Abstract
Hepatitis C virus (HCV) of genotype 1 is the most resistant to interferon (IFN) therapy. Here, we have analyzed the response to IFN of the human cell line UHCV-11 engineered to inducibly express the entire HCV genotype 1a polyprotein. IFN-treated, induced UHCV cells were found to better support the growth of encephalomyocarditis virus (EMCV) than IFN-treated, uninduced cells. This showed that expression of the HCV proteins allowed the development of a partial resistance to the antiviral action of IFN. The nonstructural 5A (NS5A) protein of HCV has been reported to inhibit PKR, an IFN-induced kinase involved in the antiviral action of IFN, at the level of control of protein synthesis through the phosphorylation of the initiation factor eIF2α (M. Gale, Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze, Mol. Cell. Biol. 18:5208–5218, 1998). Accordingly, cell lines inducibly expressing NS5A were found to rescue EMCV growth (S. J. Polyak, D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch, Hepatology 29:1262–1271, 1999). In the present study we analyzed whether the resistance of UHCV-11 cells to IFN could also be attributed to inhibition of PKR. Confocal laser scanning microscopy showed no colocalization of PKR, which is diffuse throughout the cytoplasm, and the induced HCV proteins, which localize around the nucleus within the endoplasmic reticulum. The effect of expression of HCV proteins on PKR activity was assayed in a reporter assay and by direct analysis of the in vivo phosphorylation of eIF2α after treatment of cells with poly(I)-poly(C). We found that neither PKR activity nor eIF2α phosphorylation was affected by coexpression of the HCV proteins. In conclusion, expression of HCV proteins in their biological context interferes with the development of the antiviral action of IFN. Although the possibility that some inhibition of PKR (by either NS5A or another viral protein) occurs at a very localized level cannot be excluded, the resistance to IFN, resulting from the expression of the HCV proteins, cannot be explained solely by inhibition of the negative control of translation by PKR.
Hepatitis C virus (HCV), a member of the family Flaviviridae, is a major causative agent of chronic hepatitis that can progress to cirrhosis and hepatocellular carcinoma (44). Interferon (IFN) therapy, in combination with ribavirin, is used worldwide as the best treatment so far for HCV infection. However, IFN treatment is of limited long-term efficacy, and selection of viral variants resistant to IFN can occur from the onset of infection. By complete direct sequencing and comparison of HCV genotype 1b (HCV-1b) genomes in three transient nonresponder patients before and during treatment, Enomoto et al. (11) identified clusters of nucleotide differences in the central part of the NS5A gene, more specifically in the region encoding NS5A amino acids (aa) 2209 to 2248, which was named the interferon sensitivity-determining region (ISDR). Since this observation, extensive studies of the NS5A sequence have been performed by different groups, including ours (25); discrepancies among studies conducted on patient cohorts of Japanese or European origin have brought into question the importance of the ISDR region. Sequencing of larger regions extending into the carboxy-terminal domain of NS5A revealed in addition a differential variability between IFN-sensitive and IFN-resistant HCV strains (10, 25). All of these studies, however, implicate NS5A as being a factor in the resistance of HCV to IFN. Interestingly, NS5A of HCV genotypes 1a and 1b was reported to interact with and inhibit the double-stranded RNA-dependent protein kinase PKR (13, 14). PKR is a serine/threonine protein kinase which is present in most cells at basal levels and which can be induced by IFN treatment (7). Its best-characterized substrate is the α subunit of the eukaryotic initiation factor eIF2, whose phosphorylation leads to inhibition of protein synthesis (18). PKR was shown to phosphorylate eIF2α in vitro (for a review, see reference 20) and in vivo, either in yeast cells (6) or during a viral infection in murine clones stably expressing PKR (33). The use of these two in vivo systems firmly established that PKR-mediated phosphorylation of eIF2α was directly responsible for a strong cellular growth inhibition and for inhibition of viral growth, two properties related to the action of IFN. In addition, PKR was reported to function as a tumor suppressor (2, 26, 31). From a series of studies involving two-hybrid assays, both in the yeast Saccharomyces cerevisiae and in bacteria, and different deletion mutants, NS5A was reported to inhibit the action of PKR and a direct interaction was suggested to exist between the aa 2209 to 2274 region of NS5A, including the ISDR, and the central part of PKR, which is necessary for its dimerization and subsequent activation as a kinase (13, 14). Disruption of the ISDR conformation due to mutations has been suggested to restore PKR function, probably because of abrogation of the interaction between PKR and NS5A. The ability of some viral strains to resist IFN action, and therefore, to lead to malignant transformation and to hepatocellular carcinoma, has been attributed, at least in part, to the ability of PKR and NS5A to interact, depending on variations in the ISDR sequence. This possibility is reminiscent of the situation observed with other viruses, such as human immunodeficiency virus (HIV), influenza virus, and reovirus, which have been reported to encode proteins that inhibit PKR (7). Recently, another viral HCV protein, E2, has been reported to behave as an inhibitor of PKR, emphasizing the importance of PKR in the development of the cellular antiviral response (43).
The studies conducted by Gale et al. showing that PKR and NS5A interact were based on NS5A proteins of genotypes 1a and 1b expressed either in S. cerevisiae or in mammalian cells as well as on in vitro coprecipitation analyses. However, in a natural cycle of HCV infection, NS5A, which is processed from the HCV polyprotein, presumably exists in the cell as a complex with other HCV proteins. As in the case of the pestiviruses, it is thought to establish a molecular complex with the other nonstructural proteins to form the replication complex. It is therefore of importance to determine the functional interactions of PKR and NS5A in the biological context in which all HCV proteins are expressed.
