Abstract
We have developed a novel helper-virus-free reverse genetic system to genetically manipulate influenza A viruses. The RNPs, which were purified from the influenza A/WSN/33 (WSN) virus, were treated with RNase H in the presence of NS (nonstructural) cDNA fragments. This specifically digested the NS RNP. The NS-digested RNPs thus obtained were transfected into cells together with the in vitro-reconstituted NS RNP. The NS-digested RNPs alone did not rescue viruses; however, cotransfection with the NS RNP did. This protocol was also used to rescue the NP transfectant. We obtained two NS1 mutants, dl12 and N110, using this protocol. The dl12 NS gene contains a deletion of 12 amino acids at positions 66 to 77 near the N terminus. This virus was temperature sensitive in Madin-Darby bovine kidney (MDBK) cells as well as in Vero cells. The translation of all viral proteins as well as cellular proteins was significantly disrupted during a later time of infection at the nonpermissive temperature of 39°C. The N110 mutant consists of 110 amino acids which are the N-terminal 48% of the WSN virus NS1 protein. Growth of this virus was significantly reduced at any temperature. In the virus-infected cells, translation of the M1 protein was reduced to 10 to 20% of that of the wild-type virus; however, the translation of neither the nucleoprotein nor NS1 was significantly interfered with, indicating the important role of NS1 in translational stimulation of the M1 protein.
Influenza A virus contains eight species of negative-sense viral RNAs as the genome. The viral RNAs, together with the nucleoprotein (NP) and three polymerase proteins, form RNP complexes (for reviews, see references 13 and 14). The RNPs have biological activity, and transfection of the RNPs into cells rescue viruses. Previously, a technique to genetically manipulate infectious influenza A viruses had been developed (6, 7, 18), in which RNPs were reconstituted in vitro by transcribing RNAs from cloned cDNA in the presence of NP and the three polymerase proteins. The in vitro-reconstituted RNPs were then transfected into cells which were infected with helper viruses. The transfectant viruses thus obtained had to be delicately isolated from the progeny of the helper viruses. Since the helper virus was essential in this system, this technique allowed us to obtain limited mutants. As a result, we have developed a novel helper-virus-free RNP transfection protocol.
Influenza virus NS1 protein has multiple functions including interference with splicing (9, 16, 27, 29) and interference with poly(A) addition and resultant inhibition of nuclear export (2, 20, 25, 26) of the cellular mRNAs of the infected cells. The NS1 protein is also involved in translational regulation of the viral mRNAs (3, 5, 23), as well as inactivation of the RNA-dependent protein kinase PKR (17). To understand the function and structure of the NS1 protein in virus-infected cells, we have isolated NS1 mutants by using this system. Previously, some naturally occurring NS1 deletion mutants had been isolated. The influenza virus temperature-sensitive (ts) mutant CR43-3, derived by recombination from the A/Alaska/6/77 and the cold-adapted and ts A/Ann Arbor/6/60 viruses, has been shown to have a 36-nucleotide deletion mutation in the NS1 gene (1, 28). However, the mechanism of ts phenotype during virus replication had not been characterized. A mutant A/Turkey/Oregon/71 virus has been shown to have a long carboxyl-terminal deletion resulting in an NS1 protein of only 124 amino acids, which lacks a predicted effector domain (22). Such naturally occurring mutants might have some compensatory mutations in the same or other genes. Therefore, we decided to use our RNP transfection system to isolate the NS1 transfectant containing similar mutations in the genetic background of the well-characterized influenza A/WSN/33 (WSN) virus. In this report, we demonstrate the important role of the NS1 protein in translational regulation by characterizing these transfectants.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). Vero cells were maintained in serum-free AIM-V medium (GIBCO BRL). WSN virus and transfectant viruses were grown in MDBK cells in DMEM containing 0.2% bovine serum albumin (BSA).
Preparation of plasmids.
