Abstract
Hepatitis B virus (HBV) replicates by reverse transcription of an RNA intermediate, the pregenomic RNA. The first step of HBV genome replication is the encapsidation of the pregenomic RNA encoding the encapsidation signal, termed ɛ, into the core particles, which is preceded by recognition and binding of HBV DNA polymerase to ɛ. The pregenomic RNA contains two identical ɛ elements due to its terminal redundancy: one near the 5′ end and another near the 3′ end. Despite the fact that both ɛ elements have an identical sequence, only the 5′ ɛ, but not the 3′ ɛ, is functional for encapsidation. To understand the molecular nature of this position effect, we made a series of lacZ RNA expression plasmids which contain the ɛ element at various positions from the 5′ end of the transcripts. Following transfection, the lacZ RNAs in cytoplasmic core particles were measured by RNase protection assay for encapsidation. The results indicated that the lacZ RNAs with ɛ positioned up to 65 nucleotides from the 5′ end were encapsidated, whereas the lacZ RNAs with ɛ positioned further downstream were not. Interestingly, the cap-free lacZ RNA transcribed by T7 RNA polymerase was not encapsidated, implying that the 5′ cap structure is required for encapsidation of the pregenomic RNA. We hypothesized that HBV DNA polymerase must somehow recognize the cap structure and/or its associated factors, as well as the 5′ ɛ, for encapsidation to occur.
Hepatitis B virus (HBV), a causative agent of chronic hepatitis in human, is the prototype member of the Hepadnaviridae. Related members of the hepadnavirus family include Woodchuck hepatitis virus, Ground squirrel hepatitis virus, and Duck hepatitis B virus (DHBV) (10). Although HBV has a DNA genome, it replicates through reverse transcription of an RNA intermediate, the pregenomic RNA (pgRNA) (27). Despite the general similarities to retroviruses, many steps in its replication are distinct (18, 20). One difference is shown in the mechanisms of the encapsidation process. In contrast to retroviruses, the viral polymerase rather than the core protein is responsible for specific recognition of the encapsidation signal, ɛ, of the pgRNA (2, 6).
The first step of HBV genome replication is the encapsidation of the pgRNA into core particles. Core particle assembly involves the interactions of the structural proteins, core (C) and polymerase (P), with the pgRNA (1, 2). The pgRNA serves as template for reverse transcription as well as for translation of the C and P proteins. Incorporation of P protein as well as pgRNA into assembling core particles is essential for viral DNA synthesis (1, 11). Studies suggested a model for viral assembly in which the P protein first interacts with the pgRNA, and then the P protein-pgRNA complex is recognized by the core protein (2, 22). A direct evidence for the P protein-pgRNA interaction was obtained with the related duck hepatitis B virus (DHBV) (22, 28). It is intriguing that the P protein plays a role in encapsidation, in addition to its catalytic role for the viral genome synthesis (1, 11).
The cis-acting element for encapsidation, termed ɛ, has been defined within 85 nucleotides (nt) near the 5′ end of pgRNA, which is necessary and sufficient to direct encapsidation of heterologous RNA sequences into the viral core particle (12, 14, 21). The ɛ element can fold into a stem-loop structure, which is highly conserved among hepadnaviruses (14, 21). Genetic analysis of HBV and DHBV has shown that disruption of the stem-loop structure interferes with encapsidation, thereby confirming the functional role of this folded structure in vivo (7, 15, 21, 22). Structural probing with specific RNases has revealed the presence of this stem-loop structure in core particles isolated from transfection study (21), as well as in in vitro-transcribed RNAs (15). The pgRNA has two identical ɛ elements, at both the 5′ and 3′ ends due to its terminal redundancy. However, only the 5′ ɛ element, but not its 3′ copy, is functional in the encapsidation process (12, 14). These observations suggested that there could be additional factors which enable the 5′ ɛ element to be a functional encapsidation signal (21). However, the reason why only the 5′ ɛ element is functional has not been elucidated.
To further understand the molecular nature of the position effect, a series of the lacZ-ɛ plasmids were constructed from which lacZ transcripts having the ɛ element at various positions can be transcribed. Interestingly, there was a gradual decrease in encapsidation efficiency as the ɛ element was positioned farther from the 5′ end. These observations led us to speculate that the 5′ cap structure as well as the 5′ ɛ is essential for encapsidation. Indeed, it was found that the cap-free lacZ RNA transcribed by T7 RNA polymerase was not encapsidated, indicating the requirement of the 5′ cap structure for the encapsidation. The results led us to hypothesize that for efficient encapsidation of its pgRNA, HBV DNA polymerase must recognize simultaneously the 5′ cap structure as well as the 5′ ɛ element.
MATERIALS AND METHODS
Cell culture.
Huh7 cells were used for transfection (17). Huh7 cells were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL) supplemented with 10% fetal bovine serum (GIBCO-BRL) and 10 μg of gentamicin per ml (GIBCO-BRL) at 37°C in 5% CO2 and were split every third day.
Transfection.