The single-stranded positive-sense RNA genome of HCV encodes a polyprotein of 3,010 to 3,033 aa which is processed co- and posttranslationally into structural and nonstructural proteins (29). Recently, a continuous human cell line inducibly expressing the structural and nonstructural proteins derived from the prototype HCV-H strain (genotype 1a) was established (34). It provided a good approach to study the NS5A-PKR interaction since no efficient cell culture system for HCV infection is available yet.
Here we show that expression of HCV proteins in their context allows the cells to develop partial resistance to the antiviral action of IFN. We found no evidence, however, of inhibition of PKR activity as a result of the expression of the HCV proteins. In agreement with this, confocal-microscopy analysis showed different patterns of localization of PKR and the HCV proteins in the cytoplasm. Therefore, the development of resistance to the antiviral action of IFN in cells expressing the HCV proteins may involve either interaction of PKR with these proteins at a very localized level or another mechanism.
MATERIALS AND METHODS
Plasmids.
The plasmid pcDNA1/Amp expressing PKR has been previously described (32). The plasmid pHIV1 LTR-Luc, corresponding to the AvaI-HindIII region of the HIV type 1 long terminal repeat (LTR) (9), was provided by N. Israel. The plasmid pcDNA1/Amp (TRBP2) was constructed by inserting the BamHI fragment from pBS TRBP2 encoding the full-length TAR RNA binding protein 2 (TRBP2) (16).
Cell lines.
The tetracycline-regulated UHCV-11 cell line (34) was cultured in GlutaMAXI-Dulbecco's modified Eagle medium (DMEM) (with 110 mg of sodium pyruvate/liter and 1,000 mg of glucose/liter; GIBCO BRL) containing 10% heat-inactivated fetal bovine serum, 50 U of penicillin G/ml, 50 μg of streptomycin/ml, 500 μg of G418 (Geneticin; Sigma)/ml, 1 μg of puromycin (Sigma)/ml, and 1 μg of tetracycline (Sigma)/ml. Simian Vero VC10 cells were grown in GlutaMAXI-DMEM (without sodium pyruvate but with 4,500 mg of glucose/ml and 4 mg of pyridoxine/liter) supplemented with penicillin-streptomycin (as described above) and 5% fetal calf serum. Human Daudi cells were grown in GlutaMAXI-RPMI 1640 (GIBCO) supplemented with penicillin-streptomycin (as described above), 10% fetal calf serum, and 10 μM 2-mercaptoethanol.
Antibodies.
Monoclonal antibodies (MAbs) directed against PKR (MAb 71/10) or the p69 (MAb 56/3) or p100 (MAb 25/11) isoform of the 2-5A synthetase have been previously described (21, 28). Rabbit polyclonal antibodies directed against NS5A were a gift from Hoffmann-La Roche (Basel, Switzerland). Sera of chronically HCV-infected patients were used as a source of antibodies as well. Rabbit polyclonal antibodies directed against a synthetic 13-residue phosphorylated rat eIF2α peptide were obtained from Research Genetics (Huntsville, Ala.). Mouse polyclonal antibodies directed against eIF2 were a gift of C. Proud. Anti-mouse and anti-rabbit polyclonal antibodies conjugated to either fluorescein isothiocyanate or Texas red were obtained from Caltag (San Francisco, Calif.).
Immunoprecipitation and immunoblotting.
The anti-PKR MAb 71/10 was incubated for 60 min at room temperature with protein A- and protein G-agarose (Protein A/GPlus-Agarose; Santa Cruz Biotechnologies) (10 μl of MAb per 500 μl of beads) in 2 ml of buffer I (20 mM Tris-HCl [pH 7.6], 50 mM KCl, 400 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5 mM 2-mercaptoethanol, 0.05% aprotinin, 0.2 mM phenylmethylsulfonyl fluoride, 20% glycerol). The antibody-bead mixture was then washed twice with buffer I and distributed into Eppendorf tubes at 30 μl of bead mixture per tube. UHCV-11 cell extracts were prepared in buffer I and incubated with the antibody-bead mixture overnight at 4°C. After three cycles of centrifugation (4°C, 3,000 × g) and resuspension in buffer I, the samples were processed for in vitro phosphorylation. For immunoblotting, crude extracts were resuspended in an equal volume of 2× protein electrophoresis buffer (75 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 100 mM 2-mercaptoethanol, 20% glycerol and bromophenol blue as tracking dye) and the proteins were separated by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE). The proteins were then transferred to Immobilon polyvinylidene difluoride membranes (Millipore). The membranes were saturated for 60 min at 37°C in phosphate-buffered saline (PBS) containing 5% nonfat dry milk and incubated overnight at 4°C with the primary antibodies in PBS containing 5% fetal bovine serum and 0.1% Tween-20. The membranes were then washed successively with PBS, PBS–0.5% NP-40, and PBS again and saturated (60 min, room temperature) in PBS containing 5% nonfat dry milk and 0.4% Tween 20. The membranes were then washed three times in PBS–0.5% Tween 20 and incubated for 1 h at room temperature in the same buffer with the appropriate secondary antibodies coupled to horseradish peroxidase (dilution, 1/2,000; Amersham, Arlington Heights, Ill.). After extensive washes in PBS–0.4% Tween 20, the membranes were processed for enhanced chemiluminescence according to the Amersham protocol.
