Abstract
Togavirus nucleocapsids have a characteristic icosahedral structure and are composed of multiple copies of a capsid protein complexed with genomic RNA. The assembly of rubella virus nucleocapsids is unique among togaviruses in that the process occurs late in virus assembly and in association with intracellular membranes. The goal of this study was to identify host cell proteins which may be involved in regulating rubella virus nucleocapsid assembly through their interactions with the capsid protein. Capsid was used as bait to screen a CV1 cDNA library using the yeast two-hybrid system. One protein that interacted strongly with capsid was p32, a cellular protein which is known to interact with other viral proteins. The interaction between capsid and p32 was confirmed using a number of different in vitro and in vivo methods, and the site of interaction between these two proteins was shown to be at the mitochondria. Interestingly, overexpression of the rubella virus structural proteins resulted in clustering of the mitochondria in the perinuclear region. The p32-binding site in capsid is a potentially phosphorylated region that overlaps the viral RNA-binding domain of capsid. Our results are consistent with the possibility that the interaction of p32 with capsid plays a role in the regulation of nucleocapsid assembly and/or virus-host interactions.
Rubella virus (RV) is a positive-strand RNA virus of the family Togaviridae. Despite routine vaccination programs which have been in place for 30 years, the virus persists in the human population and remains an important human pathogen (11). The most serious medical consequences of RV infection occur when seronegative women contract the virus during the first trimester of pregnancy. RV is highly teratogenic and causes a characteristic pattern of defects in the fetus which are collectively known as congenital rubella syndrome. The molecular basis of RV pathology remains poorly understood, but several recent reports have shown that the virus induces apoptosis in a cell type-dependent manner (9, 19, 33, 45). This pattern of apoptosis could potentially explain the organ-specific malformations observed in congenital rubella syndrome.
Virions contain three structural proteins that are translated from a 24S subgenomic RNA; two membrane-spanning glycoproteins (E2 and E1) and a capsid protein (38). The capsid protein is multifunctional and is involved in several different types of intermolecular interactions. First, it contains an RNA-binding domain and is responsible for packaging the genomic RNA into nucleocapsids (11, 28). Second, by analogy with other togaviruses, capsid must engage in homo-oligomeric (capsid-capsid) interactions during nucleocapsid formation. Finally, it must also interact with E2 and/or E1 during budding (37, 40).
The nucleocapsids of togaviruses have a characteristic icosahedral structure which has been extensively studied in alphaviruses (41). Although the overall structures of RV and alphavirus capsids are similar, their assembly pathways are quite different. Whereas alphavirus capsids are released into the cytosol after self-catalyzed cleavage from the structural protein precursor (34), the RV capsid does not possess protease activity and remains largely membrane associated after a signal peptidase-mediated cleavage from the E2-E1 precursor (16, 52). Moreover, the nucleocapsids of alphaviruses form in the cytoplasm of infected cells well before budding occurs (51), while RV nucleocapsid assembly occurs on the surface of intracellular membranes and is coincident with virus budding (11). Recent studies indicate that RV capsid can oligomerize in the absence of other viral and mammalian proteins: (i) RV nucleocapsid assembly can be partially reconstituted in vitro using lysates from Escherichia coli that are expressing capsid (39), and (ii) capsid-capsid interactions can be demonstrated using the yeast-two hybrid system (4). These data are consistent with our hypothesis that assembly of RV nucleocapsids is regulated by interaction with host cell proteins. Such interactions with host cell proteins have been shown to modulate the nucleocapsid assembly pathways of other, unrelated viruses (26, 27).
To identify host cell proteins that interact with the RV capsid, we screened a CV1 cDNA library using the yeast two-hybrid method. Multiple capsid-binding clones were isolated which contained the cDNA corresponding to a previously identified human protein known as p32. Interestingly, p32 has been shown to bind a variety of other virus phosphoproteins that complex with nucleic acids. In addition, p32 has a putative role in apoptosis through regulation of the mitochondrial permeability transition pore (21). We have confirmed the interaction between p32 and capsid using in vitro and in vivo methods and provide evidence that the interaction between these two proteins occurs at the mitochondria.
MATERIALS AND METHODS
Reagents.
Reagents and supplies were from the following sources. Protein A- and G-Sepharose and glutathione-Sepharose were purchased from Pharmacia (Alameda, Calif.). Phenylmethylsulfonyl fluoride, fibronectin, sodium dodecyl sulfate (SDS), bovine serum albumin, glucose oxidase, and cyanogen bromide-activated Sepharose 4B were purchased from Sigma Chemical Co. (St. Louis, Mo.). Promix [35S]methionine-cysteine (1,000 Ci/mmol), translation grade [35S]methionine (1,000 Ci/mmol), and 14C-labeled protein standards were purchased from Amersham Corp. (Arlington Heights, Ill.). Minimal essential medium lacking cysteine and methionine was purchased from ICN Biomedicals (Irvine, Calif.). Sf-900 II serum-free medium, OptiMEM, and fetal bovine serum were obtained from Life Technologies Inc. (Gaithersburg, Md.). Fugene 6 transfection reagent and Pwo polymerase were purchased from Roche Molecular Biochemicals (Laval, Quebec, Canada). Rabbit antiserum to p32 was a gift from Willie Russell (University of St. Andrews, St. Andrews, United Kingdom). Monoclonal antibody to HSP 60 was purchased from StressGen (Victoria, British Columbia, Canada). Monoclonal antibodies to RV capsid (H15 C22) and E1 (B2) were gifts from John Safford (Abbott Laboratories, North Chicago, Ill.) and Barbara Pustowoit, (University of Leipzig, Leipzig, Germany), respectively. Double-labeling-grade Texas Red-conjugated goat anti-mouse immunoglobulin G (IgG) and fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, Pa.). Mitotracker Red CXMRos was from Molecular Probes (Eugene, Oreg.). Vero and COS cells were obtained from the American Type Culture Collection (Manassas, Va.). The M33 strain of RV was obtained from Shirley Gillam (University of British Columbia). Recombinant baculovirus (OB504-1) encoding capsid was a gift of Christian Oker-Blom (VTT Biotechnology and Food Research, Espoo, Finland).
