Abstract
Two monoclonal antibodies (MAbs) against the ORF2 protein of the SAR-55 strain of hepatitis E virus (HEV) were isolated by phage display from a cDNA library of chimpanzee (Pan troglodytes) γ1/κ antibody genes. Both MAbs, HEV#4 and HEV#31, bound to reduced, denatured open reading frame 2 (ORF2) protein in a Western blot, suggesting that they recognize linear epitopes. The affinities (equilibrium dissociation constants, Kd) for the SAR-55 ORF2 protein were 1.7 nM for HEV#4 and 5.4 nM for HEV#31. The two MAbs also reacted in an enzyme-linked immunosorbent assay with recombinant ORF2 protein from a heterologous HEV, the Meng strain. Each MAb blocked the subsequent binding of the other MAb to homologous ORF2 protein in indirect competition assays, suggesting that they recognize the same or overlapping epitopes. Radioimmunoprecipitation assays suggested that at least part of the linear epitope(s) recognized by the two MAbs is located between amino acids 578 and 607. MAbs were mixed with homologous HEV in vitro and then inoculated into rhesus monkeys (Macaca mulatta) to determine their neutralizing ability. Whereas all control animals developed hepatitis (elevated liver enzyme levels in serum) and seroconverted to HEV, those receiving an inoculum incubated with either HEV#4 or HEV#31 were not infected. Therefore, each MAb neutralized the SAR-55 strain of HEV in vitro.
Hepatitis E is an acute disease endemic in many countries throughout developing parts of the world, in particular on the continents of Africa and Asia, where epidemics have also occurred. The causative agent, hepatitis E virus (HEV), is transmitted via the fecal-oral route, predominantly through contaminated water (45). HEV is an RNA virus with a positive-sense genome approximately 7.5 kb in length. The genome contains three open reading frames (ORFs); ORF2 encodes the putative capsid protein (45). Hepatitis E has a low mortality rate in young adults (38). However, mortality rates can reach 20% in women in the third trimester of pregnancy (32, 58). Surprisingly, in industrialized countries such as the United States, where hepatitis E is not endemic, a significant proportion of healthy individuals within the general population are seropositive (over 20% in some areas; 39, 50). However, clinical hepatitis E is rare in these countries and usually is seen in individuals who acquired the infection during travel to a region in which HEV is endemic or epidemic.
It has been suggested that animals serve as reservoirs for HEV and that human infections may, in part, be zoonoses. There have been several reports of HEV-specific (anti-HEV) antibody in animals (1, 9, 28, 29). Furthermore, an HEV-like virus was recently isolated from naturally infected swine in the United States (40). Although four genotypes of HEV have been identified, to date, only one serotype of HEV has been recognized. Therefore, it may be possible to produce a broadly protective vaccine. Passively transferred anti-HEV serum significantly reduced virus shedding in feces and abrogated disease in nonhuman primates challenged with a high dose of HEV (55). Furthermore, a recombinant 55-kDa ORF2 protein of the SAR-55 strain that was used to vaccinate macaques against HEV elicited a humoral immune response that correlated with protection from hepatitis E, even when a high challenge dose of homologous or heterologous HEV was given intravenously (54, 55). These data suggest that immunoglobulin (Ig) preparations similar to those used for protection against hepatitis A would be efficacious against hepatitis E. Field studies in India performed with pools of normal serum Ig collected from regions where HEV is endemic did not show significant protection from HEV infection or disease (27, 31, 63). However, it is likely that the titer of HEV-specific antibodies in these preparations was too low to be effective. Therefore, pooled normal human Ig is unlikely to be useful as an immunoprophylactic reagent against HEV. As an alternative, an Ig preparation consisting of HEV-neutralizing monoclonal antibodies (MAbs) might protect against hepatitis E.
Phage display of antibody libraries has provided a powerful tool for the isolation of human MAbs to important viral pathogens (4, 7, 12, 16, 24, 51). Large repertoires of antibodies can be displayed on the surface of filamentous phage particles (e.g., M13), and antibodies with desired specificity can be isolated by panning on the antigens of interest (6, 13, 61).
In this report, we describe a cDNA library of γ1/κ antibody genes from lymphocytes derived from chimpanzee bone marrow. The chimpanzee previously had been experimentally infected with all five recognized hepatitis viruses (A to E). Two MAbs specific for the HEV capsid protein, encoded by ORF2, were isolated and characterized.
MATERIALS AND METHODS
Animals.
Chimpanzee 1441 had been experimentally infected with HEV strain SAR-55 (Pakistan). Bone marrow was aspirated from the iliac crest of this animal 3 years postinfection. Three weeks prior to the bone marrow aspiration, the animal had been boosted with the purified SAR-55 ORF2 protein expressed from baculovirus. Chimpanzee 5835 previously had been experimentally infected with swine HEV (Meng strain) and immunized twice with the Meng ORF2 protein. Plasma samples obtained prior to infection and after immunization served as negative and positive controls, respectively, for neutralization of HEV. Ten young adult rhesus monkeys obtained from a domestic breeding colony were used to study the neutralization of HEV. All of the animals were maintained in an approved facility under conditions that met or exceeded all requirements for animal use.
Construction of γ1/κ antibody phage library.
The bone marrow lymphocytes were purified on a Ficoll gradient and stored as a viable single-cell suspension in 10% dimethyl sulfoxide–10% fetal calf serum–RPMI 1640 medium (BioWhittaker) in liquid nitrogen. Total RNA was extracted from ∼108 bone marrow lymphocytes (RNA Isolation Kit; Stratagene), and mRNA was reverse transcribed into cDNA using an oligo(dT) primer (Gibco/BRL). The cDNAs were amplified by PCR using rTth DNA polymerase (Perkin-Elmer). Thirty cycles of 94°C for 15 s, 52°C for 50 s, and 68°C for 90 s were performed. Chimpanzee κ-chain genes were amplified using primers specific for the human κ-chain genes. Fd segments (variable and first constant domains) of the chimpanzee γ1-chain genes were amplified with nine family-specific human VH primers recognizing the 5′ end of the genes (3, 43) and a chimpanzee γ1-specific 3′ primer (5′-GCATGTACTAGTTGTGTCACAAGATTTGGG-3′) (54).
The amplified κ chains were cloned into the pComb3H phage display vector as described by Williamson et al. (60). The amplified γ1 chains were cloned into the pGEM-T cloning vector (Promega), transformed into Escherichia coli XL-1 Blue (Stratagene), and expanded into a volume of 2 liters by solid-phase amplification as described by Glamann et al. (25). The γ1–pGEM-T library was digested with XhoI and SpeI (Boehringer Mannheim) and ligated into the κ-chain pComb3H library, also digested with XhoI and SpeI. The ligated products were transformed into E. coli XL-1 Blue. Transformants were expanded into a volume of 2 liters by solid-phase amplification. The final library of 1.9 × 107 clones was stored in 12.5% glycerol–Luria-Bertani (LB) broth at −80°C until use.