In vitro phosphorylation of PKR.
Immunoprecipitated PKR (30 μl of beads) which had been washed with buffer I was further washed twice with buffer II (20 mM Tris-HCL [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 0.05% aprotinin, 20% glycerol) and once in buffer III (buffer II supplemented with 2 mM MgCl2). For the phosphorylation reaction, each sample was incubated with 40 μl of buffer III supplemented with 2 mM MnCl2 and 10 μl of [γ-32P]ATP solution (buffer II plus 1.25 mCi of [γ-32P]ATP [3,000 Ci/mmol; ICN], 10 μM ATP, and 1 mM MgCl2). The reaction was performed in the absence or in the presence of heparin (Sigma) or poly(I)-poly(C) (Amersham Pharmacia Biotech). After incubation for 15 min at 30°C, 2 μl (165 ng) of a pure preparation of rabbit eIF2 complex (a gift of C. Proud) was added and the reaction was continued for another 15 min. An equal volume of 2× protein electrophoresis buffer was added, and the products were analyzed by SDS–12.5% PAGE.
PKR functional assay by microtransfection.
Eighteen to 24 h before transfection, the cells were seeded at a density of 20,000/well in 96-well microplates (Costar) (200 μl of complete medium per well). Three hours before the transfection was performed, the medium was aspirated and replaced with fresh medium. The desired amounts of plasmids, adjusted to the same final concentration by addition of the empty vector pcDNA1/Amp, were transfected into the cells by a calcium chloride precipitation-glycerol shock technique. Forty-eight hours after the transfection, the culture medium was aspirated and the cells were washed twice with PBS. The cells in each well were lysed in 140 μl of Luc lysis buffer (25 mM Tris-phosphate [pH 7.6], 8 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100, 15% glycerol). A 100-μl volume of each sample was analyzed for luciferase activity in a luminometer (Lumat; Wallac) by being automatically mixed with 100 μl of Luc lysis buffer solution containing 0.25 mM luciferin (Sigma), 1 mM ATP, and 1% bovine serum albumin. The protein contents of all samples were measured in 96-well microplates, using a Bio-Rad Bradford kit, and the absorbance at 590 nm was determined (PR 2100 microplate reader; Sanofi Pasteur). Each transfection was performed in four different wells, and the results presented below are averages of the four values obtained.
Viral yield assay.
UHCV-11 cells were plated at a density of 5 × 105 per well in six-well plates. After 18 h of incubation at 37°C, cells were left untreated or were treated with 500 U of human alpha IFN (IFN-α; Hayashibara Biochem Labs, Okayama, Japan)/ml; in both cases the cells were incubated in the presence or absence of tetracycline (1 μg/ml). After 24 h, the cells were washed once with serum-free medium and then infected with encephalomyocarditis virus (EMCV) or vesicular stomatitis virus (VSV) at the desired multiplicity of infection (MOI) by incubation in 0.3 ml of serum-free medium for 60 min at 37°C (in order to have an accurate MOI, each virus was first titrated directly in UHCV-11 cells). The virus-containing medium was removed, and the cells were further incubated in complete medium with or without IFN and tetracycline as before. Eighteen hours after infection, the plates of cells were frozen and thawed once and the supernatants were collected and centrifuged to eliminate debris. The virus yields from all wells were determined on Vero VC10 cells. The different dilutions of the virus in serum-free medium were allowed to adsorb to the cells for 60 min, and the cells were covered with DMEM containing 5% serum and 0.6% agarose. After 48 h of incubation at 37°C, the cells were fixed in 10% trichloroacetic acid for 30 min at 4°C, the agarose-containing medium was removed under water, and the plaques were visualized by staining with a solution of 0.1% crystal violet in 20% ethanol.
Confocal microscopy.
For laser scanning confocal-microscopy experiments, UHCV-11 cells were cultured in eight-well chamber slides (Lab-Tek II; Nalge Nunc International, Naperville, Ill.) in the presence of tetracycline. Twenty-four hours after being seeded, the cells were further incubated in the presence or absence of tetracycline and in the presence or absence of IFN-α. After 24 h of incubation, the cells were fixed with paraformaldehyde and permeabilized with 0.5% Triton X-100 in order to maintain the integrity of cellular structures. They were then stained with appropriate antibodies. Primary antibodies were diluted 1/100 prior to use. Anti-rabbit antibodies linked to fluorescein isothiocyanate or anti-mouse antibodies linked to Texas red (Caltag) were also diluted 1/100 prior to use. The localization of PKR, NS5A, and the 2-5A synthetases were analyzed by laser scanning confocal microscopy (Leica TCS4D). Green fluorescence and red fluorescence were collected simultaneously and then separated digitally. Colocalization of proteins resulted in a merging of red and green fluorescence to produce a yellow signal.
Measurement of 2-5A synthetase activity.
Extracts from control and IFN-treated UHCV-11 or Daudi cells were immunoprecipitated with antibodies directed against the p69 or p100 isoform of 2-5A synthetase, which had been previously coated on protein A-Sepharose. After being washed, the purified proteins (on 50 μl of protein A-Sepharose) were incubated in 100 μl of 2-5A reaction mixture, containing 20 mM HEPES, 50 mM KCl, 25 mM magnesium acetate, 7 mM 2-mercaptoethanol, 5 mM ATP, 10 mM creatine phosphate, 0.16 mg of creatine kinase/ml, 100 μg of poly(I)-poly(C)/ml, and [α-32P]ATP. The pH of the reaction mixture was adjusted to 6.5 or 7.5 for p69 or p100 activity assays, respectively. The reaction was stopped by applying heat (95°C, 5 min), and the 2-5A products were analyzed by electrophoresis on SDS-20% polyacrylamide gels containing 7 M urea (30).