The pUC119-derived plasmid pT7/WSN-NS expressing the WSN virus NS gene under the T7 phage polymerase promoter and terminal HgaI restriction enzyme site was constructed by inserting a PCR product into XbaI and EcoRI sites. The PCR product was obtained using WSN NS cDNA as a template and primers 5′-GCGCGCTCTAGACGCCCGGGAGCAAAAGCAGGGTGACAAAGACAT-3′ and 5′-GCGCGCGAATTCTTAATACGACTCACTATAAGTAGAAACAAGGGTGTTTTTTATTA-3′. Plasmid pT7/NHE-NS, containing the NheI site in the WSN NS gene, was obtained by inserting two PCR products into the SalI and EcoRI sites of pUC119. The first PCR product was obtained using pT7/WSN-NS as a template and primer pair M13RV (5′-CAGGAAACAGCTATGAC-3′) and NHENS1 (5′-GGTACGCTAGCCATGGTCATTTTG-3′) and was digested with SalI and NheI. The second PCR product was obtained using primers NHENS2 (5′-GTTTTCCCAGTCACGAC-3′) and M13M4 (5′-GTTTTCCCAGTCACGAC-3′) and was digested with NheI and EcoRI. Plasmid pT7/DL12-NS was constructed by inserting two PCR products into the SalI and EcoRI sites of plasmid pUC119. The first PCR product was obtained using primers 5′-GGTACGCTAGCCATGGTCATTTTCACTATTTGCTTTCCAGCACGGGTG-3′ and M13RV and was digested with SalI and NheI. The second PCR product, obtained using NHENS2 and M13M4, was digested with NheI and EcoRI. Plasmid pT7/N110-NS was constructed by inserting two PCR products into the XbaI and EcoRI site of pUC119, where the first PCR product, obtained using primers M13RV and 5′-GATACCTCGAGGGCCTTACTATTTCTGCTTGGGCATGAGCATG-3′, was digested with XbaI and XhoI. The second PCR product was obtained using primers 5′-GGCCCTCGAGGTATCAGAATGGACCAGGCG-3′ and M13M4, and it was digested with XhoI and EcoRI. Plasmid pT7/XHO-NP was constructed by inserting two PCR products into the SalI and EcoRI sites of plasmid pUC119. The PCR product was obtained using 5′-CGCGCGTCGACTCTTCGAGCAAAAGCAGGGTAGATAATCACT-3′ and 5′-TCCTGGGATCCATTCCTGTGCGAACAAGAGCTCGAGTCCTCTGGTAA-3′ as primers and WSN NP cDNA as a template, and it was digested with SalI and BamHI. The PCR product obtained by using primers 5′-TTACCAGAGGACTCGAGCTCTTGTTC-3′ and 5′-CGCGCGAATTCTAATACGACTCACTATAAGTAGAAACAAGGGTATTTTTCTTTA-3′ was digested with BamHI and EcoRI.
RNP transfection.
The coding region of the NS or NP cDNA was amplified by PCR using primers 5′-ATGGATCCAAACACTGTGTC-3′ and 5′-AATAAGCTGAAACGAGAAAG-3′ for NS or ATGGCGTCTCAAGGCACCAA and 5′-ATTGTCGTACTCCTCTGCAT-3′ for NP. The PCR products were partially digested with 0.05 U of RQ DNase I (Promega) per μg of DNA for 5 min at 37°C. This digestion condition was determined using different amounts of RQ DNase I. Characterization by RNA gel electrophoresis of the digested RNP showed that this condition gave the optimum length of cDNA fragments for the minimum background without increasing nonspecific RNA digestion. These cDNA fragments were purified and used for RNase H reaction.