Plasmid construction. (i) Two-hybrid constructs.
The cDNA encoding amino acids 1 to 277 of capsid was amplified by PCR using the gene-specific primers Capsid-F (5′-CGCGAATTCATGGCTTCCACTACCC-3′) and C-E2SP-R (5′-ACTGAGATCTAGCGGATGCGCCAAGGATG-3′), containing an in-frame stop codon. The EcoRI (GAATTC) and BglII (AGATCT) sites are underlined in all of the primers. The PCR product was digested with EcoRI and BglII and ligated into the EcoRI and BamHI sites of pGBT9 (Clontech) to create pGBT9-capsid. Capsid deletion mutant cDNAs generated by PCR were also digested with EcoRI and BglII and subcloned into the EcoRI and BamHI sites of pGBT9 or pGBKT7 as indicated. The primers used for each construct were as follows: pGBT9-C1-88, Capsid-F and CM11 (5′-GATCAGATCTCTAGCGACTTTCTTGCCGCTC-3′); pGBT9-C87-171, CM12 (5′-GATCGAATTCAGTCGCTCCCAGACTCCG-3′) and CM13 (5′-GATCAGATCTCTAGTCGACGCGGTAGAAGAC-3′); pGBT9-C167-277, CM14 (5′-GATCGAATTCTACCGCGTCGACCTG-3′) and C-E2SP-R; pGBKT7-C1-45, Capsid-F and CR45 (5′-GGTCAGATCTCTAGGAGTCGCGCTGTCGC-3′); and pGBKT7-C46-89, Capsid-46 (5′-GGTCGAATTCAGCACCTCCGGAGATGAC-3′) and CR89 (5′-GGTCAGATCTCTAGGAGCGACTTTCTTGCCGC-3′).
(ii) Constructs for in vitro transcription-translation.
The capsid cDNA was amplified by PCR from pCMV5-24S (17) using the primers AV11 (5′-TACGGTGGGAGGTCTATATAGC-3′) and C-E2SP-R, and the PCR product was blunt-end ligated into the EcoRV site of pBluescript KS(+) (Stratagene). The resulting plasmid, pBLU-capsid, encoded RV capsid under the transcriptional control of the T3 promoter. A plasmid encoding green fluorescent protein (GFP) under the control of the T3 promoter, pBLU-GFP, was created by excising GFP from pEGFP-1 (Clontech) with EcoRI and NotI and ligating it into pBluescript KS(+).
(iii) Constructs for expression in bacteria.
p32 was cloned in frame into the vector pGEX-4T1 (Pharmacia). The coding sequence for p32 was amplified from clone 10 using the gene-specific primer P32F2 (5′-GATCGAATTCATGCTACCTCTGCTGCGC-3′) and the vector-specific primer GAD-PRV (5′-GCATCGTGCACCATCTCAA-3′). The PCR product was digested with EcoRI and NotI and ligated into pGEX4T1 to create pGEX-p32.
(iv) Constructs for expression in mammalian cells.
The RV expression plasmids pCMV5-24S and pCMV5-E2E1 have been described previously (7, 16). PCB6+p32 was created by excising the EcoRI/NotI fragment from pGEX-p32 and ligating it into pCB6+ (42).
Yeast two-hybrid screening.
Yeast strain HF7c was sequentially transformed with pGBT9-capsid and then with a CV1 cDNA library in the vector pGAD10 (Clontech). Approximately 7 × 106 transformants were screened. Plasmids were isolated from His+ LacZ+ colonies and then retransformed with or without pGBT9-capsid into yeast strain SFY526, followed by testing for β-galactosidase activity. Plasmid clones that activated LacZ only in the presence of pGBT9-capsid were characterized by DNA sequencing or restriction endonuclease mapping. Interactions between capsid deletions (subcloned into pGBT9 or pGBKT7) and p32 (subcloned into pGAD10) were assayed by cotransfecting AH109 cells and testing for the ability to grow on medium lacking adenine, histidine, leucine, and tryptophan. β-Galactosidase filter assays were also used to confirm the interactions. All media, screening techniques, and β-galactosidase assays were performed according to the protocols described in the Clontech MATCHMAKER system.
In vitro binding interactions.
Capsid and GFP were synthesized using coupled transcription-translation systems (Promega or Ambion) in the presence of [35S]methionine. The 35S-labeled proteins were incubated with Sepharose beads coated with glutathione S-transferase (GST), GST-p32, or glucose oxidase on a rotating device overnight at 4°C. The beads were washed three times with phosphate-buffered saline (pH 8.0) (PBS) containing 0.1% Triton-X100. The bound proteins were eluted by boiling in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 5 min. Samples were analyzed by SDS-PAGE and autoradiography as described previously (12).
Generation of polyclonal antibodies to capsid.
Capsid protein containing a His6 sequence was expressed in Sf9 cells using baculovirus as described previously (47). The His6-tagged capsid was purified by SDS-PAGE and electroelution (6). Rabbits were immunized with 70 μg of capsid protein, followed by booster injections (70 μg) at 4-week intervals. The antiserum used in this study is 7W7.
Transient transfection of cell cultures.