RESULTS
Regulated expression of HCV proteins in the UHCV-11 cell line.
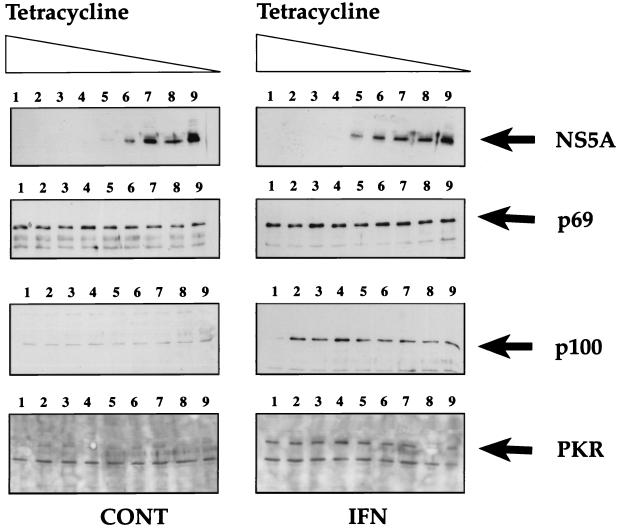
The tetracycline-regulated osteosarcoma cell line UHCV-11 has been recently characterized. Upon induction, these cells express all of the HCV structural and nonstructural proteins, which colocalize at the level of the endoplasmic reticulum (ER) (34). In a preliminary assay, the regulated expression of HCV proteins in UHCV-11 cells was examined by using antibodies directed against NS5A. The cells were cultured in the presence of decreasing concentrations of tetracycline, and the resulting cell extracts were analyzed by immunoblotting. As shown in Fig. 1 (top panels), expression of NS5A was completely repressed at tetracycline concentrations ranging from 1,000 to 5 ng/ml. Below this concentration of tetracycline, NS5A began to be detectable in the cells, with maximal expression level being attained in the absence of tetracycline.
FIG. 1.
Regulation of HCV NS5A protein expression in UHCV-11 cells and response to IFN with respect to PKR and 2-5A synthetase induction. UHCV-11 cells were seeded in four six-well tissue culture plates at a density of 300,000/well in the presence of tetracycline. After 24 h, they were washed three times with PBS to remove tetracycline and then incubated in culture medium containing different concentrations of tetracycline (1,000, 500, 100, 50, 10, 5, 1, 0.5, or 0 ng/ml [from left to right, lanes 1 to 9]). One set of cells was incubated in the absence of IFN (control [CONT]), and the other set was incubated in the presence of IFN at 500 U/ml. After 18 h of incubation, the cells were lysed in 300 μl of low-salt buffer I (containing 40 mM of NaCl instead of 400 mM) as described in Materials and Methods. The proteins contained in 50 μl of each extract (equivalent to 100,000 cells) were separated by SDS–12.5% PAGE and analyzed by immunoblotting for the presence of NS5A, p69 2-5A synthetase, p100 2-5A synthetase, and PKR.
Antiviral action of IFN against EMCV is affected in UHCV-11 cells induced for the expression of the HCV polyprotein.
One of the characteristics of HCV is resistance to the antiviral action of IFN. Here we analyzed whether the expression of HCV proteins in UHCV-11 cells interfered with the action of IFN on the growth of two different viruses, EMCV and VSV. UHCV-11 cells were induced, or not induced, for the expression of the polyprotein and then treated, or not treated, with IFN. The cells were infected with EMCV or VSV at MOIs of 0.01 and 0.1. The virus yields were then determined on Vero VC10 cells. No reproducible rescue effect could be seen for VSV (rescue was in the range of 0.4- to 3-fold, regardless of the MOI (data not shown). In contrast, we observed that expression of the HCV proteins allowed higher yields of EMCV in the presence of IFN (Table 1). This rescue could be clearly attributed to a specific inhibition of the antiviral action of IFN, since no increase in EMCV titer was observed in UHCV cells as a result of the sole expression of HCV proteins. In fact, in the latter case, the EMCV yields were even lower than those in uninduced cells. The reason for this is not known but may be related to the observation that high-level expression of HCV proteins can affect cell metabolism (34). Our rescue data (11-fold rescue at an MOI of 0.1 and 90-fold rescue at an MOI of 0.01) are consistent with those of Polyak et al., who found that cell lines inducibly expressing NS5A could partially rescue (40-fold) EMCV growth but could barely rescue (3- to 4-fold) VSV growth (37). Interestingly, these authors found that EMCV rescue occurred only when the cell lines were expressing NS5A from HCV genotype 1b and not when they were expressing genotype 1a NS5A, whereas, in contrast, UHCV-11 cells, which express HCV genotype 1a, could rescue EMCV. This suggests that there may be an enhanced effect of NS5A when it is expressed in the context of the entire HCV polyprotein or that HCV proteins other than NS5A play a role in the mechanism of resistance to IFN.
TABLE 1.