The day before transfection, cells were plated at a confluency of 75%. On the following day, the cells were washed twice with phosphate-buffered saline (PBS) and fed with fresh medium. After 2 h, cells were transfected with 10 μg of supercoiled plasmid DNA per 60-mm-diameter plate by the calcium phosphate coprecipitation technique as described previously (23). In brief, 10 μg of plasmid DNA was mixed with 250 mM CaCl2 to a final volume of 250 μl. This solution was added drop by drop to 250 μl of 2× HEPES-buffered saline (280 mM NaCl, 50 mM HEPES acid, 1.5 mM Na2HPO4 [pH 7.1]) and was incubated for 20 to 30 min at room temperature. The resulting solution with fine white precipitates was slowly dropped over a plate of cells. After incubation at 37°C for 18 h in a 5% CO2 incubator, the culture medium was replaced with fresh medium. The transfected cells were grown until harvest.
HBV expression plasmid construction.
HBV sequences were numbered starting at the unique EcoRI site of the HBV ayw subtype, according to the method of Galibert et al. (9). Nucleotide numbers indicate the HBV sequence number, otherwise indicated. In this numbering system, the 5′ end of the pregenomic RNA is at nt 1820 (29).
pCH9/3091 and pCH3142 were the generous gifts of M. Nassal. pCH9/3091 is a pgRNA expression plasmid, from which a wild-type pgRNA is transcribed from a cytomegalovirus (CMV) promoter (14). pCH3142 was used as a helper plasmid that provides C and P proteins, but lacks the functional ɛ element (see Fig. 2A) (14).
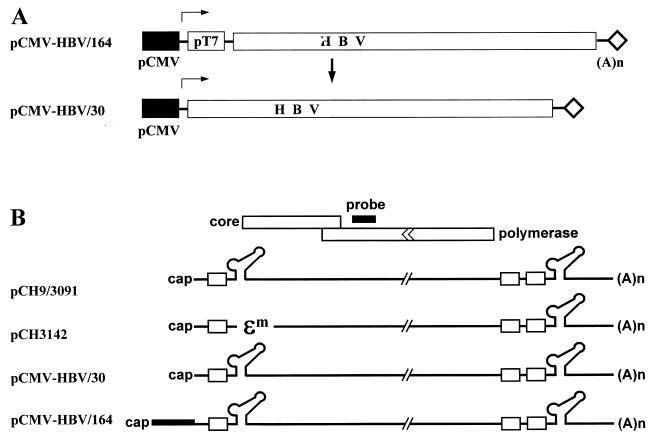
FIG. 2.
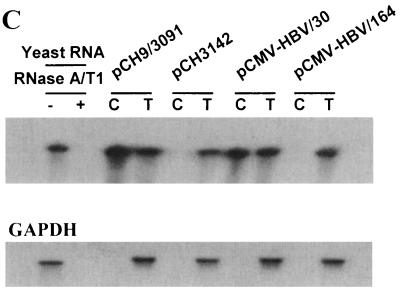
Analysis of encapsidation of HBV pgRNAs with the 5′ ɛ elements at various positions by RPA. (A) Schematic representation of the HBV pgRNA expression plasmids, in which HBV pgRNA is transcribed under the control of the CMV immediate-early promoter. In the pCMV-HBV/164, an extra 134 nt derived from vector plasmid pcDNA1/amp was included. Subsequently, the vector-derived sequence was deleted by a PCR-mediated procedure (13). T7 promoter sequence derived from the pcDNA1/amp is indicated. CMV promoters are indicated by the solid boxes. Transcription start sites are indicated by the rightward arrows. (B) Map of HBV pgRNAs made from the HBV RNA expression plasmids. pCH9/3091 is a pgRNA expression plasmid, which transcribes the pgRNA with its 5′ end mapping at nt 1820 (19). pCH3142 is a helper plasmid that expresses both C and P protein, but lacks the 5′ ɛ element (14). pCMV-HBV/30 was made such that the 5′ ends of its transcripts were identical with those of the wild-type HBV pgRNA (i.e., the 5′ ɛ element is positioned approximately 30 nt from the 5′ end). pCMV-HBV/164 was made such that the 5′ ɛ element is positioned 164 nt from the 5′ end of the transcript. The vector-derived sequences are indicated by a boldface line in pCMV-HBV/164. The HBV riboprobe is depicted by the solid box (probe) above the C and P open reading frame. (C) Analysis of encapsidation of HBV pgRNAs by RPA. Huh7 cells were transfected by the indicated plasmids. Three days after transfection, total cytoplasmic RNAs (T) and core-associated RNA (C) were extracted and measured by RPA. Glyceraldehyde-3-phosphate dehydrogenase RNA was measured by RPA as a loading control. Only one-third of the RNAs were analyzed for total cytoplasmic RNA relative to core-associated RNA for comparison. Yeast RNA was used as a negative control.
pCMV-HBV/164 was made by inserting the greater-than-genome-length FspI (nt 1804) and XbaI (nt 1992) fragment into pcDNA1/amp (Invitrogen) between the EcoRV and XbaI sites. The HBV transcript made from this plasmid has a vector-derived 134 nt at the 5′ end relative to that of wild-type pgRNA. Thus, pCMV-HBV/30 was made by removing this vector-derived 134 nt by a PCR-mediated method (13). Briefly, a fragment was made by PCR using a forward primer of the sequence 5′-CCCGAGCTCTCTGGCTAACTAACTTTTTCACCTCTGCC-3′ (SacI site underlined) and a reverse primer of the sequence 5′-CCCAAGCTTCTATTGTTCCCAAGAATATGG-3′ (nt 2839 to 2822) with pCMV-HBV/164 as a template. The resulting PCR fragment was digested by SacI and BspEI and then inserted between the SacI (nt 2894 of pcDNA1/amp) and BspEI (nt 2331) sites of pCMV-HBV/164.
lacZ-ɛ plasmid construction.