COS and Vero cells were transiently transfected using Fugene 6 transfection reagent as described by the manufacturer.
RV infection of Vero cells.
Vero cells were infected with the M33 strain of RV (multiplicity of infection of 1) for 2 h. The virus inoculum was removed, and cells were then incubated at 35°C for 2 days before use in radioimmunoprecipitation experiments.
Metabolic labeling and coimmunoprecipitation.
Transfected COS cells or infected Vero cells were metabolically labeled with [35S]methionine-cysteine and radioimmunoprecipitated as described previously (18). Immune complexes were washed with PBS containing 0.1% Triton X-100 to preserve protein-protein interactions.
Immunofluorescence and confocal microscopy.
Vero cells grown on coverslips were processed for indirect immunofluorescence microscopy 24 h after transfection. To visualize mitochondria, Mitotracker Red CXMRos (Molecular Probes) was added to the cell culture medium at a final concentration of 30 ng/ml and incubated for 20 min at 37°C prior to fixation. Cells were fixed in 3% paraformaldehyde for 20 min, followed by quenching with PBS containing 50 mM ammonium chloride. The samples were washed two times with PBS containing Ca2+ and Mg2+. Plasma membranes were permeabilized by treatment with PBS containing 25 μg of digitonin per ml for 5 min followed by washing with PBS containing Ca2+ and Mg2+. To permeabilize the intracellular membranes, cells were treated with PBS containing 0.1% Triton X-100 for 10 min before incubation with primary and secondary antibodies. Samples were examined using a Zeiss Axioskop 2 instrument, and images were captured using a digital camera (Diagnostic Laboratories, Inc., Sterling Heights, Mich.). In some cases, samples were examined using a Zeiss 510 confocal microscope. Images from optical sections (0.8 μm) were processed using Adobe Photoshop 5.0.
DNA sequencing.
Plasmids were sequenced using core facilities within the departments of Cell Biology and Biochemistry (University of Alberta).
Nucleotide sequence accession number.
The GenBank accession number for the simian p32 cDNA sequence is AF238300.
RESULTS
Identification of capsid-binding proteins.
The yeast two-hybrid assay was used to identify potential host cell capsid-binding proteins. This method was chosen since it is able to detect both transient and stable interactions between proteins (14). Vero cells are one of the few types of cultured cells in which RV is able to establish a productive infection, but two-hybrid Vero cDNA libraries are not commercially available. Since we wanted to screen cDNA from a cell type as close to Vero as possible, we chose to use a cDNA library prepared from CV1 cells, which, like Vero cells, are derived from African green monkey kidney. Bait (capsid) and prey (CV1 cDNA) plasmids were sequentially transformed into the yeast strain HF7c. Plasmids were isolated from transformants that grew on medium lacking leucine, tryptophan, and histidine. These plasmids were retransformed into SFY526 and assayed for β-galactosidase activity as a secondary test. Of 32 positive clones obtained from the screen, 16 were characterized in further detail. Fifteen of these plasmid clones were found to encode the full-length cDNA for a previously characterized protein, p32 (20, 23). The simian p32 cDNA encodes a protein of 282 amino acids which is 95% identical to the human p32 protein. The normal physiological function of this protein is not known, but is has been shown to interact with numerous cellular and virus proteins. The 16th clone encoded the carboxy-terminal region of Par-4 (49). Par-4 is up-regulated in cells committed to apoptosis and interacts with atypical isoforms of protein kinase C (8, 48). The nature of the interaction between capsid and Par-4 is currently under investigation in our laboratory and will not be discussed further in this paper.
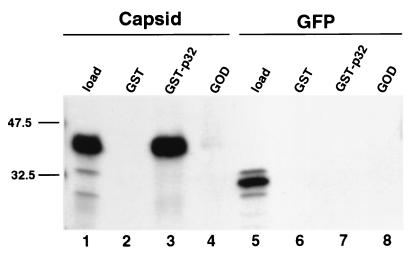
Previous studies showed that the majority of p32 localizes to the mitochondrial matrix (35). We were therefore initially surprised to find that the capsid would interact with this type of protein. However, a recent report by Lee et al., which was published during the course of this work, demonstrated that a significant proportion of RV nucleocapsids are associated with mitochondria during infection (25). To demonstrate that the capsid-p32 interaction was not limited to the yeast two-hybrid system, we employed an in vitro binding assay to measure the interaction of radiolabeled capsid with p32 immobilized on Sepharose beads. 35S-labeled capsid or GFP was incubated with either GST or GST-p32 prebound to glutathione-Sepharose beads. The beads were washed, and bound proteins were eluted and analyzed by SDS-PAGE and fluorography. In this assay 35S-labeled capsid was bound by GST-p32 but not by GST (Fig. 1, lanes 2 and 3). 35S-labeled GFP, which served as a negative control, was bound by neither GST-p32 nor GST (Fig. 1, lanes 6 and 7). We were concerned that the interaction between the capsid, which contains a large basic region, and p32, which is acidic, might be the result of nonspecific electrostatic interactions. This was shown not to be the case, since capsid did not bind to glucose oxidase, which, like p32, is an acidic soluble protein (Fig. 1, lane 4). Together, these results demonstrate that the capsid interaction with p32 is specific.
FIG. 1.
Capsid binds to p32 in vitro. Sepharose beads coated with GST (lanes 2 and 6), GST-p32 (lanes 3 and 7), or glucose oxidase (GOD) (lanes 4 and 8) were mixed with 35S-labeled capsid (lanes 2 to 4) or GFP (lanes 6 to 8). The beads were washed, and bound proteins were eluted and visualized by SDS-PAGE and fluorography. Five percent of the capsid and GFP in vitro translation reaction mixtures were loaded in lanes 1 and 5, respectively.