The WSN virus RNP was purified from virions as described previously (18). Briefly, the purified virion was digested with lysophosphatidylcoline and Triton N-101 in the presence of human placenta RNase inhibitor (RNsin; 500 U/ml; Takara, Kyoto, Japan). The RNsin was not used in the previous system (18) but was essential in this protocol. The RNP was then purified through glycerol centrifugation. This RNP preparation contained approximately 0.1 μg of RNA/μl. This RNP (10 μl) was incubated for 5 min at 37°C in the presence of 0.6 μl of 5 M NaCl (final concentration, 0.4 M) and 1 to 2 μg of NS or NP cDNA fragments. This RNP was then diluted with 40 μl of 12.5 mM Tris-HCl (pH 8.0), 5 mM MgCl2, and 1.25 mM dithiothreitol. Thirty units of RNase H (Takara) was then added, and the reaction mixture was incubated for 5 min at 37°C. The cDNA fragments in the reaction were then completely digested with 4 U of RQ DNase I together with 25 U of RNsin for 5 min at 37°C. Meanwhile, the NS and NP RNPs were reconstituted in vitro as described previously (6) by in vitro transcription in the presence of influenza virus NP and polymerase proteins in a 50-μl reaction. The RNase H-treated and in vitro-reconstituted RNPs were immediately cooled on ice after the reaction and then purified together through a Quick Spin Column Sephadex G-50 Fine (Boehringer Mannheim, Tokyo, Japan) equilibrated with phosphate-buffered saline (PBS) containing 0.01% gelatin. The eluate (approximately 100 μl) was immediately transfected into MDBK cells (approximately 20 to 30% confluency) in a 35-mm-diameter dish for 1 h as described previously (18). MDBK cells (106) in 2 ml of DMEM containing 10% FCS were then added into this dish, and the cells were incubated for 1 to 2 h at 37°C. Then the medium was removed, and the cells were overlaid with 2 ml of DMEM containing 0.2% BSA and 0.6% agarose. Viral plaques were formed during 3 to 4 days at 34 or 37°C. Viruses were purified and amplified in the MDBK cells.
Analysis of protein synthesis.
MDBK cells (106) in a 35-mm-diameter dish were infected with influenza WSN, dl12, or N110 virus (multiplicity of infection [MOI] of 3) for 30 min at room temperature. After incubation in DMEM containing 0.2% BSA at 39°C, the cells were washed twice with prewarmed PBS and were labeled for 30 min in 0.5 ml of DMEM containing 50 μCi of [35S]methionine-cysteine (NEN EXPRE35S35S protein labeling mix).
Labeled proteins were immunoprecipitated with protein G-Sepharose and with either anti-WSN virus rabbit serum supplemented with anti-NP and anti-M1 antibody or anti-NS1 antibody for the analysis of the NS1 protein. Proteins were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10 to 20% gradient (ATTO, Tokyo, Japan).
Slot blot hybridization analysis of the viral RNAs in infected cells.
Poly(A)+ and poly(A)− RNAs were purified from infected whole cells or the cytoplasmic fraction of the cells by using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech). These RNAs were diluted with 0.05 N NaOH and applied to a Hybond-N+ membrane (Amersham Pharmacia Biotech) using a Bio-Dot SF microfiltration instrument (Bio-Rad). Hybridization was performed using a 32P-labeled plus- or minus-sense probe as described previously (5). The M1-specific probe (nucleotide positions 290 to 427, a region which does not contain M2 sequence) or the NS1-specific probe (nucleotide positions 256 to 360, a region which does not contain NS2 sequence) was used for hybridization of the M1 or NS1 mRNA.
Indirect immunofluorescence of the NS1 protein.
The WSN, dl12, and N110 viruses were used to infect MDBK cells in Nunc eight-well chamber slides (Inter Med Japan) at an MOI of 3. The infected cells were incubated for 3 or 6 h at 39°C and then fixed in acetone for 20 min at −20°C. Immunofluorescence was examined using anti-NS1 antibody followed by fluorescein isothiocyanate-labeled second antibody.
RESULTS
A novel RNP transfection system.