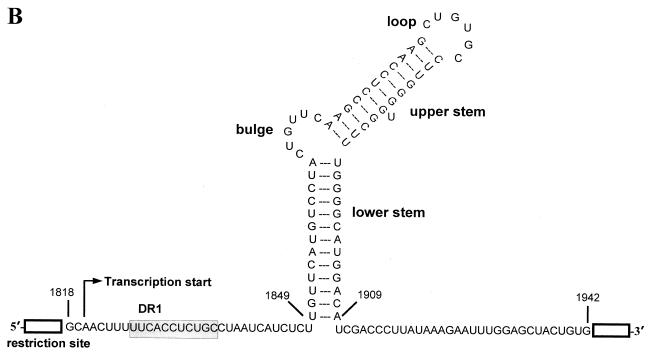
The lacZ sequences were numbered starting at the unique HindIII site of the pCH110 plasmid (Pharmacia). All lacZ-ɛ plasmids were constructed by inserting an ɛ element into the lacZ gene of the pcDNA-lacZ. First, pcDNA-lacZ was made by inserting the HindIII-EcoRI fragment of pCH110 (nt 1 to 3286 of the lacZ gene) into pcDNA1/amp. Then, the fragments encoding ɛ (nt 1818 to 1942) were inserted at various positions of the lacZ gene by using a unique restriction enzyme site (Fig. 1B). These fragments were prepared by PCR of pCH9/3091 as a template. A pair of primers was designed to contain a restriction enzyme site at each end; this site is a unique restriction site in the lacZ gene. The typical primers are read as the 5′ restriction enzyme sequence AAGCTTTTTCACCTCTGCCTA-3′ (HindIII site underlined) and the 5′ restriction enzyme sequence CACAGTAGCTCCAAATTC-3′. pLZ-ɛ-91, pLZ-ɛ-298, pLZ-ɛ-1485, pLZ-ɛ-1718, pLZ-ɛ-2312, and pLZ-ɛ-3136 were made by inserting the ɛ element at HindIII, KpnI, EcoRV, BclI, SacI, and AccI sites (nt 1, 208, 1395, 1628, 2222, and 3046, respectively, of the lacZ gene) of the pcDNA-lacZ, respectively. The orientation of the insert on the lacZ plasmid was confirmed by PCR (data not shown). Next, pLZ-ɛ-91/ΔAUG was made by deleting the sequences between two HindIII sites of the pLZ-ɛ-298 (nt 1 of lacZ gene to HindIII site generated by the forward PCR primer mentioned above). Finally, pLZ-ɛ-30 was made by replacing the greater-than-genome-length BglII-XbaI fragment (nt 1986 to 1992) of pCMV-HBV/30 to the 3.2-kb fragment between BsaAI (nt 133 of lacZ) and XbaI (multiple cloning site of pcDNA1/amp) of pcDNA-lacZ. The numbers at the end of the plasmid name indicate the distance from the 5′-end to the ɛ element of each lacZ-ɛ transcript. To determine the most distal position of the ɛ element, pLZ-ɛ-46, pLZ-ɛ-55, pLZ-ɛ-65, and pLZ-ɛ-82 were made by replacing the transcription starting region of pLZ-ɛ-30 with the fragments produced by PCR, which have an additional 16, 25, 35, and 52 nt derived from pcDNA1/amp at the transcription start site, respectively.
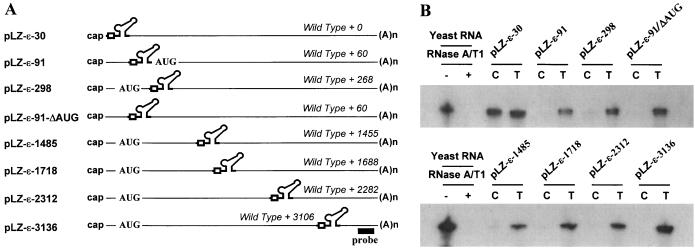
FIG. 1.
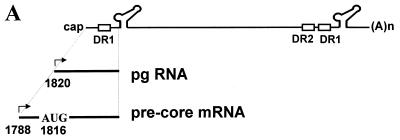
Structure of HBV pgRNAs. (A) Structure of HBV genomic RNA. The genomic RNA include the true pgRNA as well as minor additional transcripts (indicated here by pre-core mRNA) with start sites up to 32 nt (29). The minor transcripts encoding the pre-core initiation codon are not encapsidated due to translational suppression (19). They function only as mRNAs for a secreted variant of the C protein known as the pre-core. Both the DR1 and the ɛ sequence are present twice on the pgRNA due to its terminal redundancy. The ɛ elements are represented by the stem-loop structure. The direct repeat elements, DR1 and DR2, are indicated by open boxes. The sequence at the 5′ end has been enlarged below the pgRNA. The translation initiation codon of the pre-C gene is indicated by AUG. Transcription start sites are indicated by arrows. The nucleotide numbers are indicated by the numbering system of Galibert et al. (9). (B) Nucleotide sequences and predicted structure of the ɛ cassette inserted in the pLZ-ɛ plasmids. DR1 and the stem-loop structure are indicated. Restriction enzyme sites at both ends of the ɛ insertion cassette are indicated by the open boxes.
pT7-HBV plasmid construction.