Panning and ELISA reagents.
HEV ORF2 proteins (55 kDa) from a human HEV strain (Pakistani strain SAR-55) and a swine HEV strain (U.S. strain Meng) were expressed from baculovirus in insect cells and purified as described by Robinson et al. (46). The full-length ORF2 protein has a predicted molecular mass of 72 kDa and is 660 amino acids in length. However, in insect cells, the recombinant ORF2 protein is proteolytically processed to yield a major polypeptide of approximately 55 kDa which begins at amino acid (aa) 112 and ends at aa 607 (46, 56). In all panning and enzyme-linked immunosorbent assay (ELISA) experiments, recombinant ORF2 protein was diluted to 1.0 μg ml−1 in 50 mM sodium carbonate buffer (pH 9.6) and adsorbed to enzyme immunoassay-radioimmunoassay A/2 (ELISA) plates (Costar) overnight at 4°C. Fabs were detected with goat anti-human IgG heavy- and light-chain-specific antibody (Pierce). This was used to coat microtiter wells at a dilution of 1:1,000 in 50 mM sodium carbonate buffer (pH 9.6) as described above.
Library screening.
Screening of the combinatorial library was carried out as described by Barbas et al. (3) and Williamson et al. (60). Approximately 109 bacteria from the library stock were inoculated into LB broth (Gibco/BRL) supplemented with ampicillin at 100 μg ml−1 and 1% (vol/vol) glucose (Sigma), amplified, and then infected with helper phage VCS M13 (Stratagene) at a multiplicity of infection of 50 to produce the library displayed on the surfaces of phage particles. Phage were panned on SAR-55 ORF2-coated ELISA wells; in all, four rounds of panning were performed. After amplification of the selected library, the phagemid DNA was extracted and the vector was modified by restriction enzyme digestion to remove the bacteriophage coat protein III-encoding region of the phage (4). The phagemid DNAs were religated and transformed into E. coli XL-1 Blue to produce soluble Fabs. Colonies were inoculated into LB broth in individual wells of a microtiter plate and incubated at 30°C overnight. Fab production was induced as described by Glamann et al. (25), and the bacterial supernatants were tested by ELISA for reactivity with HEV ORF2 and for the presence of Fab.
Fab production and purification.
Fab purification was facilitated by modification of the vector pComb3H to encode a six-histidine tail at the end of the soluble Fab fragment (25). Bacterial culture and Fab fragment purification were carried out as described by Glamann et al. (25). Protein concentrations were determined both by dye binding assay (Bio-Rad) and by spectroscopy at A280 (using the extinction coefficient of 1.4 optical density [OD] units is equivalent to 1.0 mg ml−1). Fab purity was determined by polyacrylamide gel electrophoresis (PAGE), followed by colloidal Coomassie brilliant blue staining (Sigma).
ELISA analysis of Fab reactivity and cross-reactivity.
Protein antigens were used to coat ELISA microtiter plates at 1.0 μg ml−1 (HEV ORF2 protein) or 10.0 μg ml−1 (thyroglobulin, lysozyme, or cytochrome c [Sigma]). Antigen-coated wells were blocked for 1 h at room temperature with 3% bovine serum albumin (BSA)-phosphate-buffered saline (PBS) and washed twice with PBS-Tween 20 (0.05% [vol/vol]), and 50 μl of crude or purified Fab was added to the wells. After 1 h of incubation at 37°C, the plates were washed six times with PBS-Tween 20. Bound Fab was detected with a 1:1,500 dilution of a goat anti-human F(ab′)2 alkaline phosphatase-labeled secondary antibody (Pierce). The assay color was developed using p-nitrophenyl phosphate (Sigma) at 1 mg ml−1 in diethanolamine buffer (Pierce). OD was determined at 405 nm with a reference wavelength of 650 nm.
Nucleic acid analysis of HEV-specific Fab clones.
Nucleic acid sequencing was performed with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit by using Ampli-Taq DNA Polymerase (Perkin-Elmer) and the following sequencing primers: heavy chain, 5′-ATTGCCTACGGCAGCCGCTGG-3′ (HC1) and 5′-GGAAGTAGTCCTTGACCAGGC-3′ (HC4); κ chain, 5′-ACAGCTATCGCGATTGCAGTG-3′ (LC1) and 5′-CACCTGATCCTCAGATGGCGG-3′ (LC4) (25). Sequences were analyzed with the GeneWorks (Oxford Molecular Group) software package. Sequence similarity searches were performed with the V-BASE program, which is a compilation of all of the available human variable-segment Ig germ line sequences (10). For BstNI (New England Biologicals) fingerprinting, 1 μg of plasmid DNA was digested with 1 U of enzyme overnight at 65°C. The restriction digests were analyzed on a 3% agarose gel.
Western blotting.
The SAR-55 ORF2 protein was heated in 2× Laemmli buffer (33) and electrophoresed in a single-well 10% polyacrylamide gel (Novex). Electrophoretic transfer of the protein to a nitrocellulose membrane was carried out at 126 mA for 1 h at 4°C. The membrane was blocked for 30 min with 5% skim milk in PBS and then cut into strips prior to overnight incubation at 4°C with equal concentrations of purified HEV#4 or HEV#31 or a 1:100 dilution of chimpanzee 1441 serum. After six washes of 10 min each, an anti-human IgG (Fab-specific, alkaline phosphatase [Pierce]-labeled) secondary antibody was added at a dilution of 1:5,000 in 5% skim milk–PBS. After 1 h, the blots were washed and nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (BCIP) substrate (Pierce) was added.
Affinity determinations using BIAcore.
Gold-coated CM-5 sensor chips are coated with a carboxylated dextran polymer matrix to which the SAR-55 ORF2 protein was amine coupled (Pharmacia Biosensor). The carboxyl groups on the dextran surface were activated with 35 μl of a 50:50 (vol/vol) solution of N-hydroxysuccinimide and N-ethyl-N′-(3-diethylaminopropyl)carbodiimide. The SAR-55 ORF2 protein was diluted in 10 mM sodium acetate (pH 4.5) prior to coupling. The sensor surface was washed with HBS (10 mM HEPES [pH 7.5], 0.15 M NaCl, 3.4 mM EDTA, 0.05% Tween 20), and then the remaining active binding sites on the chip were blocked by the addition of ethanolamine hydrochloride.