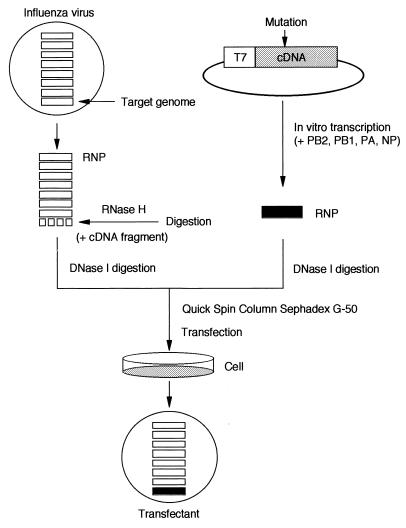
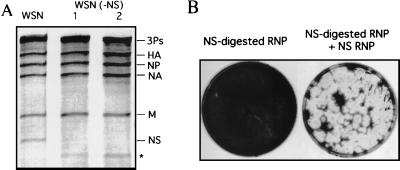
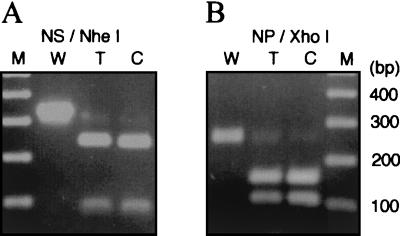
A schematic protocol of the novel RNP transfection is illustrated in Fig. 1. During the establishment of this protocol, we first determined the optimum condition which specifically digests the NS RNP without inactivating other RNPs. We considered NaCl concentration, annealing temperature, and the amount and length of the cDNA fragments as well as the following RNase H concentration (data not shown). In addition, we purified highly active RNPs (transfection of 0.1 μl of the RNP rescued >102 plaques in a 35-mm-diameter dish). Consequently, we obtained the optimum condition which specifically digests the specific RNP. Data for digestion of the NS RNP are shown in Fig. 2A. Some shorter bands were visible after digestion, indicating that some RNase H-sensitive regions might be present, but we have not characterized them further. The NS-digested RNPs thus obtained were then transfected into MDBK cells. The NS-digested RNPs alone did not rescue viruses; however, cotransfection with the in vitro-reconstituted NS RNP did (Fig. 2B). At the optimum condition, >102 plaques were obtained in a 35-mm-diameter dish by infectious center assay. The genetic markers of rescued viruses were then characterized by reverse transcription-PCR (RT-PCR) of the viral RNA (vRNA) followed by restriction enzyme NheI or XhoI digestion for NS or NP transfectants, respectively (Fig. 3). The data indicate that this protocol is generally applicable to the influenza virus genome. In addition, the protocol may be modified in some respects; for example, the transfected cells can be incubated with serum-free medium, and the rescued viruses can be isolated from the supernatant.
FIG. 1.
Scheme for the novel RNP transfection protocol. WSN virus RNPs purified from the virion were treated with the cDNA fragment for the target genome and RNase H as described in Materials and Methods. The RNP was reconstituted in vitro by the conventional method (7), i.e., by transcribing from the cloned cDNA in the presence of PB1, PB2, PA, and NP. These RNPs were purified and transfected together into MDBK cells. Transfectant viruses were obtained by infectious center assay.
FIG. 2.
Rescue of transfectant virus using the novel transfection protocol. (A) WSN RNP (10 μl) was treated with 1 or 2 μg of NS cDNA fragment in the presence of 0.4 M NaCl for 5 min at 37°C; then it was diluted and treated with 30 U of RNase H for 5 min at 37°C as described in Materials and Methods. The purified RNA was loaded onto a 3.2% polyacrylamide gel containing 7.7 M urea and visualized by silver staining as described previously (6). An asterisk indicates one of the shorter bands which may be digested fragments. (B) The NS-digested RNPs were transfected into MDBK cells with or without reconstituted NS RNP. Viral plaques were formed by infectious center assay. 3Ps, PB1, PB2, and PA.