We modified the pGEM-4Z plasmid (Promega) in two ways to generate pGEM-4Z/T7, a cap-free RNA expression vector. First, T7 promoter sequence (5′-TCTCCCTATAGTGAGTCGTATTA-3′) was changed to T7m (5′-TCTCCGTATAGTGAGTCGAATTA-3′) to reduce spurious transcription by host RNA polymerase II as described previously (16). Second, the T7 RNA polymerase terminator derived from the pTM1 (8) was inserted between unique EcoRI and NdeI restriction sites of pGEM-4Z. pT7m-HBV/42 was created in a stepwise fashion by first inserting a genome-length HindIII (multiple cloning site of pcDNA1/amp)-to-FspI (nt 1804) fragment of pCMV-HBV/164 into the HindIII and EcoRI fill-in site of pGEM-4Z/T7. Subsequently, the HindIII-EcoRI (nt 1804 to 3182) site was replaced by a PCR-generated HindIII-EcoRI fragment lacking 77 nt between the transcription start site and the 5′ ɛ element. As a result, the 5′ ɛ element is positioned 42 nt from the 5′ end in the transcript made from pT7m-HBV/42. For comparison, pCMV-HBV/42 was made by replacing the transcription start region of pCMV-HBV/30 with a PCR-produced fragment containing an additional 12 nt derived from pGEM-4Z at the transcription start site.
Isolation of total RNA.
Three days after transfection, cells were washed twice with cold PBS, and total RNA was prepared from transfected cells by the standard guanidinium isothiocyanate method as described previously (5).
Isolation of core RNA.
Three days after transfection, cells were washed twice with cold PBS and lysed with 1 ml of cold lysis buffer (10 mM Tris-Cl [pH 7.9], 1 mM EDTA, 1% NP-40, 50 mM NaCl) at 37°C for 5 min. The lysate was transferred to a 1.5-ml tube and centrifuged for 2 min at 12,000 × g to pellet nuclei. To isolate core-associated RNA, the supernatant was treated with RNase-free DNase I (10 U/ml; Sigma) and micrococcal nuclease (30 U/ml; Calbiochem) for 15 min at 37°C. A 4× concentration of PNE buffer (26% polyethylene glycol, 1.4 M NaCl, 40 mM EDTA) was added, and the mixture was incubated for 1 h on ice. The sample was centrifuged at 12,000 × g for 15 min to pellet core particles, and the resulting pellet was resuspended with 50 μl of DNase I buffer (10 mM Tris-Cl [pH 7.9], 6 mM MgCl2). RNA was extracted by the guanidinium isothiocyanate method as described previously (5).
RPAs.
The RNase protection assay (RPA) was performed as previously described (24). Briefly, samples of total RNA (30 μg) or core-associated RNA were hybridized with 105 cpm of [α-32P]UTP (3,000Ci/mmol; Amersham)-labeled probe (see below) at 42°C overnight. RNase digestions were carried out with a mixture of RNase A and RNase T1 (Ambion) at 37°C for 0.5 h. The digested products were separated on a 5% acrylamide–8 M urea gel. The RNA probe used to detect HBV-specific RNA was generated by in vitro transcription of a pGEM-4Z-SE after cleavage with BamHI (nt 2906); it comprises sequence from nt 3182 to nt 2906. The pGEM-4Z-SE was generated by insertion of the SphI-EcoRI fragment (nt 1238 to 3182) of pCMV-HBV/30 into the same site of pGEM-4Z (Promega). The RNA probe used to detect lacZ-specific RNA was generated by in vitro transcription of pLZ-ɛ-30 after cleavage with AccI (nt 3046 of lacZ gene); it comprises sequence from nt 3286 to 3046 of lacZ.
Vaccinia virus infection.
A vaccinia virus strain expressing T7 RNA polymerase, vvT7-3, was employed to generate cap-free RNAs (8). Two hours before transfection, the cells (approximately 3 × 106) were washed with PBS twice and infected by a vaccinia virus (6 × 106 PFU). Subsequently, transfection was carried out as described above. After 48 h, total and core-associated RNAs were harvested as described above.
RESULTS
Analysis of the encapsidation process by using HBV constructs.
We wanted to examine why or how only the 5′ ɛ element, but not the 3′ ɛ element, functions as an encapsidation signal. To address this question, we tested whether the 5′ proximal location of the ɛ element is essential for encapsidation. A number of HBV pgRNA expression plasmids were made. Initially, the pCMV-HBV/164 plasmid was made by inserting the greater-than-genome-length FspI (nt 1804) and XbaI (nt 1992) fragments of the HBV dimer (ayw subtype) into pcDNA1/amp (Invitrogen) between the EcoRV and XbaI sites (Fig. 2A). The HBV RNA made from this construct has an extra 134 nt at the 5′ end relative to a wild-type pgRNA derived from the vector sequences. As a result, the ɛ element is positioned 164 nt from the 5′ end of the transcripts. Subsequently, the pCMV-HBV/30 construct was made by removing the vector-derived 134 nt as described in Materials and Methods (Fig. 2A). This construct was designed such that RNA transcripts made from pCMV-HBV/30 would be identical to the 3.5-kb pgRNA of the HBV with respect to the 5′ end; i.e., the ɛ element positioned approximately 30 nt from the 5′ end (Fig. 1A). To examine encapsidation efficiency, Huh7 cells were transfected with these plasmids. Following transfection, we isolated either the total cytoplasmic RNA or the RNA contained within purified cytoplasmic core particles. Then, the extracted RNAs were subjected to RNase protection analysis with a riboprobe derived from HBV sequence (Fig. 2C). As expected, the RNA made from a helper plasmid, pCH3142, lacking the functional ɛ element was not encapsidated, while the transcripts from pCH9/3091, which generates the wild-type pgRNA (i.e., the 5′ ɛ element located approximately 30 nt from the 5′ end) were encapsidated (Fig. 2C) (14, 19). On the other hand, the transcript from pCMV-HBV/164 was not encapsidated, whereas that of pCMV-HBV/30 was encapsidated. These results were consistent with the reported findings that the position of the 5′ ɛ element is a critical parameter for the encapsidation of the HBV pgRNA (14).