Capsid and p32 interact in vivo.
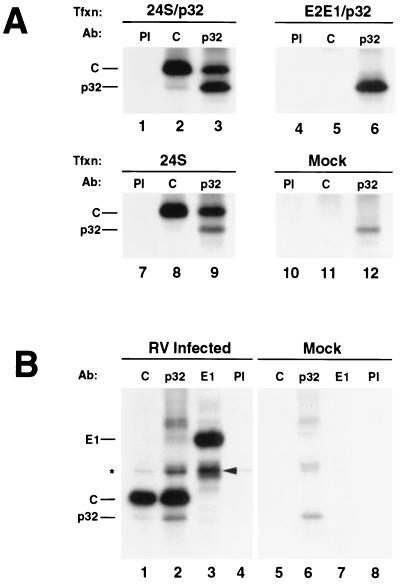
Coimmunoprecipitation studies were used to determine if capsid interacts with p32 in vivo. COS cells were cotransfected with expression vectors encoding p32 and 24S (capsid, E2, and E1) or E2-E1 (E2 and E1). Alternatively, cells were transfected with the 24S or p32 expression vectors alone. At 40 h posttransfection, cells were biosynthetically labeled with [35S]methionine-cysteine and cell lysates were prepared using nondenaturing conditions. Lysates were subjected to radioimmunoprecipitation with antibodies specific for capsid or p32, and the immune complexes were analyzed by SDS-PAGE and fluorography. Capsid-p32 complexes were coimmunoprecipitated from cells expressing all three RV structural proteins, using antibodies to capsid or p32 (Fig. 2A, lanes 2, 3, and 9). Neither capsid nor p32 were immunoprecipitated by preimmune rabbit serum (Fig. 2A, lanes 1 and 7). For negative controls, the immunoprecipitations were performed on E2-E1- or p32-transfected and mock-transfected COS cells (Fig. 2A, lanes 4 to 6 and 10 to 12). Capsid was not immunoprecipitated from these lysates, and p32 was immunoprecipitated only using anti-p32 (Fig. 2A, lane 6). To confirm that the capsid-p32 interaction is relevant in the context of viral infection, immunoprecipitations were repeated using lysates prepared from RV-infected Vero cells. Similar to the results obtained using transfected cells, capsid and p32 were coimmunoprecipitated using antibodies specific for capsid or p32 in a reciprocal fashion from infected cells but not from mock-infected cells (Fig. 2B, lanes 1, 2, 5, and 6). In addition, neither capsid nor p32 was immunoprecipitated by preimmune rabbit serum or a monoclonal antibody specific for E1 (Fig. 2B, lanes 3, 4, 7 and 8). Although capsid and p32 coimmunoprecipitated in a reciprocal fashion, the amount of p32 immunoprecipitated with anticapsid antibodies was much lower than the amount of capsid immunoprecipitated with anti-p32 antibodies. One possibility is that binding of the polyclonal antibodies to capsid results in disruption of the capsid-p32 complexes. However, given that very small amounts of p32 were also coimmunoprecipitated when the experiments were performed using several different anticapsid monoclonal antibodies (data not shown), this scenario seems unlikely.
FIG. 2.
Capsid and p32 associate in vivo. (A) COS cells were transfected (Tfxn) with expression vectors encoding the RV structural proteins (24S) and p32 (lanes 1 to 3), the RV envelope proteins (E2-E1) and p32 (lanes 4 to 6), or 24S alone (lanes 7 to 9) or were mock transfected (lanes 10 to 12). At 24 h posttransfection the cells were biosynthetically labeled with [35S]methionine-cysteine, lysed and then subjected to immunoprecipitation with preimmune serum (PI) (lanes 1, 4, 7, and 10), or antibodies (Ab) specific for capsid (lanes 2, 5, 8, and 11) or p32 (lanes 3, 6, 9, and 12). Immune complexes were subjected to SDS-PAGE and fluorography. (B) Vero cells were infected or mock infected with RV at a multiplicity of infection of 1. At 48 h postinfection, the cells were biosynthetically labeled with [35S]methionine-cysteine. Immunoprecipitations were performed using lysates from infected cells (lanes 1 to 4) and mock-infected cells (lanes 5 to 8) as described above, using antibodies specific for capsid (lanes 1 and 5), p32 (lanes 2 and 6), E1 (lanes 3 and 7), or preimmune serum (lanes 4 and 8). The positions of E1, capsid, and p32 are indicated to the left of the gels. E2 which coprecipitates with E1 (lane 3) is indicated with an arrowhead. The asterisk indicates an unknown protein that comigrates with E2 in lanes 1, 2, and 4.