FIG. 3.
Analysis of transfectant viral genome. (A) RT-PCR and NheI digestion of the NS transfectant vRNA (T). Plasmid pT7/NHE-NS DNA (C) or WSN vRNA (W) was used as the control. (B) RT-PCR and XhoI digestion of the NP transfectant vRNA (T). Plasmid pT7/XHO-NP DNA (C) or WSN vRNA (W) were used as the control.
Rescue of the NS1 mutants.
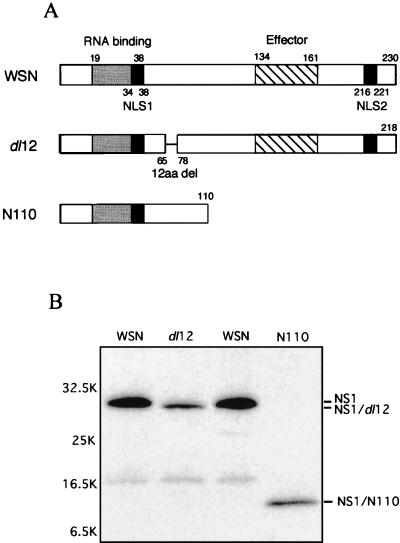
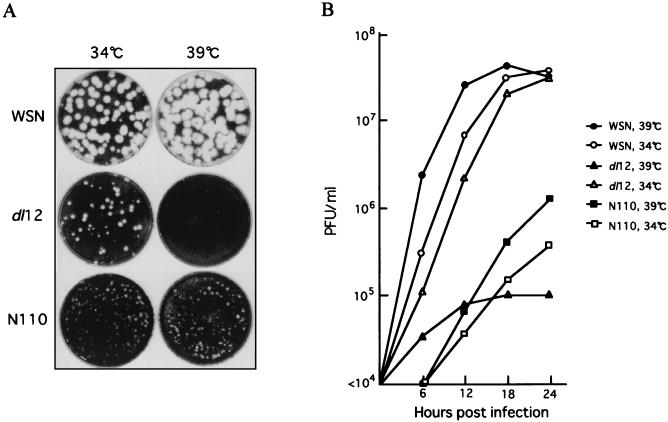
We then applied this protocol to rescue NS1 mutants. WSN virus-derived transfectant dl12 contains a deletion of 12 amino acids near the predicted RNA binding domain of the NS1 protein (25) (Fig. 4A, dl12). Another virus contains 52% deletion of the C terminus of the NS1 protein, which deletes the effecter domain (25) (Fig. 4A, N110). The resulting NS1 protein consists of 110 amino acids. The mutations in the rescued viruses were confirmed by nucleotide sequencing of the viral genome (data not shown). The viruses expressed the expected size of the proteins in vivo (Fig. 4B). The growth of the viruses were then characterized. The dl12 virus was ts (Fig. 5A), and infectious virus formation was less than 10−2 of wild-type virus formation at 39°C (Fig. 5B). Previously, it was reported that the NS1 protein was not essential in Vero cells (10). Therefore, we examined the growth of the dl12 virus in Vero cells, but the virus was ts (data not shown). On the other hand, growth of the N110 virus was attenuated at any temperature, but it was not ts (Fig. 5).
FIG. 4.
Rescue of NS1 mutants. (A) Genome structure of the NS1 mutants. The dl12 NS1 contains a 12-amino-acid deletion (12aa del) at amino acid positions 66 to 77. N110 NS1 contains the amino-terminal 110 amino acids. The predicted RNA binding domain (25), NLS1 and NLS2 (11), and predicted effector domain (25) are indicated. (B) Protein expression of the NS1 mutants in vivo. MDBK cells in a 35-mm-diameter dish were infected with virus at an MOI of 3. After 4 h of incubation at 34°C, cells were labeled with [35S]methionine-cysteine (100 μCi/ml) for 1 h. The cell lysates were immunoprecipitated with anti-NS1 antibody and analyzed by SDS-PAGE (10 to 20% gel).