Analysis of the encapsidation process by using the lacZ-ɛ constructs.
Based on the results presented above, we speculated that viral polymerase recognizes the ɛ element only when it is located in close proximity to the 5′ end. To address this notion, we employed a heterologous lacZ RNA to exclude the possibility that the viral RNA sequence or its secondary structure other than the ɛ element modulates the encapsidation efficiency. Specifically, we made a series of lacZ RNA expression plasmids by inserting the ɛ cassette at various positions of the lacZ gene (Fig. 1B). The ɛ insert cassettes include DR1 as well as ɛ sequence (nt 1818 to 1942). Seven lacZ RNA expression constructs (i.e., pLZ-ɛ) were made in which the position of each ɛ element from the 5′ end was indicated as a suffix (Fig. 3A). Each pLZ-ɛ plasmid was transfected to Huh7 cells, and RNAs were extracted. In these transfections, a helper plasmid, pCH3142, was cotransfected to provide the HBV polymerase (P) and core (C) proteins that are essential for the encapsidation (2, 14). To examine whether the transcripts of pLZ-ɛ plasmids were encapsidated into core particles, the extracted RNAs were subjected to RPA analysis with the lacZ-specific probe as described in Materials and Methods (Fig. 3B). The lacZ RNAs were detected in the total cytoplasmic fraction in all transfected cells as expected (Fig. 3B). In contrast, in cytoplasmic core particles, the RNA was detected only in pLZ-ɛ-30 transfection, but was not detected in others. These results indicated that (i) the 5′ proximal ɛ element is necessary and sufficient for encapsidation and (ii) the position of the ɛ element from the 5′ end is a critical parameter for encapsidation. We speculated that the ɛ element could be recognized by the encapsidation machinery (which probably involves HBV polymerase) only if it is located in close proximity to the 5′ end.
FIG. 3.
Analysis of encapsidation of the lacZ-ɛ RNAs with 5′ ɛ at various positions by RPA. (A) Map of transcripts made from the various lacZ-ɛ plasmids. The number at the end of the plasmid name indicates the position of the ɛ element from the 5′ end. The position of the 5′ ɛ element relative to that of the wild type is indicated on the transcripts. The ɛ cassettes encoding the DR1 and ɛ elements (nt 1818 to 1942) are depicted by the open box and the stem-loop structure, respectively. The AUG codon derived from the lacZ gene is indicated by AUG. The pLZ-ɛ-91/ΔAUG lacks the lacZ AUG codon. The probe used for the RPA analysis is indicated by the solid box. (B) Analysis of encapsidation of the lacZ-ɛ RNA by RPA. Huh7 cells were transfected by the plasmids indicated above each lane, along with a pCH3142. Three days after transfection, total RNAs (T) and core-associated RNA (C) were extracted and measured by RPA. Only one-third of RNAs were analyzed for total RNA relative to core-associated RNA for comparison.
On the other hand, close examination of the pLZ-ɛ plasmids revealed that most of the unencapsidated lacZ transcripts encoded the translation initiation codon (AUG codon derived from the lacZ open reading frame) upstream of the inserted ɛ element: pLZ-ɛ-298, pLZ-ɛ-1485, pLZ-ɛ-1718, pLZ-ɛ-2312, and pLZ-ɛ-3136 (Fig. 3A). Relevantly, it was reported that the AUG codon (i.e., pre-core AUG codon of the HBV pgRNA) located upstream of the ɛ element suppresses the encapsidation of the pgRNA (Fig. 1A) (i.e., translational suppression of encapsidation) (19). Thus, to examine the possibility that the AUG codon present in the pLZ-ɛ-298 suppressed the encapsidation, pLZ-ɛ-91/ΔAUG was made, in which the fragment containing (nt 1 to 208) the lacZ AUG codon was deleted (Fig. 3A). As a result, the ɛ element is positioned 91 nt from the 5′ end. Nevertheless, the lacZ transcript derived from pLZ-ɛ-91/ΔAUG was not encapsidated (Fig. 3B). Therefore, the possibility that the presence of upstream AUG in the lacZ transcript blocked the encapsidation by translational suppression was excluded.