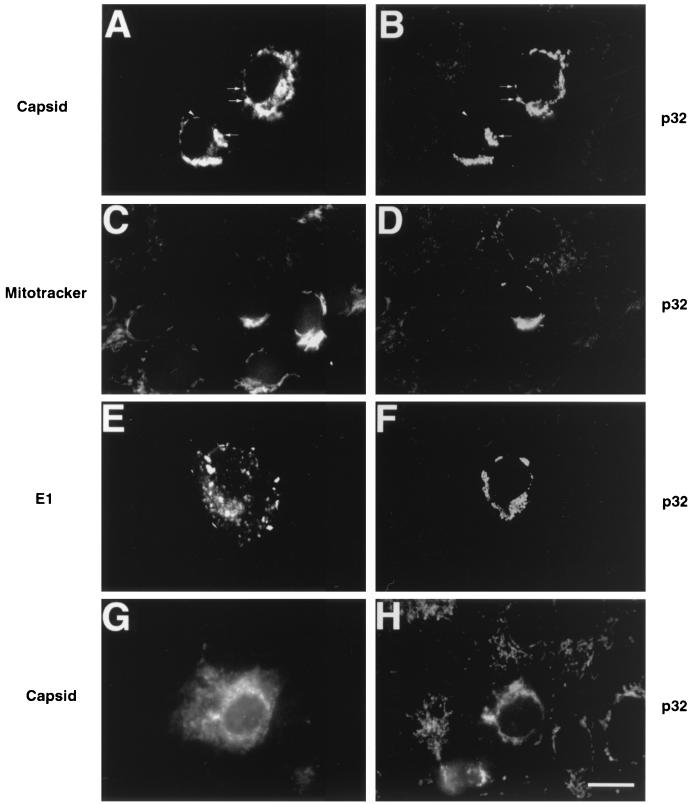
Indirect immunofluorescence microscopy was used to determine the intracellular site(s) of capsid-p32 interaction in transiently transfected cells. In cells expressing RV structural proteins and p32, a significant proportion of capsid colocalized with p32 to cytoplasmic vesicular structures in the perinuclear region (Fig. 3A and B, arrows). These structures were identified as mitochondria, since p32 colocalized with HSP60, a resident mitochondrial protein (data not shown), and the mitochondrion-specific dye Mitotracker Red CXMRos (Fig. 3C and D). No colocalization was observed between p32 and the RV envelope protein E1 (Fig. 3E and F). Capsid also colocalized with p32 to the mitochondria in cells transfected with 24S alone, showing that these results are not due to overexpression of p32 (Fig. 3G and H). Interestingly, the mitochondria in cells expressing RV structural proteins were more spherical and were clustered in the perinuclear region, in contrast to those in nontransfected cells, which were more lacy and peripherally localized (Fig. 3B, F, and H). This effect was specific for RV structural proteins, since cells overexpressing p32 alone did not exhibit this phenotype (data not shown).
FIG. 3.
Capsid colocalizes with p32 at the mitochondria. Vero cells were transfected with expression vectors encoding the RV structural proteins (24S) and p32 (A to F) or 24S alone (G and H). Cells were incubated with antibodies to capsid (A and G), p32 (B, D, F, and H) or E1 (E). For samples shown in panels E and F, mitochondria were labeled prior to fixation with Mitotracker Red (C) followed by staining with anti-p32. Primary antibodies were detected with Texas Red-conjugated donkey anti-mouse IgG and FITC-conjugated donkey anti-rabbit IgG. The Texas Red channel is shown on the left (A, C, E, and G), and the FITC channel is shown on the right (B, D, F, and H). Areas of colocalization are shown by arrows, whereas the arrowhead indicates a perinuclear pool of capsid which does not overlap with p32. Bar, 20 μm.
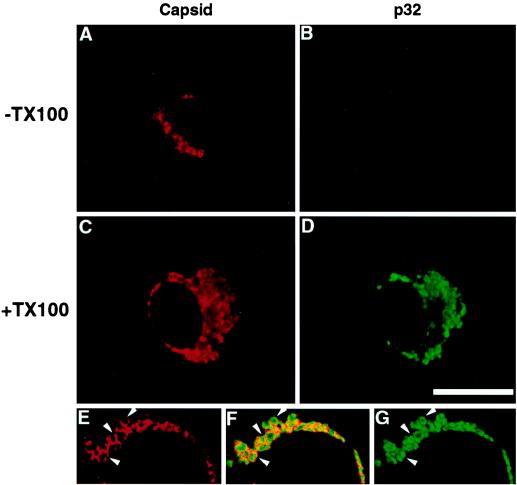
To determine whether p32 and capsid interact on the cytoplasmic surface or within the matrix of the mitochondria, cells were fixed and treated with digitonin to permeabilize only the plasma membrane or with digitonin followed by Triton X-100 to permeabilize intracellular membranes. Using this method, mitochondrial staining of capsid was observed in both digitonin and digitonin- plus Triton X-100-permeabilized cells (Fig. 4A and C). In contrast, p32 was only visible in digitonin- and Triton X-100-treated cells. These results indicate that p32, but not capsid, is translocated into the mitochondria, and they can potentially explain why the distributions of these two proteins are slightly different (Fig. 3 and 4). This conclusion is supported by confocal microscopy analysis, which showed that anticapsid and anti-p32 antibodies stained different regions of the mitochondria (Fig. 4E, F, and G). Capsid staining was often localized to the periphery of mitochondria (Fig. 4E), whereas p32 staining was more central and extended into the interior of these structures (Fig. 4G).
FIG. 4.
Capsid is associated with the cytoplasmic side of the mitochondria. Vero cells were transfected with expression vectors encoding the RV structural proteins (24S) and p32, and fixed with 3% paraformaldehyde. (A to D) The plasma membranes were permeabilized with digitonin (A and B). To permeabilize intracellular membranes, cells (C and D) were treated with Triton X-100 (+TX100). Cells were double labeled with antibodies specific for capsid (A and C) and p32 (B and D). Primary antibodies were detected with Texas Red-conjugated donkey anti-mouse IgG and FITC-conjugated donkey anti-rabbit IgG. The Texas Red channel is shown on the left (A and C), and the FITC channel is shown on the right (B and D). Bar, 20 μm. (E to G) Confocal images of Triton X-100-treated cells. The arrowheads in panel E indicate capsid staining at the periphery of mitochondria which does not coincide with p32 staining (G). Panel F is a merge of the images shown in panels E and G.
The amino terminus of capsid binds p32.