FIG. 5.
Growth of the mutants viruses. (A) Plaque morphology of the mutant viruses. MDBK cells in a 35-mm-diameter dish were infected with approximately 102 PFU of the WSN, dl12, or N110 virus, and plaques were formed for 3 days at 34 or 39°C. (B) MDBK cells in a 35-mm-diameter dish were infected with the WSN, dl12, or N110 virus at an MOI of 3. The culture was incubated for the indicated times at 34 or 39°C. Infectious viruses in the supernatant were determined by plaquing on the MDBK cells at 34°C.
Translational defects of the mutant viruses.
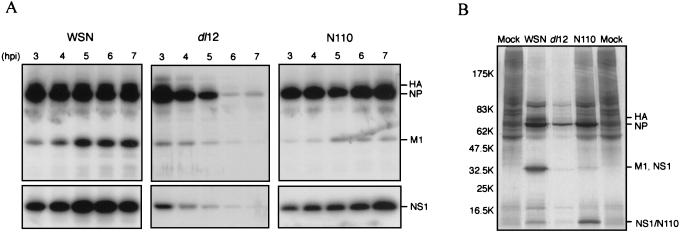
We then characterized the translation, transcription, and replication of these viruses in vivo. In the dl12 virus-infected cells, translation of all viral proteins was inhibited during the later stages of the infection (at 6 to 7 h postinfection [hpi]) at the nonpermissive temperature of 39°C (Fig. 6A, dl12). In the N110 virus-infected cells, translation of the late viral proteins including HA (hemagglutinin) and M1 was lower (10 to 20% of that of wild-type virus) at any time point; however, that of the early proteins including NP and NS1 was not significantly altered (Fig. 6A, N110). This result was consistent with another recent study (4, 19). Meanwhile, the NS1 protein was shown to be involved in the translational shutoff of host mRNAs. Therefore, we analyzed whole protein synthesis of the infected cells (Fig. 6B). Translation of all viral and cellular proteins was extremely disrupted in the dl12 virus-infected cells. On the other hand, the N110 NS1, which lacks an essential effector domain, had a defect in the shutoff of host protein synthesis as expected.
FIG. 6.
Protein synthesis in mutant virus-infected cells. (A) MDBK cells in a 35-mm-diameter dish were infected with the WSN, dl12, or N110 virus at an MOI of 3 then incubated for the indicated times at 39°C. Cells were labeled with [35S]methionine-cysteine (100 μCi/ml) for 30 min. Proteins were immunoprecipitated with anti-WSN rabbit serum supplemented with anti-NP and anti-M1 or with anti-NS1 antibody. Proteins were analyzed by SDS-PAGE (10 to 20% gel). (B) MDBK cells in a 35-mm-diameter dish were infected with the WSN, dl12, or N110 virus at an MOI of 3. Cells were labeled with [35S]methionine-cysteine (100 μCi/ml) for 30 min at 7 hpi. Virus-infected or mock-infected whole cell extracts were analyzed by SDS-PAGE (10 to 20% gel).