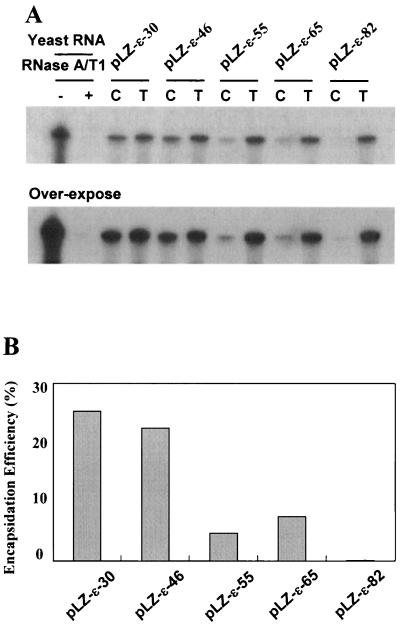
The results in Fig. 3 showed that the lacZ RNA with ɛ positioned 91 nt away from the 5′ end was not encapsidated, whereas the RNA with ɛ positioned 30 nt from the 5′ end was encapsidated (Fig. 3B). Next, we investigated the maximal distal position of the ɛ element that can be packaged. Thus, four additional constructs were made from which the RNAs transcribed have the ɛ element located 46, 55, 65, and 82 nt away from the 5′ end of the lacZ RNA, respectively. These constructs were transfected, and the extracted RNAs were analyzed by RPA to determine their encapsidation efficiency as described above (Fig. 4A). We found that the RNAs from pLZ-ɛ-46 were encapsidated as efficiently as that of pLZ-ɛ-30. In contrast, the RNA from pLZ-ɛ-82 was not detected in core particles. Moreover, the gradual decrease in encapsidation efficiency was apparent as the ɛ element was located distal to the 5′ end, with 30 to 46 nt being the positions required for maximal efficiency (Fig. 4B). This result was interpreted to indicate that the distance between the 5′ cap and the 5′ ɛ element is critical to be recognized by HBV polymerase for encapsidation. The correlation between the encapsidation efficiency and the distance between the 5′ cap and the 5′ ɛ element led us to speculate that the 5′ cap is involved in the encapsidation process. Thus, we hypothesized that both the 5′ cap structure and the 5′ ɛ element are essential for recognition of pgRNA by HBV polymerase for encapsidation.
FIG. 4.
Analysis of encapsidation of the lacZ-ɛ RNAs to determine the most distal position of the functional ɛ element. (A) RPA of the lacZ-ɛ RNAs. Huh7 cells were transfected with each lacZ-ɛ plasmid along with pCH3142. Total RNA (T) and core-associated RNA (C) were extracted and subjected to RPA analysis. Only one-third of the RNAs were analyzed for total RNA relative to core-associated RNA. The overexposed film was included for comparison. (B) Encapsidation efficiency of the lacZ-ɛ RNAs. The percentage of the core-associated RNA to total RNA was quantified with a Fuji Phosphoimager. A gradual decrease in encapsidation efficiency was noted because the ɛ element is positioned distal to the 5′ end.
Cap structure is required for encapsidation.
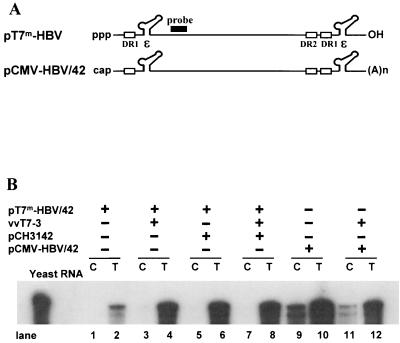
To substantiate the notion that both the 5′ cap structure and the 5′ ɛ element are essential, we examined whether cap-free RNA would be encapsidated. We employed the T7 RNA polymerase transcription system for de novo synthesis of cap-free RNA in cells. Thus, the pT7m-HBV/42 plasmid (i.e., the 5′ ɛ element positioned 42 nt from the 5′ end) was made so that the HBV pregenomic RNA can be transcribed under the control of the T7 promoter (Fig. 5A). On the other hand, it has been reported that RNA could be made at low levels from the T7 promoter by host RNA polymerase II (16). To minimize the spurious transcription by host RNA polymerase II, the T7 promoter was further mutated as described previously (16). Presumably, HBV pgRNAs made from pT7m-HBV/42 by T7 RNA polymerase were virtually cap free. Huh7 cells were transfected, and the extracted RNAs were analyzed by RPA for encapsidation (Fig. 5B). A vaccinia virus strain expressing T7 RNA polymerase, vvT7-3, provided T7 RNA polymerase in this experiment (8). As expected, only a small amount of the transcripts made from pT7m-HBV/42 was detected in the total cytoplasmic fraction, unless T7 RNA polymerase was provided (Fig. 5B, lanes 1 and 2). The RNAs, presumably transcribed from the spurious promoter present in the vector sequences by host RNA polymerase, were not encapsidated. It is most likely that the 5′ ends of the majority of these spurious transcripts were positioned beyond 65 nt upstream of the ɛ element. The lack of encapsidation of these transcripts is in line with our conclusion shown in Fig. 4, indicating that the 65-nt segment is the most distal position of the 5′ ɛ element. When T7 RNA polymerase was provided by coinfection with vaccinia virus vvT7-3, the transcripts were detected in the total cytoplasmic fraction, but not in the core particle fraction (Fig. 5B, lanes 3 and 4). In contrast, the identical transcripts made from the CMV promoter (pCMV-HBV/42) were encapsidated, as expected (Fig. 5B, lanes 9 and 10). The data suggested that the 5′ cap is essential for encapsidation.
FIG. 5.