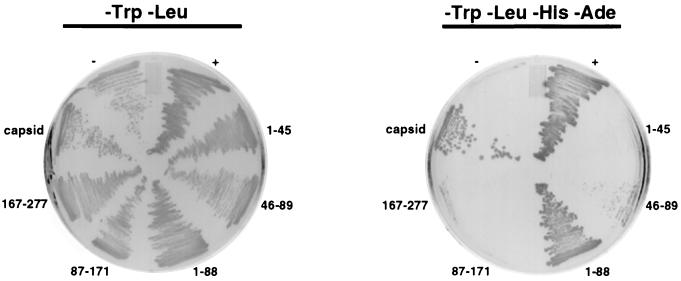
We used the yeast two-hybrid system to identify the region of capsid that binds p32. Three different but similarly sized regions of capsid cDNA were amplified by PCR and subcloned into GAL4 DNA binding domain plasmids (pGBKT7 or pGBT9). The capsid constructs were cotransfected into AH109 cells and tested for the ability to grow on medium lacking adenine, leucine, tryptophan, and histidine. These interactions were further tested using the β-galactosidase filter assay. Full-length capsid and the amino-terminal region of capsid (amino acids 1 to 88) interacted with p32 in these assays (Fig. 5). In contrast, the middle and carboxy-terminal regions of capsid (amino acids 87 to 171 or 167 to 277, respectively) did not interact with p32. The amino-terminal 88 amino acids of capsid were subdivided into two smaller regions and tested using the same assays. A weak interaction between amino acids 46 to 89 of capsid and p32 occurred as evidenced by the presence of smaller, slow-growing colonies on medium lacking adenine, leucine, tryptophan, and histidine (Fig. 5). In contrast, no growth was observed when a construct encoding capsid residues 1 to 45 was used (Fig. 5). These results indicate that the minimal region of capsid which can interact with p32 is located within amino acids 46 to 89.
FIG. 5.
The p32-binding site is located in the amino-terminal region of capsid. AH109 yeast cells were transformed with pGAD-p32 and GAL4 DNA binding domain plasmids encoding full-length capsid (pGBKT7-capsid) or portions of capsid. The capsid constructs are named according to the amino acids in capsid which they encode. For example, 1–45 is pGBKT7 plus the coding region for amino acids 1 to 45 of capsid. The transformants were plated onto medium lacking tryptophan and leucine (-Trp-Leu) or medium lacking tryptophan, leucine, histidine, and adenine (-Trp-Leu-His-Ade). The positive control (+) is a Clontech system control that utilizes two strongly interacting proteins, p53 (in pGBKT7) and simian virus 40 large T antigen (in pGAD). As a negative control (−), AH109 was transformed with two plasmids which do not interact, pGBKT7-p53 and pGAD-ABP280. Growth on the -Trp-Leu-His-Ade plates is indicative of a strong interaction between the two proteins being tested.
DISCUSSION
In this study we have shown that RV capsid interacts with the mitochondrial protein p32. A number of in vitro and in vivo methods were used to confirm the interaction between these two proteins. Our data indicate that the interaction takes place at the mitochondria. In light of the recent discovery that RV nucleocapsids associate with mitochondria in infected Vero cells (25), it seems quite likely that capsid-p32 interactions may be important for this process. However, not all of the capsid was associated with p32, since a significant proportion of this protein was not localized to the mitochondria (Fig. 3). This is to be expected, since much of the capsid will be engaged in productive virus assembly, in which case it must be recruited to the site of virus budding, the Golgi (17).
p32 was originally identified through its association with the human mRNA splicing factor ASF/SF2 (23). The physiological function of p32 is not known, but it has been shown to complex with other cellular and viral proteins (for a complete list, see reference 32). The binding of p32 to several of these proteins has been shown to regulate their intermolecular interactions by modulating their phosphorylation states. For example, p32 has an inhibitory effect on the phosphorylation of ASF/SF2 which in turn regulates its binding to both RNA and other proteins (43). In addition, p32 is thought to regulate interactions between members of the lamin B receptor complex in a manner which is also dependent on phosphorylation of lamin B receptor (36). The common feature among p32-binding proteins is that they are phosphoproteins, which in many cases bind to nucleic acids and other proteins. RV capsid is also a phosphoprotein (12, 30), and the p32-binding region (amino acids 45 to 89) contains several potential phosphorylation sites as predicted using the PROSITE algorithm (1). This region of capsid also contains part of the genomic RNA-binding domain (28). Interestingly, the RNA-binding activity of hepatitis B virus capsid protein is modulated by phosphorylation (22). By analogy, p32 may regulate the binding of genomic RNA to RV capsid by modulating its level of phosphorylation. This in turn may have an effect upon capsid oligomerization and, ultimately, virus assembly.
Previous studies are consistent with the possibility that p32 is able to leave the mitochondria and bind to other proteins in such places as the nucleus and the cell surface (13, 32). Indeed, numerous other mitochondrial proteins have been shown to leave this organelle under certain conditions (50). However, in the present study we observed that the majority of p32 is localized to the mitochondria and that the interaction with capsid occurs at this location. p32 contains a 74-amino-acid amino-terminal domain which is required for targeting to the mitochondria (35). During translocation into the mitochondrial matrix, this amino-terminal domain is cleaved to yield the mature form of p32. Our data suggest that capsid associates with the mature form of p32 but is not translocated into the mitochondria. This observation is consistent with those of Lee et al. (25), who showed that capsid antibodies stain the cytoplasmic face of mitochondrial membranes in RV-infected cells. Accordingly, we hypothesize that capsid binds to the carboxy terminus of newly synthesized p32 in the cytoplasm, leaving the amino-terminal region of p32 available to target the complex to mitochondria. Upon docking at this organelle, the bulk of p32 is translocated into mitochondria, where cleavage of the leader peptide occurs. Capsid remains bound to the cytoplasmic side of the mitochondria still attached to the carboxy terminus of p32. Alternatively, capsid may associate with mitochondria independently where it binds to a pool of p32 as it leaves this organelle. This scenario seems unlikely, since there is no obvious mechanism to target capsid to this organelle independently of p32.