Affinity measurements were made for three different coating concentrations of the SAR-55 ORF2 protein, one for each flow cell on the chip, with the fourth flow cell left uncoated and blocked as a control. The affinity measurements were initiated by passing HBS over the sensor surface for 100 s at 10 μl min−1, and then 50 μl of Fab was injected at the same flow rate. The kinetic analysis was performed twice. The first chip was coated with 37, 203, and 334 resonance units of the SAR-55 ORF2 protein. The second chip was coated with 363, 225, and 106 resonance units of the SAR-55 ORF2 protein. Serial dilutions of HEV#4 or HEV#31 were made in HBS buffer. Eight dilutions of each MAb were passed over the sensor surfaces; the dilutions ranged from 0.5 to 200 nM and from 1.0 to 400 nM for HEV#4 and HEV#31, respectively. Between MAb binding phases, the sensor surface was regenerated with a 1-min pulse of regeneration buffer (1 M NaCl, 50 mM NaOH). Regeneration of the ORF2 protein did not affect subsequent MAb binding.
Fab biotinylation and indirect competition ELISA.
Prior to biotinylation, purified HEV#4 or HEV#31 was dialyzed against PBS overnight at 4°C. Conjugation with biotin was carried out in accordance with the manufacturer's (Pierce) instructions. The biotinylated Fabs were titrated on ORF2-coated wells to determine a dilution that was subsaturating and gave an OD reading of approximately 1.0 at 405 nm. For the competition assay, threefold dilutions of unlabeled and nonpurified Fab were incubated in ORF2 protein-coated wells for 1 h at 37°C and then washed four times with PBS-Tween 20. A single dilution of biotinylated Fab was added to the wells, and they were incubated for 1 h at 37°C. After four washes with PBS-Tween 20, streptavidin-alkaline phosphatase (Pierce) was added at a 1:500 dilution and the wells were incubated for 1 h at 37°C. The color was developed as described above.
In vitro neutralization of HEV with in vivo monitoring in rhesus monkeys.
Ten rhesus monkeys that were anti-HEV antibody negative (<1:100) by a sensitive ELISA (56) were divided into five groups of two animals each. A 10% stool suspension of Pakistani HEV strain SAR-55 previously titrated for infectivity in monkeys (57) was diluted to contain 64 50% monkey infectious doses (MID50) per inoculum. Sixty-four MID50 was used because of previous experience with this dose in in vitro neutralization experiments (22, 23). This inoculum was incubated with either purified Fab or chimpanzee plasma. HEV#4, HEV#31, or the irrelevant Fab HBV#8 (D. J. Schofield et al., unpublished data) was diluted to 1.9 mg ml−1 in 10% BSA–PBS. The concentration of Fabs used was the highest attainable for all three MAbs (HEV#4, HEV#31, and HBV#8). Monovalent Fab fragments have been shown to be up to 1,000-fold less efficient at neutralization than their parent whole IgG (reviewed in references 14 and 15). Therefore, we tested for neutralization with the highest possible concentration of Fab. Chimpanzee 5835 preimmune and hyperimmune plasma samples were heat inactivated (56°C for 30 min) prior to being diluted to 10% solutions in 10% BSA–PBS. The virus and antibody were mixed and incubated for 1 h at room temperature and then overnight at 4°C. The inoculum was diluted with 1 ml of ice-cold PBS prior to intravenous inoculation. Serum samples were collected prior to inoculation and for 20 weeks thereafter. Sera were assayed for levels of alanine aminotransferase (ALT) with a commercially available test (Metpath Inc.). The anti-HEV ELISA was performed as described elsewhere (56). Seroconversion to HEV was used as the criterion for infection.
Construction of clones expressing truncated SAR-55 ORF2 proteins.
SAR-55 ORF2 genes truncated at the 3′ terminus were made by PCR amplification of portions of the SAR-55 ORF2-encoding gene of pHEVORF2 63.2 (56). Truncated genes were amplified with a common 5′ primer (5′ SAR-55 aa112 [5′-ATGGCGGTCGCTCCGGCCCATGACACCC-3′]) and one of 3′ SAR-55 aa308 (5′-CTATTAGCGGAACTCAAGTTCGAGGGCAAAGTC-3′), 3′ SAR-55 aa408 (5′-CTATTAAGTCGGCTCGCCATTGGCTGAGACGAC-3′), 3′ SAR-55 aa508 (5′-CTATTACTGCGCGCCGGTCGCAACATTAACCAA-3′), 3′ SAR-55 aa578 (5′-CTATTACCGATGCCCAGCGGCATTCTCAACG-3′), or 3′ SAR-55 aa607 (5′-CTATTATAGCACAGAGTGGGGGGCTAAAACA-3′). Amino acids 112 through 607 comprise the 55-kDa protein used previously in vaccination studies (54, 55). Amino acids 112 through 578 represent a recombinant 53-kDa protein which readily formed virus-like particles (R. H. Purcell, unpublished data). The 5′ SAR-55 aa112 primer was phosphorylated with T4 polynucleotide kinase prior to PCR amplification, and the products were cloned into mammalian expression vector pCR3.1 (Unidirectional TA Cloning Kit; Invitrogen).
Radioimmunoprecipitation of 35S-labeled ORF2 proteins.
In vitro transcription and translation of truncated SAR-55 ORF2 clones aa308 to aa607 were carried out in accordance with the manufacturer's (Promega) protocol (T7 TNT in vitro transcription/translation) using [35S]methionine (Redivue; Amersham) as the radiolabeled. The five truncated ORF2 products were visualized by 10 to 20% PAGE, followed by autoradiography, and then pooled. Five microliters of pooled products was mixed with 1 μl of antibody and 5 μl of 2× native RIPA buffer (0.5 M NaCl, 5% glycerol, 0.2 M Tris-HCl [pH 8.0], 1.0% Tween 20, 2 mM EDTA) and incubated with rocking overnight at 4°C. For chimpanzee 1441 pre- or postimmune serum, precipitations were performed with the addition of recombinant protein G-coupled agarose beads (Gibco) and incubation with rocking on ice for 1 h. For Fab HEV#4, HEV#31, or HBV#8, 1 μl of goat anti-human IgG [F(ab′)2]-specific antibody was used in addition to protein G-coupled agarose beads. The beads were pelleted and then washed three times in 1× RIPA buffer and once with distilled, deionized H2O. Samples were then resuspended in 15 μl of 2× Laemmli buffer and incubated for 10 min at 95°C prior to 10 to 20% PAGE (Novex). After 1 h at 126 V, the gel was fixed in a solution of 10% acetone and 10% methanol for 20 min, washed twice in distilled, deionized H2O, and then incubated in Amplify solution (Amersham) for 20 min. After drying, the gel was exposed to X-ray film at −70°C.