Antiviral action of interferon against EMCV in the UHCV cell line depends on expression of the HCV polyproteina
| MOI | Polyprotein expressed | EMCV yield (log PFU/ml)
|
ΔLog PFU/ml | Virus rescueb | |
|---|---|---|---|---|---|
| CONT | IFN | ||||
| 0.1 | No | 8.27 ± 0.39 | 4.47 ± 0.38 | 3.79 ± 0.49 | |
| Yes | 7.61 ± 0.41 | 4.89 ± 0.50 | 2.73 ± 0.31 | 11.68c | |
| 0.01 | No | 8.33 ± 0.33 | 3.31 ± 0.27 | 5.02 ± 0.53 | |
| Yes | 7.60 ± 0.33 | 4.54 ± 0.43 | 3.06 ± 0.23 | 90.40d | |
UHCV-11 cells were infected at MOI of 0.01 or 0.1 PFU/cell in the absence (control [CONT]) or presence of IFN at 500 U/ml and under conditions by which expression of the HCV polyprotein was repressed or induced. After 18 h of infection, the virus yields (expressed as mean log PFU/ml ± standard deviation [n = 6]) were titrated on Vero VC10 cells as described in Materials and Methods. The antiviral effect of IFN is given by the difference (Δlog) between the virus yields of CONT and IFN-treated cells.
Virus rescue indicates the ability of the cells induced for the expression of the polyprotein to develop resistance to the antiviral action of IFN. This rescue was calculated as 10ΔΔ, where ΔΔ represents the difference between the Δlog values of cells repressed or induced for the expression of the polyprotein. The reproducibility of the data was analyzed statistically by one-way analysis of variance (Duncan's multirange test), using the computer program Statview 4.5.
P = 0.02.
P = 0.003.
Expression of PKR and 2-5A synthetases in the tetracycline-regulated UHCV-11 cell line.
IFN induces the expression of a number of cellular proteins. Among them, the two double-stranded RNA-dependent enzymes PKR and 2-5A synthetase are known to play a major role in the antiviral action of IFN against EMCV (5, 33). Particular attention is directed here toward a role for PKR in the antiviral action of IFN against HCV since PKR was reported to interact with NS5A (13, 14). In the present study, we showed that IFN could induce the synthesis of PKR in UHCV-11 cells. In addition, we determined the effect of IFN on the synthesis in UHCV-11 cells of the p100 and p69 isoforms of 2-5A synthetase (30) (Fig. 1). Whereas PKR and the p100 2-5A synthetase were fully induced by IFN, the levels of the p69 2-5A synthetase were found to be already high in control cells and only slightly increased after IFN treatment. Treatment of the UHCV-11 cells with IFN did not affect the tetracycline-dependent induction of NS5A. Reciprocally, induction of the HCV proteins did not affect the levels of expression of 2-5A synthetase and PKR. The small reduction in the detection of PKR observed upon high-level induction of the polyprotein (Fig. 1; bottom right panel, lane 8) may have been due to the preparation of cell extracts and was not reproduced in subsequent experiments (see Fig. 2B).
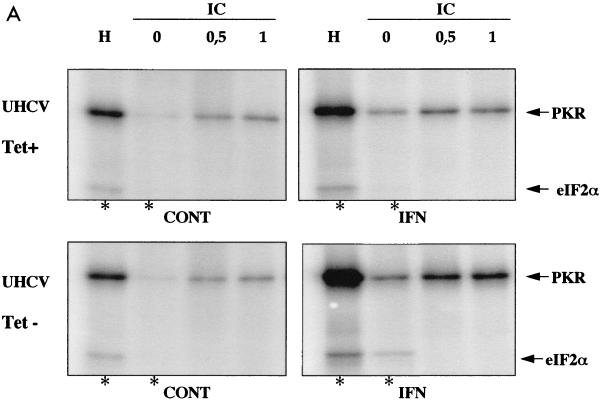
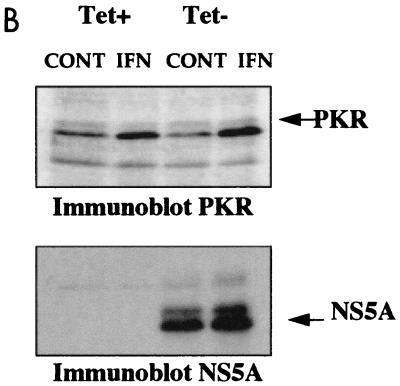
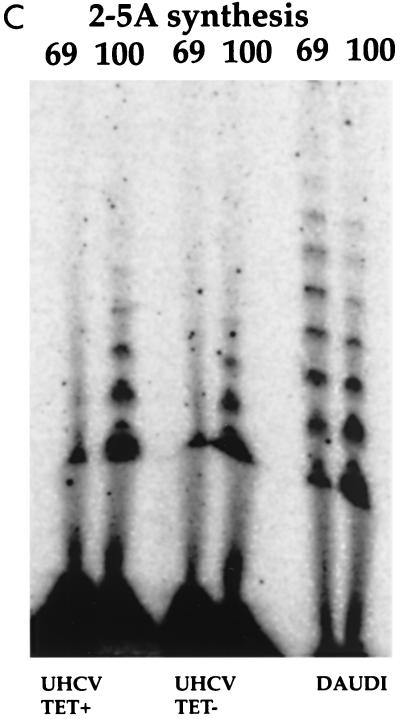
FIG. 2.