The localization of capsid to the mitochondria and association with a mitochondrial protein is intriguing in light of previous studies which document a link between RV replication complexes and mitochondrial function and localization (2, 3, 24, 25). In RV-infected cells, clustering of mitochondria occurs and electron-dense plaques form between the membranes of apposing mitochondria and between mitochondria that associate with the endoplasmic reticulum. Moreover, the mitochondria cluster around RV replication sites which are modified endosomes or lysosomes and are located in the perinuclear region (29). This phenomenon is unique to RV and does not occur with other togaviruses. Lee et al. proposed that recruitment of the mitochondria to RV replication sites could provide the energy required for virus replication (24). It follows that the capsid-p32 interactions may play a role in the formation of these plaques and/or association of mitochondria with RV replication sites. In the present study we showed that clustering of mitochondria in the perinuclear region requires expression of RV structural proteins but does not require virus replication.
Interactions between viral capsids and host cell proteins can also have important consequences for viral pathogenesis and the host immune response (5, 10, 31, 53, 54). Several recent studies have shown that RV is able to induce apoptosis in certain types of cultured cells (9, 19, 33, 45). Interestingly, similar to expression of RV structural proteins, translocation of proapoptotic proteins such as truncated Bid to mitochondria causes clustering of these organelles in the perinuclear region prior to breakdown of mitochondrial membrane integrity (44). This is particularly intriguing in light of the hypothesis that p32 regulates opening of the permeability transition pore of the mitochondrial inner membrane (21), a process which is known to have a critical role in apoptosis (15). In this regard, p32 may act as a calcium buffer that modulates the concentration of divalent metal ions and consequently the permeability of mitochondria. Accordingly, binding to capsid may affect the ability of p32 to act as a calcium buffer. At this point it is unclear whether the interaction between p32 and capsid is pro- or antiapoptotic, and resolving this issue may not be straightforward given that numerous virus proteins have been shown to have both pro- and antiapoptotic functions (46). Nevertheless, if capsid-p32 complexes are involved in apoptosis, a more detailed study of how these two proteins interact should provide important insight into how RV initiates persistent infections or congenital defects.
ACKNOWLEDGMENTS
We thank Barbara Pustowoit, Shirley Gillam, Christian Oker-Blom, John Safford, and Willie Russell for their generous gifts of reagents. We also thank Bruce Stevenson for critical reading of the manuscript.
This work was supported by a grant from the Medical Research Council of Canada. M.D.B. is the recipient of a graduate studentship award from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Appel R D, Bairoch A, Hochstrasser D F. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem Sci. 1994;19:258–260. doi: 10.1016/0968-0004(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 2.Bardeletti G. Respiration and ATP level in BHK21/13S cells during the earliest stages of rubella virus replication. Intervirology. 1977;8:100–109. doi: 10.1159/000148884. [DOI] [PubMed] [Google Scholar]
- 3.Bardeletti G, Gautheron D C. Phospholipid and cholesterol composition of rubella virus and its host cell BHK 21 grown in suspension cultures. Arch Virol. 1976;52:19–27. doi: 10.1007/BF01317861. [DOI] [PubMed] [Google Scholar]
- 4.Beatch M, Hobman T C. Proceedings of the Fifth International Symposium on Positive Strand RNA Viruses. 1998. Association of rubella virus capsid protein with host cell proteins; pp. 1–39. [Google Scholar]
- 5.Chen C-M, You L-R, Hwang L H, Lee Y-H W. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-β receptor modulates the signal pathway of the lymphotoxin-β receptor. J Virol. 1997;71:9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cikaluk D E, Tahbaz N, Hendricks L C, DiMattia G E, Hansen D, Pilgrim D, Hobman T C. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic messenger RNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Meco M T, Municio M M, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- 9.Duncan R, Muller J, Lee N, Esmaili A, Nakhasi H L. Rubella virus-induced apoptosis varies among cell lines and is modulated by Bcl-XL and caspase inhibitors. Virology. 1999;255:117–128. doi: 10.1006/viro.1998.9562. [DOI] [PubMed] [Google Scholar]
- 10.Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7:160–165. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 11.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garbutt M, Law L M, Chan H, Hobman T C. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J Virol. 1999;73:3524–3533. doi: 10.1128/jvi.73.5.3524-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghebrehiwet B, Lim B L, Peerschke E I, Willis A C, Reid K B. Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J Exp Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarente L. Strategies for the identification of interacting proteins. Proc Natl Acad Sci USA. 1993;90:1639–1641. doi: 10.1073/pnas.90.5.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiden M G V, Thompson C B. Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis? Nat Cell Biol. 1999;1:E209–E216. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 16.Hobman T C, Gillam S. In vitro and in vivo expression of rubella virus E2 glycoprotein: the signal peptide is located in the C-terminal region of capsid protein. Virology. 1989;173:241–250. doi: 10.1016/0042-6822(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 17.Hobman T C, Lundstrom M L, Gillam S. Processing and intracellular transport of rubella virus structural proteins in COS cells. Virology. 1990;178:122–133. doi: 10.1016/0042-6822(90)90385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobman T C, Woodward L, Farquhar M G. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J Cell Biol. 1993;121:269–281. doi: 10.1083/jcb.121.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann J, Pletz M W, Liebert U G. Rubella virus-induced cytopathic effect in vitro is caused by apoptosis. J Gen Virol. 1999;80:1657–1664. doi: 10.1099/0022-1317-80-7-1657. [DOI] [PubMed] [Google Scholar]