RESULTS
Isolation of HEV ORF2-specific Fabs.
Total RNA was extracted from bone marrow lymphocytes obtained from chimpanzee 1441. mRNA was reverse transcribed using an oligo(dT) primer to generate cDNA. Amplification of the cDNA was carried out by PCR using both κ-chain and γ1-chain primers specific for the human antibody genes (see Materials and Methods for details). The amplified κ- and γ1-chain genes were purified and cloned into a phage display vector. The resultant Fab-phage library was then selected against the baculovirus-expressed SAR-55 ORF2 protein. The enriched library was expressed as soluble Fabs in E. coli following restriction enzyme digestion of the phage display vector. The specificity of the Fabs was determined in an ELISA with the SAR-55 ORF2 protein and a panel of unrelated protein antigens (data not shown). A total of 144 clones were screened, of which 7 were SAR-55 ORF2 specific.
BstNI restriction digest analysis.
The restriction enzyme BstNI cuts frequently in the human γ1 heavy chain (37). BstNI restriction digestion was used to screen for different heavy-chain sequences among the Fab clones. Two distinct BstNI restriction patterns representing five and two clones each were observed (Fig. 1a).
FIG. 1.

(a) Restriction fragment analysis of seven HEV-specific Fab clones. Plasmid DNA was digested with BstNI, and an aliquot was electrophoresed in a 3% agarose gel. (b) Sequence data for the γ1 chains of HEV ORF2-specific MAbs. FR, framework region.
Sequence analysis of HEV ORF2 protein-specific Fabs.
Sequence analysis of the seven Fab clones confirmed the results of the BstNI digestion. There were two distinct γ1 heavy chains; one was represented by HEV#4-like clones, and the other was represented by HEV#31-like clones. The two γ1 chains varied markedly in all three complementarity-determining regions (CDR; Fig. 1b). The κ light-chain sequences were also divergent (data not shown).
We attempted to determine the specific germ line origin of the two MAbs by conducting a sequence similarity search of all of the known human Ig genes. The two γ1 heavy-chain sequences exhibited the most identity with the human VH3 family of germ line segments. HEV#4 was most closely related to the DA-8 (11) VH gene segment, with 89.4% overall identity and 92% identity excluding CDR1 and CDR2. HEV#31 was most closely related to the DP-47 (52) VH gene segment, with 88.5% overall identity and 91.7% identity excluding CDR1 and CDR2. No significant similarity was found among the human D segments for either HEV#4 or HEV#31. Both the HEV#4 and HEV#31 JH segments exhibited the most identity with the human JH4b family of JH gene segments. The κ light-chain sequences exhibited the most identity with the human Vκ1 family of germ line segments. The HEV#4 Jκ segment exhibited the most identity to the human Jκ1 family, while the HEV#31 Jκ segment exhibited the most identity to the human Jκ4 family.
In a subsequent panning experiment using HEV#4 and HEV#31 to mask the epitopes that they recognize on the HEV ORF2 protein, 15 more unique MAbs were generated that represented four of the other major human VH gene families (VH1, VH4, VH5, and VH6; Schofield et al., unpublished). These are being characterized further.
Affinity determination using a biosensor.
The affinities of the two MAbs for ORF2 were determined using BIAcore. Association and dissociation kinetics were measured for the binding of both HEV#4 and HEV#31 to the SAR-55 ORF2 protein (Table 1). Both MAbs had high equilibrium dissociation constants (Kd, 1.7 nM for HEV#4 and 4.5 nM for HEV#31).
TABLE 1.
Affinity data for MAbs HEV#4 and HEV#31 determined by the BIAcore system
Kd, equilibrium dissociation constant.
ka, association rate constant.
kd, dissociation rate constant.
Western blotting with HEV-specific MAbs.
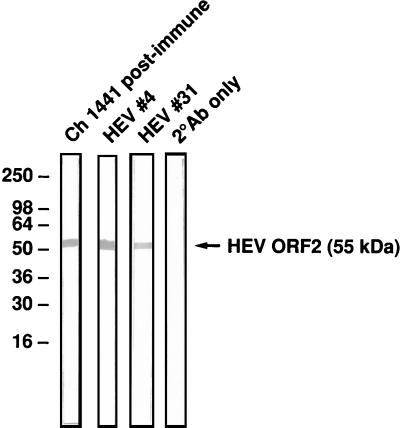
A Western blotting assay was performed to determine the nature of the epitope recognized by each of the two MAbs (i.e., linear or conformational epitope). HEV#4 and HEV#31 both recognized the reduced, denatured SAR-55 ORF2 protein (Fig. 2), suggesting that they are both directed to linear epitopes on the virus capsid.
FIG. 2.
Western blot assay of SAR-55 ORF2 protein (55 kDa) with chimpanzee (Ch) 1441 postimmune serum, HEV#4, and HEV#31. Molecular size markers (kilodaltons) are indicated. 2° Ab, secondary antibody.
Indirect competition assay to map the relative binding sites of HEV#4 and HEV#31.
An indirect competition assay was performed to determine whether the two MAbs recognize similar or overlapping epitopes on the HEV capsid. Unlabeled HEV#4 blocked the binding of biotinylated HEV#4 or HEV#31 to the SAR-55 ORF2 protein to similar extents (63 and 66% inhibition of binding). Similarly, unlabeled HEV#31 blocked the binding of biotinylated HEV#31 (81%) or HEV#4 (72%) to the SAR-55 ORF2 protein. Therefore, HEV#4 and HEV#31 probably recognized the same or overlapping epitopes on the SAR-55 ORF2 protein.
Epitope mapping.
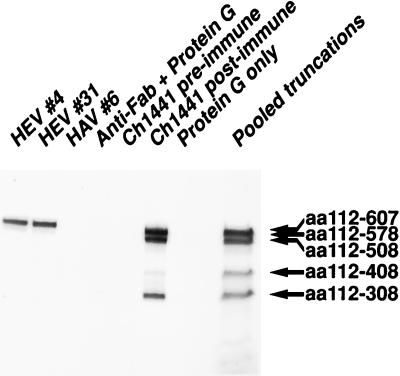
Purified HEV#4 and HEV#31 were incubated at 4°C overnight with a pool of five different 35S-labeled ORF2 translation products. Each MAb precipitated SAR-55 aa607 (Fig. 3), corresponding to the 55-kDa panning antigen. However, the shorter polypeptides were not precipitated to any significant degree by either MAb. HAV#6, an HAV-specific MAb (Schofield et al., unpublished), did not precipitate any of the ORF2 truncations, nor did the secondary antibody alone or protein G alone (Fig. 3). Chimpanzee 1441 immune serum precipitated SAR-55 aa308 to SAR-55 aa607, while the preimmune serum did not react with any polypeptide. These data were confirmed by immunoprecipitation assays with each polypeptide individually (data not shown).
