA viruses and double-stranded RNA viruses (5, 14, 26, 35, 60), which are mechanistically and structurally unrelated to the metal-independent cellular RNA triphosphatase. The fact that metazoan species encode no obvious homologues of the viral RNA triphosphatases suggests that an inhibitor of viral RNA triphosphatase should have selectivity for the viral pathogen and minimal effect on the host organism. The yeast-based genetic system we describe here provides a valuable tool for basic and applied studies of viral capping enzymes. We show that vaccinia virus RNA triphosphatase and vaccinia virus guanylyltransferase can function in lieu of the essential yeast enzymes Cet1p and Ceg1p, provided that the vaccinia virus protein is fused to the mammalian capping enzyme, which targets the viral protein to the pol II elongation complex. Hence, the capping functions of the viral enzyme can be studied in vivo independent of ancillary roles of the viral protein in the context of virus replication. We have used this system to establish a concordance between mutational effects on vaccinia virus RNA triphosphatase activity in vitro and in vivo.
We have illuminated structure-activity relationships for four essential amino acids of vaccinia virus RNA triphosphatase. Three essential glutamates, Glu37 and Glu39 in motif A and Glu192 in motif C, cannot be replaced by glutamine, suggesting that these side chains comprise the metal-binding site of vaccinia virus triphosphatase. These results are consistent with insights gained from the crystal structure of yeast RNA triphosphatase Cet1p complexed with the divalent cation manganese. Cet1p adopts a novel enzyme fold whereby an antiparallel eight-strand β barrel forms a hydrophilic “triphosphate tunnel” that is topologically closed (28). Multiple acidic side chains point into the tunnel cavity, including the essential glutamates of motifs A and C. The interior of the tunnel contains a single sulfate ion coordinated by basic side chains projecting into the tunnel. Insofar as sulfate is a structural analog of phosphate, we surmise that the side chain interactions of the sulfate reflect contacts made by the enzyme with the γ phosphate of the triphosphate-terminated RNA and nucleoside triphosphate substrates. A manganese ion within the tunnel cavity is coordinated with octahedral geometry to the sulfate, to the side chain carboxylates of the two essential glutamates in motif A, and to one of the glutamates in motif C. Glutamine substitutions for any of the three Cet1p glutamates that directly coordinate the manganese result in a complete loss of catalytic activity (37). By analogy with Cet1p, we propose that Glu37, Glu39, and Glu192 of vaccinia virus capping enzyme interact directly with a divalent cation. The active site of Cet1p may be less flexible than that of vD1(1-545)p, insofar as none of the three metal-binding glutamates of Cet1p could be functionally replaced by aspartic acid.
A second essential glutamate in motif C of Cet1p coordinates a water molecule bound to manganese. We invoke a similar role for the Glu194 in motif C of vaccinia virus capping enzyme. Coordination of solvent by polar interactions of vaccinia virus side chain 194 would be consistent with the retention of catalytic function by the E194Q mutant. Whereas glutamine may suffice for coordinating solvent bound to magnesium, phosphohydrolase activity in manganese and cobalt was strictly dependent on a glutamate side chain. Spatial differences in the coordination spheres of these metals may account for the distinct mutational effects with different metal cofactors.
Motifs A and C are located within β strands of yeast Cet1p that are widely separated in the primary structure but closely approximated in the tertiary structure (28). Motifs A and C are located on the tunnel “floor” that abuts the globular core of the protein. In motifs A and C of the fungal RNA triphosphatases, alternating charged side chains are interdigitated with alternating aliphatic or aromatic side chains (Fig. 3). This sequence pattern is reprised in motifs A and C of the poxvirus, African swine fever virus, and baculovirus RNA triphosphatases, suggesting that the metal-binding residues of the viral enzymes may also be located within β strands.
Although we propose that yeast and vaccinia virus RNA triphosphatases share a common metal-binding site, we suspect that the active sites of the yeast and viral enzymes may adopt different tertiary structures, i.e., that the vaccinia virus active site does not residue within an enclosed tunnel, because the motif A and C glutamates of vaccinia virus D1(1-545)p are accessible to limited digestion with V8 protease (60). A definitive assessment of the similarities between fungal and viral RNA triphosphatases will hinge on successful crystallization of vD1(1-545)p.