Analysis of in vitro activity of PKR and 2-5A synthetases from UHCV-11 cells. (A) PKR activity. Extracts of UHCV-11 cells that had (Tet−) or had not (Tet+) been induced to express the polyprotein and that had or had not (control [CONT]) been treated with IFN were prepared in buffer I. Immunoprecipitation of PKR and an in vitro phosphorylation assay were performed with extracts corresponding to 107 cells as described in Materials and Methods. PKR activity was assayed in the absence (0) and in the presence of poly(I)-poly(C) (IC) at 0.5 or 1 μg/ml or in the presence of heparin (H) at 10 U/ml (used as activators). Fifteen minutes after the beginning of the phosphorylation reaction, 2 μl of a purified preparation of eIF2 was added to each of the samples containing no activator or containing heparin (asterisks), and the reactions were continued for another 15 min. The reactions were stopped by addition of an equal amount of 2× SDS sample buffer, and the proteins were separated by SDS–12.5% PAGE. (B) Immunoblotting for PKR and NS5A. Ten percent of each of the crude extracts used for the immunoprecipitation detailed above and corresponding to untreated and IFN-treated UHCV cells that had or had not been induced to produce the HCV polyprotein were analyzed by immunoblotting for induction of PKR after IFN treatment (immunoblot PKR) and for induction of NS5A by tetracycline removal (immunoblot NS5A). (C) 2-5A synthetase activity. Extracts from UHCV-11 cells repressed (TET+) or induced (TET−) for the expression of the polyprotein and extracts from Daudi cells, used as controls, were immunoprecipitated with antibodies against either the p69 or the p100 form of 2-5A synthetase and analyzed for their capacity to synthesize 2-5A oligomers as described in Materials and Methods.
In vitro activity of PKR and 2-5A synthetases from the UHCV-11 cell line.
Since PKR may play a role in the antiviral action of IFN against HCV, it was necessary to control its activity in UHCV-11 cells. These cells were incubated for 18 h in the absence or presence of tetracycline and in the presence or absence of IFN. PKR was then immunoprecipitated from cell extracts, and its activity was assayed in vitro. The PKR from UHCV-11 cells was found to be active, as shown by its ability to autophosphorylate in the presence of heparin or poly(I)-poly(C) and to phosphorylate its substrate, eIF2α (21, 35) (Fig. 2A). Its activity was increased after IFN treatment of the cells, as a result of the induction of the protein (Fig. 2B) (induction of the HCV proteins by tetracycline removal is shown in parallel). We noticed some increase in the activity of PKR from the extracts induced to express the HCV proteins compared to its activity in the uninduced samples (Fig. 2A). This might have resulted from a better accessibility of PKR to its antibody due to modifications in cell extracts containing the HCV proteins. Whatever the case, our data clearly show that active PKR was recovered from the UHCV-11 cells. In parallel, the activities of the p69 and p100 isoforms of the 2-5A synthetase were also determined by measuring their ability to catalyze the incorporation of AMP into 2-5A oligomers (30) (Fig. 2C). We observed that only the p100 isoform of the 2-5A synthetase from UHCV-11 cells was active. Its activity was not modified whether the cells were cultured in the presence or absence of tetracycline. The lack of activity of the p69 isoform in extracts from UHCV-11 cells is intriguing, particularly in view of the fact that this isoform was found to be constitutively expressed in UHCV-11 cells (Fig. 1). This phenomenon was not analyzed further in the present study. Since UHCV-11 cells were derived from an osteosarcoma cell line, the occurrence of different rearrangements or mutations might explain this situation. In particular, constitutive upregulation of the p69 2-5A synthetase gene, which may result in activation of the 2-5A system and, hence, inhibition of cell growth, may have been corrected by mutations in the protein.
Subcellular localization of PKR, 2-5A synthetase, and NS5A in UHCV-11 cells.
Direct interaction of NS5A of HCV genotypes 1a and 1b with PKR has been observed in studies, involving binding of in vitro-translated PKR to glutathione S-transferase–NS5A (14). Furthermore, NS5A was also shown to inactivate PKR both in an in vitro phosphorylation assay and in a functional assay in S. cerevisiae (14). These observations were made when NS5A was expressed alone. The data presented in Fig. 2A show that the activity of PKR immunoprecipitated from UHCV cell extracts, which contain all HCV proteins, including NS5A, was not inhibited. A search for coprecipitation of PKR and NS5A in extracts of UHCV cells did not reveal any convincing interaction (data not shown). We therefore examined whether interaction of PKR and NS5A in UHCV-11 cells could be observed by immunofluorescence techniques, using confocal microscopy. In parallel, colocalization of NS5A with the 2-5A synthetases was also explored.