FIG. 3.
Radioimmune precipitation of C-terminally truncated SAR-55 ORF2 proteins. An autoradiograph of a radioimmunoprecipitation assay using pooled 35S-labeled truncations is shown. Pooled truncations served as size markers for the immunoprecipitated products. Ch, chimpanzee.
Cross-reactivity of Fabs with another HEV strain.
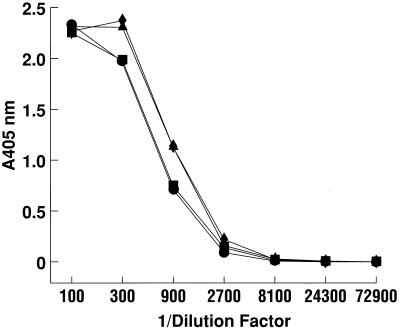
Only one serotype of HEV has been identified. However, nucleotide and amino acid sequences of known strains differ enough to justify classifying the strains into four genotypes. Two of the most divergent strains of HEV are Pakistan (SAR-55) and swine (Meng) HEV. The two Fabs were tested by ELISA for cross-reactivity with the Meng ORF2 protein. Titration curves for HEV#4 and HEV#31 were identical for the heterologous Meng and homologous SAR-55 ORF2 proteins (Fig. 4).
FIG. 4.
Titration by ELISA of HEV#4 and HEV#31 reactivity with recombinant baculovirus-expressed ORF2 protein from the Pakistani (SAR-55) and swine (Meng) HEV strains. Symbols: ■, HEV#4 with SAR-55 ORF2 protein; ●, HEV#4 with Meng ORF2 protein; ▴, HEV#31 with SAR-55 ORF2 protein; ⧫, HEV#31 with Meng ORF2 protein.
Neutralization of HEV.
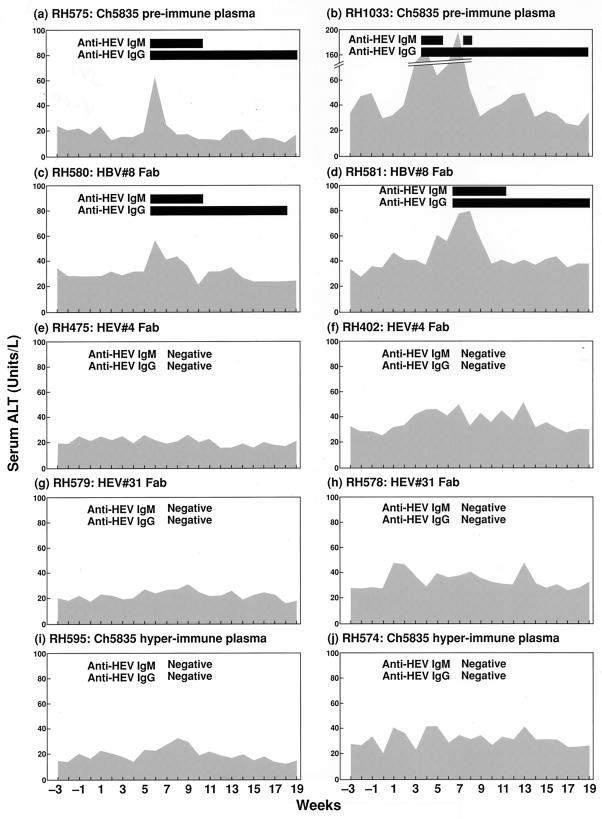
Sixty-four MID50 of the SAR-55 strain of HEV was incubated with HEV#4, HEV#31, or irrelevant Fab HBV#8 (Schofield et al., unpublished) at 1.9 mg ml−1 or with a 10% solution of either chimpanzee 5835 pre- or hyperimmune plasma prior to inoculation into rhesus monkeys. Inoculations were performed in duplicate. After intravenous inoculation, the monkeys were monitored for 20 weeks for biochemical evidence of hepatitis (serum ALT) and for seroconversion to HEV ORF2 antigen by ELISA. All of the monkeys that received HEV incubated with either chimpanzee 5835 preimmune plasma or HBV#8 developed hepatitis E, as evidenced by a rise in ALT levels and seroconversion to HEV ORF2 protein (Fig. 5a and d). In contrast, all of the monkeys receiving HEV incubated with HEV#4, HEV#31 or chimpanzee 5835 hyperimmune plasma were not infected with HEV (Fig. 5e to j).
FIG. 5.
Monitoring of rhesus monkeys inoculated with HEV incubated with chimpanzee 5835 preimmune plasma (a and b), HBV#8 Fab (c and d), HEV#4 Fab (e and f), HEV#31 Fab (g and h), and chimpanzee 5835 hyperimmune plasma (i and j). The cutoff in the anti-HEV ELISA was a titer of <1:100.
DISCUSSION
Antibodies to a wide range of viral pathogens have been isolated using combinatorial antibody libraries displayed on the surface of filamentous phage particles. In general, human donors naturally infected with specific viral pathogens have been used as the source of bone marrow cells or peripheral blood lymphocytes for the construction of these libraries. Although some “naive” libraries have been constructed from cells collected from uninfected donors (37), few antiviral antibodies have been produced from these naive libraries to date.
The advantages of using a chimpanzee as a donor for repertoire cloning are twofold. First, the chimpanzee can be infected experimentally by many of the important human viral pathogens, e.g., human immunodeficiency virus type 1, hepatitis C virus, hepatitis B virus, and respiratory syncytial virus. Second, the chimpanzee is the primate most closely related to humans. Therefore, chimpanzee antibodies could, theoretically, be used directly in the immune prophylactic treatment of diseases. A number of publications have indirectly addressed the possibility of using primate reagents in human prophylaxis and therapy by introducing human immune components into primates (18–21, 35, 41, 44). These data show that little immunogenicity is seen when human immune components are introduced into chimpanzees, compared to other primates. Human antibodies are recognized as self by the chimpanzee immune system. In a chimpanzee, the half-life of a human MAb was equivalent to the estimated half-life of IgG in humans (41). Hence, one would expect chimpanzee antibodies not to be immunogenic in humans and possibly to be useful in human immunotherapy without modification (humanization).