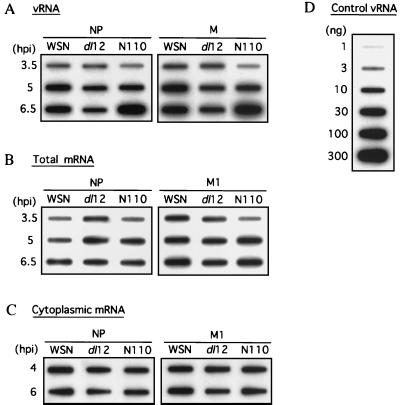
We previously reported that the NS1 protein plays an important role in viral translation but not in transcription or replication (5). Therefore, we expected that such defects may be explained during translation. We then characterized the vRNAs in the infected cells. In dl12 virus-infected cells, replication and transcription were 30 to 70% of levels for the WSN virus at 6.5 hpi (Fig. 7A to C, dl12). This reduction may be explained by the reduced expression of the viral polymerase and NP proteins, however, this moderate reduction of the mRNA level cannot explain the complete inhibition (>95%) of translation. On the other hand, RNA replication of the NP and M genomes was relatively stimulated in the N110 virus-infected cells (two- to threefold compared to that of the WSN virus at 6.5 hpi) (Fig. 7A, N110), which may be explained by the reduced expression of the M1 protein. Importantly, the transcriptional levels of NP and M1 were similar between WSN and N110 (Fig. 7B and C, N110). The data were consistent with our prediction. We also characterized the NS RNAs, and the data were the same as for NP and M RNAs (data not shown).
FIG. 7.
Replication, transcription, and cytoplasmic accumulation of mRNAs. (A) Characterization of the vRNA. MDBK cells in a 10-cm-diameter dish were infected with the WSN, dl12, or N110 virus at an MOI of 3, and the cells were incubated for the indicated times at 39°C. Poly(A)− RNAs were purified from the cells. RNAs were blotted onto a Hybond-N+ membrane and hybridized with plus-sense 32P-labeled probe. (B) Characterization of total mRNA. MDBK cells were infected as described above and then incubated for the indicated times at 39°C. Poly(A)+ RNAs were purified from the cells. RNAs were blotted onto a Hybond-N+ membrane and were hybridized with NP or M1 mRNA-specific minus-sense 32P-labeled probe. (C) Characterization of cytoplasmic mRNA. MDBK cells were infected as described above and then incubated for the indicated times at 39°C. Poly(A)+ RNAs were purified from the cytoplasmic fraction of the cells. RNAs were blotted onto a Hybond-N+ membrane and hybridized with NP or M1 mRNA-specific minus-sense 32P-labeled probe. (D) The indicated amount of vRNAs, purified from the WSN virus, was used for blotting as a control. vRNA-specific plus-sense 32P-labeled probe was used for hybridization.
Intracellular localization of the mutant NS1 proteins.
Influenza virus NS1 protein contains two nuclear localization signals, NLS1 and NLS2. Therefore, NS1 protein accumulates in the nucleus during the early stages of the infection. During the later stages, the NS1 protein is exported to the cytoplasm and is involved in the translational regulation. Previously, the effector domain was shown to be essential for this nuclear export (15). Therefore, intracellular localization of these mutant NS1 proteins was characterized by indirect immunofluorescence. The dl12 NS1 protein accumulated in the cytoplasm during virus infection at 39°C. In contrast, the N110 NS1 protein had a defect in the nuclear export and accumulated in the nucleus during the later stage of the virus infection (data not shown).
DISCUSSION
We have isolated two NS1 mutants, dl12 and N110, using a novel helper-virus-free reverse genetic system. These mutants have defects in translation in a different manner. The NS1 protein is transported to the nucleus during the early stage of the infection, and it is involved in the shutoff of cellular gene expression via interfering nuclear export of the cellular mRNAs (2, 20, 25, 26). Later during the infection, the NS1 protein is exported to the cytoplasm, where it is involved in translational regulation of the viral proteins (3, 5, 12, 23). The N110 NS1 protein accumulated in the nucleus but had a defect in host shutoff, which may be explained by the deletion of the effector domain. In addition, translation of HA and M1 was significantly reduced, which may be explained by the absence of the NS1 protein in the cytoplasm. Another possibility is that the C-terminal half of the NS1 protein may contain the functional domain for the interaction with the translational factor which is required for the regulation. Recently, GRSF-1 was found to interact with the 5′ untranslated region (UTR) of the NP and NS mRNAs and to stimulate translation (24). The NS1 protein binds the 5′ UTR of NP and M1 and stimulates its translation (5, 23). Interaction of the NS1 protein with such translational factors might be involved in the translation of late viral proteins. Interestingly, dl12 NS1 was shown to inhibit not only viral protein synthesis but also cellular translation. One possible explanation is that the mutant NS1 might bind to the host factor which is essential for virus and host translation, but the NS1 might be ts in binding to the 5′ UTR of the viral mRNAs; consequently, such host factors might be depleted in the cytoplasm, forming inactive complexes with the mutant NS1. The dl12 and N110 NS1 might be useful for identification of such host factors. In addition, we recently introduced a chloramphenicol acetyltransferase reporter gene downstream of the short NS1 gene in the N110 virus (M. Enami et al., submitted for publication). The N110 virus may be useful for developing a new vaccine vector.