Evidence that the 5′ cap is required for encapsidation. (A) Map of the HBV pgRNAs transcribed by T7 RNA polymerase. The ɛ elements are positioned 42 nt from the 5′ end in these transcripts. T7m represents a mutant T7 promoter which encodes 2 base substitutions from the wild-type T7 promoter (16). The mutant T7 promoter derives significantly smaller amounts of transcription by host RNA polymerase II relative to the wild-type T7 promoter without compromising its own promoter activity. The probe used for the RPA analysis is indicated by the solid box. DR1 and ɛ are indicated. (B) RPA for encapsidation of the HBV transcripts derived from the T7 promoter. Huh7 cells were infected by a vaccinia virus, vvT7-3, and then transfected with the indicated plasmids as described in Materials and methods. Two days after transfection, total RNA (T) and core-associated RNA (C) were extracted and subjected to RPA analysis. The probe was prepared by in vitro transcription of an HBV fragment (nt 2008 to 2504), as indicated by the solid box. Only one-third of RNAs were analyzed for total RNA relative to core-associated RNA for comparison.
Control experiments were conducted to substantiate the conclusion presented above. It is known that the 5′-end cap plays a role in efficient translation. Thus, it is possible that the amounts of C and P proteins translated from the cap-free T7 transcripts were limiting in those experiments (Fig. 5B, lanes 3 and 4). To exclude this possibility, C and P proteins were supplemented by cotransfection of a helper, pCH3142. Nevertheless, the T7 transcripts were not encapsidated (Fig. 5B, lanes 5, 6, 7, and 8). In addition, it appears that the coinfecting vaccinia virus did not substantially reduce encapsidation efficiency, since the encapsidation efficiency was similar regardless of vaccinia virus infection (Fig. 5B, lanes 9 to 12), although encapsidation efficiency was somewhat lower than that observed in Fig. 4. It should be noted that RNA was extracted 2 days after transfection in this experiment due to inevitable cytotoxicity associated with vaccinia virus infection, as opposed to extraction of RNA 3 days after transfection in all other experiments in this study.
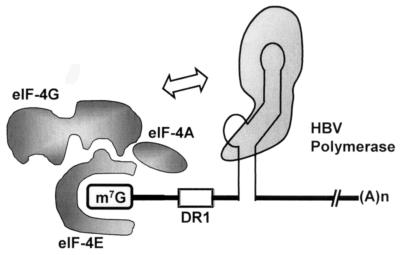
Taken together, our results clearly demonstrated that the cap-free HBV pgRNAs transcribed by T7 RNA polymerase were not encapsidated. Since the ɛ element was positioned 42 nt away from the 5′ end in the T7 transcripts, the RNA would be encapsidated otherwise. Thus, we concluded that the cap, as well as the 5′ ɛ element, is essential for the encapsidation. We hypothesized that for encapsidation to occur, HBV polymerase recognizes the 5′ cap structure or factors associated with it, such as eIF4E or eIF4G (Fig. 6).
FIG. 6.
Proposed model to account for the requirement of the 5′ cap structure for encapsidation. We hypothesized that HBV polymerase interacts with one of the cap-binding factors, such as eIF4F, during encapsidation. In the early phase of infection, the 40S ribosome subunit recognizes the pgRNA via the cap-binding factors that are associated with the 5′ cap structure. However, in the later phase of infection, HBV polymerase may compete with the 40S subunit of ribosome for the pgRNA. This competition can be envisioned as an autoregulation of C and P protein synthesis during the HBV infection cycle.
DISCUSSION
In this report, we examined the molecular nature of the selectivity observed in the encapsidation of the HBV pgRNA by use of a cotransfection system in which the lacZ-ɛ chimeric mRNA was packaged by C and P that were provided in trans by a helper plasmid as described previously (14, 15). We were interested in the mechanism that entails the position effect of the encapsidation of the pregenomic RNA. The data presented indicated that (i) the ɛ element is necessary and sufficient for the encapsidation of the pgRNA into viral core particles, (ii) the 5′ proximal ɛ element needs to be positioned within 65 nt from the 5′ cap to be encapsidated efficiently, and (iii) the 5′ cap structure is required for the encapsidation. Based on these results, we propose that HBV polymerase recognizes the cap structure itself or factors associated with it as well as the 5′ proximal ɛ element during encapsidation.
To determine the functional position of the ɛ element, we made a series of lacZ RNA expression plasmids encoding ɛ at various positions and analyzed whether the RNA is encapsidated into core particles in the transfected Huh7 cells. As shown in Fig. 3, only the lacZ RNA with the ɛ element positioned at the 5′ proximal site was encapsidated. The other six RNAs with the ɛ element positioned further downstream were not encapsidated into viral core particles (Fig. 3B). The results indicated that the 5′ proximity of the ɛ element is essential for the encapsidation. On the other hand, it was reported that one of the minor 3.5-kb RNAs, which has the ɛ element positioned 60 nt away from the 5′ end, is not packaged into the core particle (19). This RNA encodes an additional in-frame AUG codon upstream of the 5′ ɛ element (Fig. 1A). This AUG codon is used as an initiation codon for the pre-core protein synthesis. This minor 3.5-kb RNA would be encapsidated if the AUG codon is inactivated (19). Taken together, it appears that the distance limit of the functional ɛ element for encapsidation is somewhere between 65 and 91 nt apart from the 5′ end. This result is in good agreement with our data of Fig. 4. Interestingly, we noted a gradual decrease in encapsidation efficiency as the ɛ element was positioned further distal to the 5′ end (Fig. 4). These data led us to speculate that the cap structure itself is involved in the encapsidation process. Indeed, the cap-free RNAs made by T7 RNA polymerase were not encapsidated (Fig. 5). Thus, we concluded that the cap structure is essential for the encapsidation. We speculated that the cap structure itself or its associated factors are directly recognized by HBV DNA polymerase. In addition, a formal possibility that the lack of polyadenylation in the T7 transcripts prevents the transcripts from being encapsidated is not excluded.