Finally, the availability of isogenic yeast strains containing mammalian versus viral capping systems provides a means of drug discovery aimed at blocking viral cap formation. For example, any compound that is selectively cytotoxic to the vD1(1-549)-MCE1(211-597)-K294A strain but not to the MCE1 strain is a strong candidate for being a specific inhibitor of vaccinia virus capping. Identification of effective antipoxviral drugs is a reasonable goal in light of the high incidence of molluscum contagiosum infection as a sequela of AIDS and the potential threat of smallpox as a biological weapon. We anticipate extending the yeast complementation approach to the methyltransferase component of vaccinia virus capping enzyme (19, 32, 33) with the goal of generating yeast strains in which the entire capping apparatus is of viral origin. It will also be of interest to apply the yeast complementation test to the capping enzymes of other viruses that affect human health, e.g., rotavirus (7, 40). Genetic complementation by viral polypeptides fused to the mammalian delivery vehicle also provides a means to identify the capping enzymes of viruses in cases where the biochemical activities are refractory to purification.
REFERENCES
- 1.Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236:1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- 5.Bisaillon M, Lemay G. Characterization of the reovirus λ1 protein RNA 5′-triphosphatase activity. J Biol Chem. 1997;272:29954–29957. doi: 10.1074/jbc.272.47.29954. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter M S, DeLange A M. A temperature-sensitive lesion in the small subunit of vaccinia virus-encoded mRNA capping enzyme causes a defect in viral telomere resolution. J Virol. 1991;65:4042–4050. doi: 10.1128/jvi.65.8.4042-4050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D, Luongo C L, Nibert M L, Patton J T. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology. 1999;265:120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- 8.Cho E J, Rodriguez C R, Takagi T, Buratowski S. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong P, Shuman S. Covalent catalysis in nucleotidyl transfer: a KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J Biol Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 11.Cong P, Shuman S. Mutational analysis of mRNA capping enzyme identifies amino acids involved in GTP binding, enzyme-guanylate formation, and GMP transfer to RNA. Mol Cell Biol. 1995;15:6222–6231. doi: 10.1128/mcb.15.11.6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensinger M J, Martin S A, Paoletti E, Moss B. Modification of the 5′-terminus of mRNA by soluble guanylyl and methyl transferases from vaccinia virus. Proc Natl Acad Sci USA. 1975;72:2525–2529. doi: 10.1073/pnas.72.7.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst T R, Moss B. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells: importance of the 5′ untranslated leader. J Mol Biol. 1989;206:333–348. doi: 10.1016/0022-2836(89)90483-x. [DOI] [PubMed] [Google Scholar]
- 14.Gross C H, Shuman S. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J Virol. 1998;72:10020–10028. doi: 10.1128/jvi.72.12.10020-10028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino L A, Jin J, Dong W. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J Virol. 1998;72:10003–10010. doi: 10.1128/jvi.72.12.10003-10010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagler J, Shuman S. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science. 1992;255:983–986. doi: 10.1126/science.1546295. [DOI] [PubMed] [Google Scholar]
- 17.Häkansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 18.Hassett D E, Lewis J I, Xing X, DeLange L, Condit R C. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered change-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- 19.Higman M A, Christen L A, Niles E G. The mRNA (guanine-7-) methyltransferase domain of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem. 1994;269:14974–14981. [PubMed] [Google Scholar]
- 20.Ho C K, Lehman K, Shuman S. An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res. 1999;27:4671–4678. doi: 10.1093/nar/27.24.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho C K, Pei Y, Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 22.Ho C K, Schwer B, Shuman S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol Cell Biol. 1998;18:5189–5198. doi: 10.1128/mcb.18.9.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 24.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 25.Ho C K, Van Etten J L, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin J, Dong W, Guarino L A. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J Virol. 1998;72:10011–10019. doi: 10.1128/jvi.72.12.10011-10019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman K, Schwer B, Ho C K, Rouzankina I, Shuman S. A conserved domain of yeast RNA triphosphatase flanking the catalytic core regulates self-association and interaction with the guanylyltransferase component of the mRNA capping apparatus. J Biol Chem. 1999;274:22668–22678. doi: 10.1074/jbc.274.32.22668. [DOI] [PubMed] [Google Scholar]