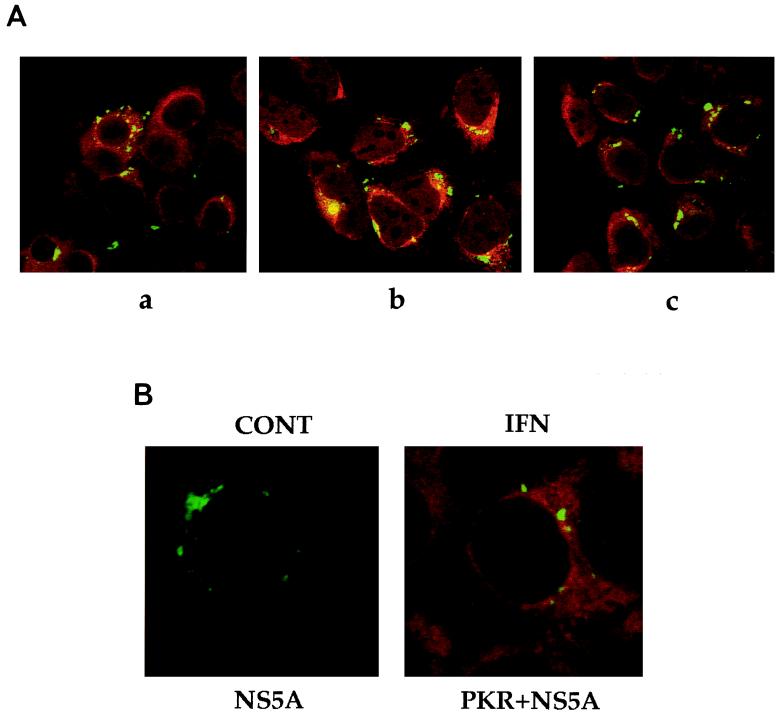
UHCV-11 cells were cultured in the absence of tetracycline to induce the synthesis of the HCV proteins and were subjected to IFN treatment to induce the expression of PKR and the p69 and p100 2-5A synthetases. The results are shown in Fig. 3A. NS5A (green fluorescence) was found to be located in the cytoplasm, predominantly around the nucleus, as previously reported (34). PKR and the two 2-5A synthetases (red fluorescence) were found throughout the cytoplasm as expected (24). The p69 2-5A synthetase was located in association with the membranes of the ER, whereas the p100 2-5A synthetase was more diffuse, being found throughout the cytoplasm, as previously reported (4, 23). No striking evidence of colocalization of NS5A with PKR or either of the two 2-5A synthetases was found. Localization of NS5A was not affected by IFN treatment (Fig. 3B). It is interesting that the majority of free PKR or 2-5A synthetase was seen in the cytoplasm, away from the HCV proteins attached to the ER structures. Our data therefore indicate that if an interaction between PKR and any of the HCV proteins occurs when they are coexpressed in vivo, it may occur at a very localized level, with the involvement of a minimal amount of PKR, leaving the rest of the PKR free and potentially able to be activated.
FIG. 3.
Subcellular localization of PKR, 2-5A synthetase, and NS5A in UHCV-11 cells. (A) UHCV-11 cells, seeded in eight-well chamber slides (Lab-Tek), were induced for the full expression of the HCV polyprotein and analyzed for the colocalization of NS5A (green fluorescence) and either PKR (a), p69 2-5A synthetase (b), or p100 2-5A synthetase (c) (red fluorescence). (B) UHCV-11 cells, seeded in eight-well chamber slides (Lab-Tek), were induced for the full expression of the HCV polyprotein in the absence (control [CONT]) or presence of IFN of and analyzed for the effect of IFN on localization of NS5A (green fluorescence) and PKR (red fluorescence).
Expression of the HCV polyprotein does not reverse PKR-mediated inhibition of protein synthesis.
We next examined whether PKR activity could be inhibited in vivo when UHCV-11 cells were induced for expression of the HCV proteins, using a functional assay for PKR.
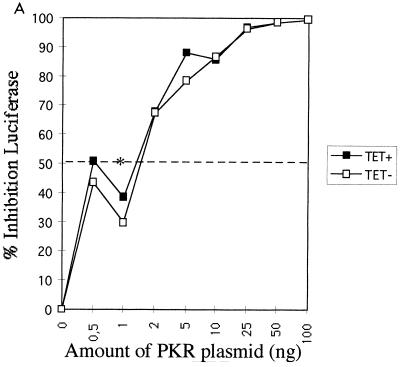
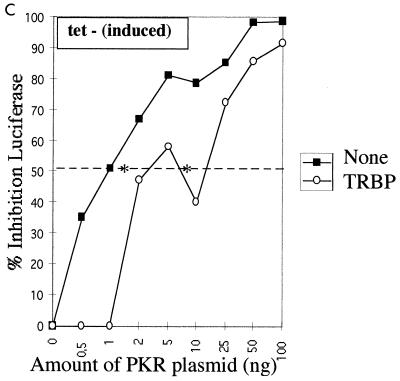
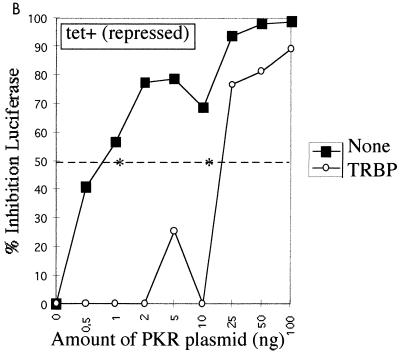
We have developed an in vivo functional assay for PKR based on this enzyme's ability to inhibit protein synthesis. In this assay, the pHIV-1 LTR-Luc vector was used both as an activator of PKR (32) and as a reporter through the expression of luciferase. In this situation, the luciferase mRNA transcribed from the LTR presents at its 5′ end the double-stranded TAR region of the LTR, which activates PKR (38). Therefore, luciferase mRNAs accumulating in the cytoplasm have the potential to activate PKR, and subsequently their own protein synthesis can be inhibited. The PKR functional assay was first performed in UHCV-11 cells repressed for the expression of the polyprotein. When transfection of the pHIV1 LTR-Luc plasmid was performed in the presence of increasing amounts of a PKR-expressing plasmid (pPKR), a concomitant progressive inhibition of luciferase expression occurred. A 50% inhibition of reporter expression was observed for concentrations of pPKR ranging from 0.8 to 1 ng, corresponding to the 50% inhibitory dose (ID50) of PKR under these experimental conditions (Fig. 4). When the transfection assay was performed in the absence of tetracycline to induce the expression of the polyprotein, we observed a general lower-level expression of the reporter than that seen with uninduced cells (data not shown). This may have been due to the cell culture conditions, as already described (34). However, no differences in the inhibition of the reporter by PKR were observed (Fig. 4A). As a positive control for PKR inhibition in this assay, pPKR and pHIV1 LTR-Luc were transfected with a plasmid expressing TRBP, a strong PKR inhibitor (3, 15). The results (Fig. 4B and C) showed that TRBP could significantly reverse the PKR-mediated inhibition of luciferase by shifting the ID50 of PKR from the 0.8- to 1-ng range to 10 ng in cells repressed for the expression of the polyprotein (Fig. 4B) and to 8-ng in cells induced to express the polyprotein (Fig. 4C).