In this study, we did not address whether sequence upstream of the 5′ ɛ element modulated the encapsidation efficiency. In fact, it was shown that deletion of these upstream sequences from the wild-type pgRNA resulted in a significant reduction in packaging efficiency (4, 21). It remains to be seen whether this reduction is mediated by specific sequences upstream of the ɛ element or by a spacing requirement between the 5′ end of the transcript and the ɛ element. We favor the latter possibility, since (i) the upstream sequences are present in all of the unencapsidated transcripts we tested in this study (Fig. 3 and 4), and (ii) a short linker-derived sequence present at the 5′ end of the transcript does not per se interfere with encapsidation (14, 15). In contrast, in a case of DHBV, in addition to the 5′ ɛ sequence, additional sequences near the middle of the pgRNA are required for the encapsidation of DHBV pgRNA (3, 12). It remains to be seen whether the 5′ cap is required for encapsidation of the pgRNA of avian hepadnavirus as well. It is unprecedented that viruses employed the host-modified structure as a subset of the encapsidation signal. Why has HBV evolved to use the 5′ cap structure as an encapsidation signal? Given that (i) all four HBV transcripts encode the 3′ ɛ element because they share a polyadenylation signal (10) and (ii) sequences surrounding the ɛ element are identical regardless of its location, and therefore it could not serve for encapsidation specificity, and (iii) based on the fact that the only sequence and/or structure that is near the 5′ ɛ element and unique to the pgRNA is the 5′ cap structure, we can deduce that HBVs have adopted the 5′ cap structure as a subset of the encapsidation signal during evolution.
The pgRNAs serve a dual function: as a template for the reverse transcription and as an mRNA for C and P protein synthesis. Consequently, the pgRNAs are subjected to either encapsidation or translation during productive viral infection. In other words, it is possible that the HBV polymerase competes with the 40S ribosomal subunit for pgRNAs for encapsidation. The notion was substantiated by the observation that the encapsidation of the 3.5-kb RNA is suppressed by translation: i.e., translational suppression of encapsidation (19). According to this scenario, the pgRNA with bound translation initiation factors could interact either with HBV polymerase for encapsidation or with the 40S ribosome subunit for translation. Apparently, HBV polymerase and the 40S subunit of ribosome might compete for the factor-bound pgRNA. In the early phase of infection, the pgRNAs are used solely for the translation of C and P proteins. As HBV polymerase accumulates at later phase, it interacts with the factor-bound pgRNA for encapsidation. Thus, the pgRNA is no longer available for the synthesis of C and P proteins. This regulation could be envisioned as autoregulation of C and P protein synthesis. Whether HBV polymerase recognizes the cap structure directly or in association with other host factors which recognize the cap structure, such as eIF4E or eIF4G (26), merits further investigation.
ACKNOWLEDGMENTS
We thank M. Nassal for providing the pCH9/3091 and pCH3142 plasmids and B. Moss and B.-Y. Ahn for sharing a vaccinia virus strain expressing T7 RNA polymerase, vvT7-3. We also thank T.-G. Lee and B.-L. Seong for critical reading of the manuscript.
This study was supported in part by research grants from the Korea Research Foundation and the Ministry of Public Health, Republic of Korea.
REFERENCES
- 1.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvert J, Summers J. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang P W, Jeng K S, Hu C P, Chang C M. Characterization of a cis element required for packaging and replication of the human hepatitis B virus. Virology. 1992;186:701–711. doi: 10.1016/0042-6822(92)90037-p. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Fallows D A, Goff S P. Hepadnaviruses: current models of RNA encapsidation and reverse transcription. Adv Virus Res. 1996;46:165–194. doi: 10.1016/s0065-3527(08)60072-x. [DOI] [PubMed] [Google Scholar]
- 7.Fallows D A, Goff S P. Mutations in the ɛ sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J Virol. 1995;69:3067–3073. doi: 10.1128/jvi.69.5.3067-3073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 10.Ganem D, Varmus H E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch R C, Loeb D D, Pollack J R, Ganem D. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J Virol. 1991;65:3309–3316. doi: 10.1128/jvi.65.6.3309-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes M J, Andrews D W. Creation of deletion, insertion and substitution mutations using a single pair of primers and PCR. BioTechniques. 1996;20:188. doi: 10.2144/96202bm06. , 192–196. [DOI] [PubMed] [Google Scholar]
- 14.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knaus T, Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber A, Sandig V, Strauss M. A mutant T7 phage promoter is specifically transcribed by T7-RNA polymerase in mammalian cells. Eur J Biochem. 1993;217:387–394. doi: 10.1111/j.1432-1033.1993.tb18257.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 18.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 19.Nassal M, Junker-Niepmann M, Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 20.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepatitis. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 21.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryu W-S, Bayer M, Taylor J. Assembly of hepatitis delta virus particles. J Virol. 1992;66:2310–2315. doi: 10.1128/jvi.66.4.2310-2315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlicht H J, Radziwill G, Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989;56:85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- 25.Seeger C, Maragos J. Identification and characterization of the woodchuck hepatitis virus origin of DNA replication. J Virol. 1990;64:16–23. doi: 10.1128/jvi.64.1.16-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonenberg N, Gingras A C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 27.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 28.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 29.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]