- 28.Lima C D, Wang L K, Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 29.Luo Y, Mao X, Deng L, Cong P, Shuman S. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J Virol. 1995;69:3852–3856. doi: 10.1128/jvi.69.6.3852-3856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao X, Schwer B, Shuman S. Mutational analysis of the Saccharomyces cerevisiae ABD1 gene: cap methyltransferase activity is essential for cell growth. Mol Cell Biol. 1996;16:475–480. doi: 10.1128/mcb.16.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit: identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 33.Mao X, Shuman S. Vaccinia virus mRNA (guanine-7-) methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photocrosslinking of the cap to the methyl acceptor site. Biochemistry. 1996;35:6900–6910. doi: 10.1021/bi960221a. [DOI] [PubMed] [Google Scholar]
- 34.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myette J R, Niles E G. Domain structure of the vaccinia virus mRNA capping enzyme: expression in Escherichia coli of a subdomain possessing the RNA 5′-triphosphatase and guanylyltransferase activities and a kinetic comparison to the full-size enzyme. J Biol Chem. 1996;271:11936–11944. doi: 10.1074/jbc.271.20.11936. [DOI] [PubMed] [Google Scholar]
- 36.Myette J R, Niles E G. Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleotide triphosphate phosphohydrolase activities: demonstration that both activities are carried out at the same active site. J Biol Chem. 1996;271:11945–11952. doi: 10.1074/jbc.271.20.11945. [DOI] [PubMed] [Google Scholar]
- 37.Pei Y, Ho C K, Schwer B, Shuman S. Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J Biol Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- 38.Pei Y, Lehman K, Tian L, Shuman S. Characterization of Candida albicans RNA triphosphatase and mutational analysis of the active site. Nucleic Acids Res. 2000;28:1885–1892. doi: 10.1093/nar/28.9.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pena L, Yanez J, Revilla Y, Vinuela E, Salas M L. African swine fever virus guanylyltransferase. Virology. 1992;193:319–328. doi: 10.1006/viro.1993.1128. [DOI] [PubMed] [Google Scholar]
- 40.Pizarro J L, Sandino A M, Pizarro J M, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J Gen Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 41.Saha N, Schwer B, Shuman S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J Biol Chem. 1999;274:16553–16562. doi: 10.1074/jbc.274.23.16553. [DOI] [PubMed] [Google Scholar]
- 42.Schwer B, Shuman S. Mutational analysis of yeast mRNA capping enzyme. Proc Natl Acad Sci USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 44.Shuman S. Functional domains of vaccinia virus mRNA capping enzyme: analysis by limited tryptic digestion. J Biol Chem. 1989;264:9690–9695. [PubMed] [Google Scholar]
- 45.Shuman S. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J Biol Chem. 1990;265:11960–11966. [PubMed] [Google Scholar]
- 46.Shuman S. Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuman S. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology. 1997;271:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 48.Shuman, S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol., in press. [DOI] [PubMed]
- 49.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc Natl Acad Sci USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuman S, Broyles S S, Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987;262:12372–12380. [PubMed] [Google Scholar]
- 51.Shuman S, Surks M, Furneaux H, Hurwitz J. Purification and characterization of a GTP-pyrophosphate exchange activity from vaccinia virions: association of the GTP-pyrophosphate exchange activity with vaccinia mRNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex (capping enzyme) J Biol Chem. 1980;255:11588–11598. [PubMed] [Google Scholar]
- 52.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 53.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme beta subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 54.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA: association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-) methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 55.Vos J C, Sasker M, Stunnenberg H G. Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J. 1991;10:2553–2558. doi: 10.1002/j.1460-2075.1991.tb07795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S P, Shuman S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J Biol Chem. 1997;272:14683–14689. doi: 10.1074/jbc.272.23.14683. [DOI] [PubMed] [Google Scholar]
- 58.Wen Y, Yue Z, Shatkin A J. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc Natl Acad Sci USA. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada-Okabe T, Doi R, Shimmi O, Arisawa M, Yamada-Okabe H. Isolation and characterization of a human cDNA for mRNA 5′-capping enzyme. Nucleic Acids Res. 1998;26:1700–1706. doi: 10.1093/nar/26.7.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu L, Shuman S. Mutational analysis of the RNA triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1996;70:6162–6168. doi: 10.1128/jvi.70.9.6162-6168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]