The cDNA library described in this report is a potential repository for antibodies to the five recognized human hepatitis viruses A to E, since the donor chimpanzee had been experimentally infected with each of these. In this first analysis of the library, two MAbs directed to the ORF2 protein of HEV were identified. The γ1 heavy chains had high degrees of identity (89.4% for HEV#4 and 88.5% for HEV#31) at the nucleotide level with two different γ1 heavy chains from the human VH3 gene family. The degree of identity between the chimpanzee and human γ-chain genes was similar to that of the only other chimpanzee antibody characterized to date, an anti-HIV gp160 MAb having 92% identity with its nearest human germ line equivalent (59).
Both HEV#4 and HEV#31 neutralized HEV. Neutralization of the SAR-55 strain of HEV by MAbs HEV#4 and HEV#31 was determined by intravenous challenge of rhesus monkeys with 64 MID50 of HEV after incubation of the virus with either of the two MAbs. None of the animals receiving HEV incubated with either HEV#4 or HEV#31 or with hyperimmune plasma from chimpanzee 5835 had biochemical or serological evidence of HEV infection. In contrast, all control animals inoculated with an irrelevant MAb or preimmune plasma were infected with HEV since they seroconverted to HEV and also had mild ALT elevations that were characteristic of infections with a small challenge dose (57). Since the Fabs are monovalent, lack of infectivity was not due to the aggregation of virus particles. Neutralization of HEV was caused by the binding of the MAbs alone, since Fab fragments lack an Fc region, and thus would not be able to neutralize the virus by an Fc-mediated function, such as antibody-dependent cell-mediated cytotoxicity.
The hyperimmune plasma from chimpanzee 5835 had been generated by infection with the swine HEV (Meng strain) and subsequent immunization with the Meng ORF2 protein. The fact that this plasma neutralized SAR-55 emphasizes that highly divergent strains comprise a single serotype of HEV.
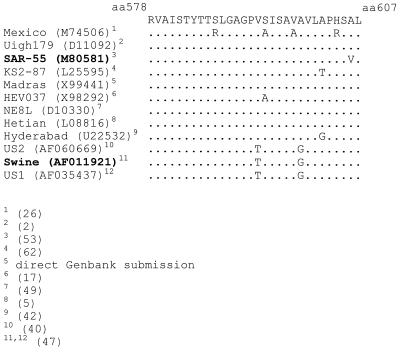
HEV#4 and HEV#31 had high affinities for the SAR-55 ORF2 protein, with Kd values in the nanomolar range. These values were comparable to Kd values determined for neutralizing Fabs to other viruses, e.g., influenza A virus (48), human immunodeficiency virus type 1 (8), and murine hepatitis virus (34). In a Western blot, both HEV#4 and HEV#31 recognized reduced, denatured ORF2, suggesting that they are directed to linear rather than to conformational epitopes on the ORF2 protein. Indirect competition assays suggested that HEV#4 and HEV#31 recognize spatially related or overlapping epitopes on the ORF2 protein since each Fab inhibited the other from binding. The approximate location of the epitope was determined by radioimmunoprecipitation studies with a set of five C-terminally truncated SAR-55 ORF2 proteins. HEV#4 and HEV#31 precipitated the polypeptide corresponding to aa 112 to 607 (55-kDa protein) but did not precipitate those terminating at or prior to aa 578. Therefore, at least part of the epitope(s) lies between aa 578 and 607 on the ORF2 protein in the antigenic region designated no. 6 by Khudyakov et al. (30). It is unclear whether the entire linear epitope(s) is encompassed by the region between aa 578 and 607 or whether part of the epitope extends N terminal of aa 578. In the latter situation, the epitope would be disrupted in the aa 578 construct, thus abrogating binding by either MAb. Interestingly, the chimpanzee serum taken at the same time as the bone marrow aspirate precipitated all five truncated ORF2 polypeptides. This suggests that antibodies to other epitopes on the ORF2 protein are present in the antibody library, although they were not isolated in this panning. The reactivity of the two MAbs with recombinant ORF2 protein from a highly divergent heterologous HEV, the swine-derived Meng strain, was determined by ELISA. The two MAbs had similar titration curves with the SAR-55 and Meng ORF2 proteins. Since the region containing this epitope(s) is relatively well conserved (Fig. 6), it is conceivable that HEV#4 and HEV#31 would neutralize most or many of the different HEV isolates. Currently, we are trying to clone ORF2 from the Mexico strain to determine if the epitope(s) recognized by the two MAbs is conserved in the strain that is most divergent in this region of ORF2.
FIG. 6.
Comparison of the sequences of aa 578 to 607 in the ORF2 proteins of different HEV strains. HEV#4 and HEV#31 reacted by ELISA with the two strains in bold type; the remaining strains have not been tested. Reference numbers are in parentheses at the bottom.
No vaccine is available for the prevention of hepatitis E. Therefore, a need exists for anti-HEV Igs for protection of individuals at high risk from HEV infection in areas of the world where HEV is endemic and where epidemics occur. Since normal immune globulin is expensive and of dubious therapeutic value in this instance, economically viable and renewable sources of potent anti-HEV IgGs would be very beneficial. Production of antibodies generated from stably transfected cell lines is prohibitively expensive for developing countries, where these antibodies would be most useful. However, new techniques, such as the expression of whole IgG molecules in plants (36), may make these antibodies cheaper to produce.
In summary, these are the first primate (chimpanzee or human) MAbs generated to the HEV ORF2 protein. These MAbs prevented infection when virus-antibody mixtures were intravenously inoculated into rhesus monkeys. Competition studies suggest that the two Fabs recognize the same epitope or overlapping epitopes on the ORF2 protein. At least part of the neutralization epitope demonstrated in this study lies between aa 578 and 607 of the capsid protein.
ACKNOWLEDGMENTS
We thank Max Shapiro and other members of Bioqual (Rockville, Md.) for providing animal care. We thank Lynn Rasmussen for nucleotide sequencing, Maxine Medaglia for work with the BIAcore system, and Ron Engle for performing the serological assays.