- 20.Honore B, Madsen P, Rasmussen H H, Vandekerckhove J, Celis J E, Leffers H. Cloning and expression of a cDNA covering the complete coding region of the P32 subunit of human pre-mRNA splicing factor SF2. Gene. 1993;134:283–287. doi: 10.1016/0378-1119(93)90108-f. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Zhang Y, Krainer A R, Xu R M. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kann M, Gerlich W H. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol. 1994;68:7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 24.Lee J Y, Bowden D S, Marshall J A. Membrane junctions associated with rubella virus infected cells. J Submicrosc Cytol Pathol. 1996;28:101–108. [PubMed] [Google Scholar]
- 25.Lee J Y, Marshall J A, Bowden D S. Localization of rubella virus core particles in vero cells. Virology. 1999;265:110–119. doi: 10.1006/viro.1999.0016. [DOI] [PubMed] [Google Scholar]
- 26.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Dai R, Tian C J, Dawson L, Gorelick R, Yu X F. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Yang D, Qiu Z, Lim K T, Chong P, Gillam S. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J Virol. 1996;70:2184–2190. doi: 10.1128/jvi.70.4.2184-2190.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 30.Marr L D, Sanchez A, Frey T K. Efficient in vitro translation and processing of the rubella virus structural proteins in the presence of microsomes. Virology. 1991;180:400–405. doi: 10.1016/0042-6822(91)90046-e. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto M, Hsieh T Y, Zhu N, VanArsdale T, Hwang S B, Jeng K S, Gorbalenya A E, Lo S Y, Ou J H, Ware C F, Lai M M. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews D A, Russell W C. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 33.Megyeri K, Berencsi K, Halazonetis T D, Prendergast G C, Gri G, Plotkin S A, Rovera G, Gonczol E. Involvement of a p53-dependent pathway in rubella virus-induced apoptosis. Virology. 1999;259:74–84. doi: 10.1006/viro.1999.9757. [DOI] [PubMed] [Google Scholar]
- 34.Melancon P, Garoff H. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J Virol. 1987;61:1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 36.Nikolakaki E, Simos G, Georgatos S D, Giannakouros T. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J Biol Chem. 1996;271:8365–8372. doi: 10.1074/jbc.271.14.8365. [DOI] [PubMed] [Google Scholar]
- 37.Nolandt O, Kern V, Muller H, Pfaff E, Theilmann L, Welker R, Krausslich H G. Analysis of hepatitis C virus core protein interaction domains. J Gen Virol. 1997;78:1331–1340. doi: 10.1099/0022-1317-78-6-1331. [DOI] [PubMed] [Google Scholar]
- 38.Oker-Blom C, Kalkkinen N, Kaariainen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostendorp R, Kuhn R, Rossman M. Proceedings of the Fifth International Symposium on Positive Strand RNA Viruses. 1998. Capsid proteins from rubella virus and hepatitis C virus: an efficient strategy for isolating proteins with high isoelectric points; pp. P1–38. [Google Scholar]
- 40.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 41.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patwardhan S, Gashler A, Siegel M G, Chang L C, Joseph L J, Shows T B, Le Beau M M, Sukhatme V P. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- 43.Petersen-Mahrt S K, Estmer C, Ohrmalm C, Matthews D A, Russell W C, Akusjarvi G. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter A G. Protein translocation in apoptosis. Trends Cell Biol. 1999;9:394–401. doi: 10.1016/s0962-8924(99)01624-4. [DOI] [PubMed] [Google Scholar]
- 45.Pugachev K V, Frey T K. Rubella virus induces apoptosis in culture cells. Virology. 1998;250:359–370. doi: 10.1006/viro.1998.9395. [DOI] [PubMed] [Google Scholar]
- 46.Roulston A, Marcellus R C, Branton P E. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt M, Tuominen N, Johansson T, Weiss S A, Keinanen K, Oker-Blom C. Baculovirus-mediated large-scale expression and purification of a polyhistidine-tagged rubella virus capsid protein. Protein Expr Purif. 1998;12:323–330. doi: 10.1006/prep.1997.0851. [DOI] [PubMed] [Google Scholar]
- 48.Sells S F, Han S S, Muthukkumar S, Maddiwar N, Johnstone R, Boghaert E, Gillis D, Liu G, Nair P, Monnig S, Collini P, Mattson M P, Sukhatme V P, Zimmer S G, Wood D P, Jr, McRoberts J W, Shi Y, Rangnekar V M. Expression and function of the leucine zipper protein Par-4 in apoptosis. Mol Cell Biol. 1997;17:3823–3832. doi: 10.1128/mcb.17.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sells S F, Wood D P, Jr, Joshi-Barve S S, Muthukumar S, Jacob R J, Crist S A, Humphreys S, Rangnekar V M. Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Differ. 1994;5:457–466. [PubMed] [Google Scholar]
- 50.Soltys B J, Gupta R S. Mitochondrial-matrix proteins at unexpected locations: are they exported? Trends Biochem Sci. 1999;24:174–177. doi: 10.1016/s0968-0004(99)01390-0. [DOI] [PubMed] [Google Scholar]
- 51.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suomalainen M, Garoff H, Baron M D. The E2 signal sequence of rubella virus remains part of the capsid protein and confers membrane association in vitro. J Virol. 1990;64:5500–5509. doi: 10.1128/jvi.64.11.5500-5509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.You L R, Chen C M, Lee Y H W. Hepatitis C virus core protein enhances NF-kappaB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J Virol. 1999;73:1672–1681. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai M M. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–3697. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]