Recently, two groups independently reported plasmid-based influenza virus reverse genetics system (8, 21). Their system may be useful to introduce mutations into multiple genes at the same time but might be inconvenient for introducing mutations into different viral strains. On the other hand, the present system may be useful to introduce mutations in a single gene for various viral strains, because this system does not require cDNA cloning of all eight genes. Recently, we have applied this system to introduce an attenuated recombinant H5 HA gene into avirulent chicken influenza virus in order to generate a vaccine strain for the virulent H5N1 virus (S. Itamura et al., submitted for publication). This may demonstrate one of the advantages of this system.
ACKNOWLEDGMENTS
We thank P. Palese for helpful discussions.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan and from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Buonagurio D A, Krystal M, Palese P, DeBorde D C, Maassab H F. Analysis of an influenza A virus mutant with a deletion in the NS segment. J Virol. 1984;49:418–425. doi: 10.1128/jvi.49.2.418-425.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Li Y, Krug R M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Luna S, Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer A, Alexandrova G, Katinger H, Muster T. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enami M, Luytjes W, Krystal M, Palese P. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci USA. 1990;87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enami M, Palese P. High-efficiency formation of influenza virus transfectants. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor E, Devenish L, Engelhardt O G, Palese P, Brownlee G G, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 11.Greenspan D, Palese P, Krystal M. Two nuclear localization signals in influenza virus NS1 nonstructural protein. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krug R M, Etkind P R. Cytoplasmic and nuclear specific proteins in influenza virus-infected MDCK cells. Virology. 1973;56:334–348. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- 13.Krug R M, Alonso-Kaplen F V, Julkunen I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- 14.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–97. [Google Scholar]
- 15.Li Y, Yamakita Y, Krug R M. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. J Virol. 1998;95:4864–4869. doi: 10.1073/pnas.95.9.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Y, Qian X Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 18.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Amplification, expression, and packaging of a foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 19.Marión R M, Aragón T, Beloso A, Nieto A, Ortín J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemeroff M E, Barabino S M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein interacts with the cellular 30kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 21.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffman E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norton G P, Tanaka T, Tobita K, Nakada S, Buonagurio D A, Greenspan D, Krystal M, Palese P. Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology. 1987;156:204–213. doi: 10.1016/0042-6822(87)90399-0. [DOI] [PubMed] [Google Scholar]
- 23.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 24.Park Y W, Wilusz J, Katze M G. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc Natl Acad Sci USA. 1999;96:6694–6699. doi: 10.1073/pnas.96.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian X-Y, Alonso-Caplen F, Krug R M. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J Virol. 1994;68:2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Y, Nemeroff M, Krug R M. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder M H, London W T, Maassab H F, Chanock R M, Murphy B R. A 36 nucleotide deletion mutation in the coding region of the NS1 gene of an influenza A virus RNA segment 8 specifies a temperature-dependent host range phenotype. Virus Res. 1990;15:69–84. doi: 10.1016/0168-1702(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Krug R M. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA. 1998;4:55–64. [PMC free article] [PubMed] [Google Scholar]