REFERENCES
- 1.Arankalle V A, Goverdhan M K, Banerjee K. Antibodies against hepatitis E virus in Old World monkeys. J Viral Hepatitis. 1994;1:125–129. doi: 10.1111/j.1365-2893.1994.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 2.Aye T T, Uchida T, Ma X Z, Iida F, Shikata T, Zhuang H, Win K M. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986–1988) of China. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas C F, III, Kang A S, Lerner R A, Benkovic S J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender E, Woof J M, Atkin J D, Barker M D, Bebbington C R, Burton D R. Recombinant human antibodies: linkage of an Fab fragment from a combinatorial library to an Fc fragment for expression in mammalian cell culture. Hum Antibodies Hybridomas. 1993;4:74–79. [PubMed] [Google Scholar]
- 5.Bi S L, Purdy M A, McCaustland K A, Margolis H S, Bradley D W. The sequence of hepatitis E virus isolated directly from a single source during an outbreak in China. Virus Res. 1993;28:233–247. doi: 10.1016/0168-1702(93)90024-h. . (Erratum, 33(i):98, 1994.) [DOI] [PubMed] [Google Scholar]
- 6.Burton D R, Barbas C F., III Human antibodies from combinatorial libraries. Adv Immunol. 1994;57:191–280. doi: 10.1016/s0065-2776(08)60674-4. [DOI] [PubMed] [Google Scholar]
- 7.Burton D R, Barbas III C F, Persson M A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton D R, Pyati J, Koduri R, Sharp S J, Thornton G B, Parren P W, Sawyer L S, Hendry R M, Dunlop N, Nara P L, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 9.Clayson E T, Innis B L, Myint K S, Narupiti S, Vaughn D W, Giri S, Ranabhat P, Shrestha M P. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am J Trop Med Hyg. 1995;53:228–232. doi: 10.4269/ajtmh.1995.53.228. [DOI] [PubMed] [Google Scholar]
- 10.Cook G P, Tomlinson I M. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237–242. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- 11.Cook G P, Tomlinson I M, Walter G, Riethman H, Carter N P, Buluwela L, Winter G, Rabbitts T H. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat Genet. 1994;7:162–168. doi: 10.1038/ng0694-162. [DOI] [PubMed] [Google Scholar]
- 12.Crowe J E, Jr, Murphy B R, Chanock R M, Williamson R A, Barbas C F, 3rd, Burton D R. Recombinant human respiratory syncytial virus (RSV) monoclonal antibody Fab is effective therapeutically when introduced directly into the lungs of RSV-infected mice. Proc Natl Acad Sci USA. 1994;91:1386–1390. doi: 10.1073/pnas.91.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Kruif J, van der Vuurst de Vries A R, Cilenti L, Boel E, van Ewijk W, Logtenberg T. New perspectives on recombinant human antibodies. Immunol Today. 1996;17:453–455. doi: 10.1016/0167-5699(96)30057-y. [DOI] [PubMed] [Google Scholar]
- 14.Dimmock N J. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 15.Dimmock N J. Update on the neutralisation of animal viruses. Rev Med Virol. 1995;5:165–179. [Google Scholar]
- 16.Ditzel H J, Parren P W, Binley J M, Sodroski J, Moore J P, Barbas C F, 3rd, Burton D R. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J Mol Biol. 1997;267:684–695. doi: 10.1006/jmbi.1997.0912. [DOI] [PubMed] [Google Scholar]
- 17.Donati M C, Fagan E A, Harrison T J. Sequence analysis of full length HEV clones derived directly from human liver in fulminant hepatitis E. In: Rizzetto M, Purcell R H, Gerin J L, Verme G, editors. Viral hepatitis and liver disease. Turin, Italy: Edizioni Minervva Medica; 1997. pp. 313–316. [Google Scholar]
- 18.Ehrlich P H, Harfeldt K E, Justice J C, Moustafa Z A, Ostberg L. Rhesus monkey responses to multiple injections of human monoclonal antibodies. Hybridoma. 1987;6:151–160. doi: 10.1089/hyb.1987.6.151. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich P H, Moustafa Z A, Harfeldt K E, Isaacson C, Ostberg L. Potential of primate monoclonal antibodies to substitute for human antibodies: nucleotide sequence of chimpanzee Fab fragments. Hum Antibodies Hybridomas. 1990;1:23–26. [PubMed] [Google Scholar]
- 20.Ehrlich P H, Moustafa Z A, Justice J C, Harfeldt K E, Gadi I K, Sciorra L J, Uhl F P, Isaacson C, Ostberg L. Human and primate monoclonal antibodies for in vivo therapy. Clin Chem. 1988;34:1681–1688. [PubMed] [Google Scholar]
- 21.Ehrlich P H, Moustafa Z A, Justice J C, Harfeldt K E, Ostberg L. Further characterization of the fate of human monoclonal antibodies in rhesus monkeys. Hybridoma. 1988;7:385–395. doi: 10.1089/hyb.1988.7.385. [DOI] [PubMed] [Google Scholar]
- 22.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geoffroy F, Sodoyer R, Aujame L. A new phage display system to construct multicombinatorial libraries of very large antibody repertoires. Gene. 1994;151:109–1130. doi: 10.1016/0378-1119(94)90639-4. [DOI] [PubMed] [Google Scholar]
- 25.Glamann J, Burton D R, Parren P W H I, Ditzel H J, Kent K A, Arnold C, Montefiori D, Hirsch V M. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J Virol. 1998;72:585–592. doi: 10.1128/jvi.72.1.585-592.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 27.Joshi Y K, Babu S, Sarin S, Tandon B N, Gandhi B M, Chaturvedi V C. Immunoprophylaxis of epidemic non-A non-B hepatitis. Indian J Med Res. 1985;81:18–19. [PubMed] [Google Scholar]
- 28.Kabrane-Lazizi Y, Meng X-J, Purcell R H, Emerson S U. Evidence that the genomic RNA of hepatitis E virus is capped. J Virol. 1999;73:8848–8850. doi: 10.1128/jvi.73.10.8848-8850.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karetnyi V, Dzhumalieva D I, Usmanov R K, Titova I P, Litvak Ia I, Balaian M S. The possible involvement of rodents in the spread of viral hepatitis E. Zh Mikrobiol Epidemiol Immunobiol. 1993;4:52–56. [PubMed] [Google Scholar]
- 30.Khudyakov Y E, Lopareva E N, Jue D L, Crews T K, Thyagarajan S P, Fields H A. Antigenic domains of the open reading frame 2-encoded protein of hepatitis E virus. J Clin Microbiol. 1999;37:2863–2871. doi: 10.1128/jcm.37.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khuroo M S, Dar M Y. Hepatitis E: evidence for person-to-person transmission and inability of low dose immune serum globulin from an Indian source to prevent it. Indian J Gastroenterol. 1992;11:113–116. [PubMed] [Google Scholar]
- 32.Khuroo M S, Teli M R, Skidmore S, Sofi M A, Khuroo M I. Incidence and severity of viral hepatitis in pregnancy. Am J Med. 1981;70:252–255. doi: 10.1016/0002-9343(81)90758-0. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Lamarre A, Talbot P J. Protection from lethal coronavirus infection by immunoglobulin fragments. J Immunol. 1995;154:3975–3984. [PubMed] [Google Scholar]
- 35.Logdberg L, Kaplan E, Drelich M, Harfeldt E, Gunn H, Ehrlich P, Dottavio D, Lake P, Ostberg L. Primate antibodies to components of the human immune system. J Med Primatol. 1994;23:285–297. doi: 10.1111/j.1600-0684.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 36.Ma J K, Hikmat B Y, Wycoff K, Vine N D, Chargelegue D, Yu L, Hein M B, Lehner T. Characterization of a recombinant plant monoclonal secretory antibody and preventive immunotherapy in humans. Nat Med. 1998;4:601–606. doi: 10.1038/nm0598-601. [DOI] [PubMed] [Google Scholar]
- 37.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 38.Mast E E, Alter M J. Epidemiology of viral hepatitis: an overview. Semin Virol. 1993;4:273–283. [Google Scholar]
- 39.Mast E E, Kuramoto P O, Favorov M O, Schoening V R, Burkholder B T, Shapiro C N, Holland P V. Prevelance of and risk factors for antibody to hepatitis E virus seroreactivity among blood donors in northern California. J Infect Dis. 1997;176:34–40. doi: 10.1086/514037. [DOI] [PubMed] [Google Scholar]
- 40.Meng X J, Purcell R H, Halbur P G, Lehman J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogata N, Ostberg L, Ehrlich P H, Wong D C, Miller R H, Purcell R H. Markedly prolonged incubation period of hepatitis B in a chimpanzee passively immunized with a human monoclonal antibody to the a determinant of hepatitis B surface antigen. Proc Natl Acad Sci USA. 1993;90:3014–3018. doi: 10.1073/pnas.90.7.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panda S K, Nanda S K, Zafrullah M, Ansari I-H, Ozdener M H, Jameel S. An Indian strain of hepatitis E virus (HEV): cloning, sequence, and expression of structural region and antibody responses in sera from individuals from an area of high-level HEV endemicity. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persson M A, Caothien R H, Burton D R. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc Natl Acad Sci USA. 1991;88:2432–2436. doi: 10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prince A M, Horowitz B, Baker L, Shulman R W, Ralph H, Valinsky J, Cundell A, Brotman B, Boehle W, Rey F, et al. Failure of a human immunodeficiency virus (HIV) immune globulin to protect chimpanzees against experimental challenge with HIV. Proc Natl Acad Sci USA. 1988;85:6944–6948. doi: 10.1073/pnas.85.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell R H. Hepatitis E virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippinscott-Raven; 1996. pp. 2831–2843. [Google Scholar]
- 46.Robinson R A, Burgess W H, Emerson S U, Leibowitz R S, Sosnovtseva S A, Tsarev S, Purcell R H. Structural characterization of recombinant hepatitis E virus ORF2 proteins in baculovirus-infected insect cells. Protein Expr Purif. 1998;12:75–84. doi: 10.1006/prep.1997.0817. [DOI] [PubMed] [Google Scholar]
- 47.Schlauder G G, Dawson G J, Erker J C, Kwo P Y, Knigge M F, Smalley D L, Rosenblatt J E, Desai S M, Mushahwar I K. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447–56. doi: 10.1099/0022-1317-79-3-447. . (Erratum, 79:2563, 1998.) [DOI] [PubMed] [Google Scholar]
- 48.Schofield D J, Dimmock N J. Determination of affinities of a panel of IgGs and Fabs for whole enveloped (influenza A) virions using surface plasmon resonance. J Virol Methods. 1996;62:33–42. doi: 10.1016/0166-0934(96)02086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas D L, Yarbough P O, Vlahov D, Tsarev S A, Nelson K E, Saah A J, Purcell R H. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J Clin Microbiol. 1997;35:1244–1247. doi: 10.1128/jcm.35.5.1244-1247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson J, Pope T, Tung J S, Chan C, Hollis G, Mark G, Johnson K S. Affinity maturation of a high-affinity human monoclonal antibody against the third hypervariable loop of human immunodeficiency virus: use of phage display to improve affinity and broaden strain reactivity. J Mol Biol. 1996;256:77–88. doi: 10.1006/jmbi.1996.0069. [DOI] [PubMed] [Google Scholar]
- 52.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 53.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsarev S A, Tsareva T S, Emerson S U, Govindarajan S, Shapiro M, Gerin J L, Purcell R H. Recombinant vaccine against hepatitis E: dose response and protection against heterologous challenge. Vaccine. 1997;15:1834–1838. doi: 10.1016/s0264-410x(97)00145-x. [DOI] [PubMed] [Google Scholar]
- 55.Tsarev S A, Tsareva T S, Emerson S U, Govindarajan S, Shapiro M, Gerin J L, Purcell R H. Successful passive and active immunization of cynomolgus monkeys against hepatitis E. Proc Natl Acad Sci USA. 1994;91:10198–10202. doi: 10.1073/pnas.91.21.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsarev S A, Tsareva T S, Emerson S U, Kapikian A Z, Ticehurst J, London W, Purcell R H. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 57.Tsarev S A, Tsareva T S, Emerson S U, Yarbough P O, Legters L J, Moskal T, Purcell R H. Infectivity titration of a prototype strain of hepatitis E virus in cynomolgus monkeys. J Med Virol. 1994;43:135–142. doi: 10.1002/jmv.1890430207. [DOI] [PubMed] [Google Scholar]
- 58.Tsega E, Hansson B G, Krawczynski K, Nordenfelt E. Acute sporadic viral hepatitis in Ethiopia: causes, risk factors, and effects on pregnancy. Clin Infect Dis. 1992;14:961–965. doi: 10.1093/clinids/14.4.961. [DOI] [PubMed] [Google Scholar]
- 59.Vijh-Warrier S, Murphy E, Yokoyama I, Tilley S A. Characterization of the variable regions of a chimpanzee monoclonal antibody with potent neutralizing activity against HIV-1. Mol Immunol. 1995;32:1081–1092. doi: 10.1016/0161-5890(95)00081-x. [DOI] [PubMed] [Google Scholar]
- 60.Williamson R A, Burioni R, Sanna P P, Partridge L J, Barbas III C F, Burton D R. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc Natl Acad Sci USA. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. . (Erratum, 91:1193, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 62.Yin S, Purcell R H, Emerson S U. A new Chinese isolate of hepatitis E virus: comparison with strains recovered from different geographical regions. Virus Genes. 1994;9:23–32. doi: 10.1007/BF01703432. [DOI] [PubMed] [Google Scholar]
- 63.Zhuang H, Cao X Y, Liu C B, Wang G M. Epidemiology of hepatitis E in China. Gastroenterol Jpn. 1991;26(Suppl. 3):135–138. doi: 10.1007/BF02779283. [DOI] [PubMed] [Google Scholar]