FIG. 4.
Expression of HCV polyprotein does not reverse the PKR-mediated inhibition of protein synthesis, whereas TRBP does. (A) UHCV-11 cells were transfected by a microtransfection technique described in Materials and Methods. To each well (20,000 cells) was added 100 ng of pHIV1 LTR-L and 0.5 to 100 ng of pcDNA1/Amp (PKR) as indicated. All samples were adjusted to contain the same amount of DNA by addition of the empty pcDNA1/Amp vector. The transfection assay was performed with cells either repressed (TET+) (closed squares) or induced (TET−) (open squares) for the expression of the HCV proteins. The results are expressed as the percentage of inhibition of reporter expression when transfection took place in the presence of PKR compared with its transfection in the absence of the PKR plasmid. (B and C) UHCV-11 cells either repressed (B) or induced (C) for the expression of the polyprotein were transfected with pHIV1 LTR-L and increasing concentrations of pcDNA1/Amp (PKR), either alone (closed squares) as for panel A or in the presence of 300 ng of pcDNA1/Amp (TRBP2) (open circles). The results are expressed as the percentage of inhibition of reporter expression by PKR. Addition of TRBP to the cells allows a shift of the ID50 of PKR from the 0.8- to 1-ng range to the 8- to 10-ng range, thus indicating its ability to reverse the PKR-mediated inhibition of protein synthesis. In each graph, the ID50 of PKR is represented by an asterisk placed where the graph crosses the 50% inhibition value (broken line).
Therefore, our results show that in contrast to the action of TRBP on PKR, induction of the HCV proteins in the cells does not affect the PKR-mediated inhibition of translation. As we have shown in Fig. 3, all HCV proteins are located around the nuclei whereas most of the PKR is diffused throughout the cytoplasm. If one protein from HCV were to have the potential to inhibit PKR, its effect might therefore be limited only to the areas where it is close enough to interact with PKR.
Induction of the HCV proteins does not affect in vivo phosphorylation of PKR and eIF2α in UHCV-11 cells.
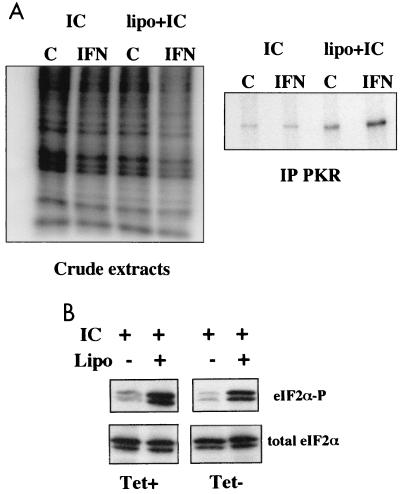
The reporter assay described above showed that coexpression of the HCV proteins did not lead to inhibition of PKR activity. However, this assay was based on the transfection of a plasmid encoding PKR in the cells and did not measure the actual activity of the endogenous PKR. We therefore chose to directly activate the endogenous PKR by transfecting poly(I)-poly(C) into the cells in order to examine the effect of induction of the HCV proteins on the state of phosphorylation of eIF2α, the PKR substrate. In a preliminary assay, 32P-phosphate labeling of the cells was used to detect poly(I)-poly(C)-mediated in vivo phosphorylation of PKR. We found a shift in the phosphorylation of PKR in the UHCV-11 cells induced to express the HCV proteins, provided that the PKR levels were first induced by IFN treatment and that poly(I)-poly(C) had been efficiently transfected into the cells in the presence of Lipofectin (Fig. 5A, right panel). In addition, this assay showed that the situation which triggered PKR phosphorylation [IFN treatment and transfection of poly(I)-poly(C)] also led to a reduced general state of phosphorylation of cellular proteins (Fig. 5A, left panel). This most probably reflects the PKR-mediated inhibition of protein synthesis and indicates that the endogenous PKR can be active in UHCV-11 cells induced to express the HCV proteins. To confirm this and to determine the effect of expression of the HCV proteins on PKR activity, we then analyzed the phosphorylation state of the PKR substrate, eIF2α, in response to poly(I)-poly(C) and Lipofectin treatment of IFN-treated UHCV-11 cells repressed or induced for expression of the HCV proteins. The results (Fig. 5B) clearly showed that the increases in eIF2α phosphorylation were similar regardless of whether the cells were expressing HCV proteins. These results confirmed the data from the reporter assay. In conclusion, therefore, we showed that coexpression of the HCV proteins in UHCV-11 cells did not eliminate the ability of PKR to inhibit gene expression, through eIF2α phosphorylation, regardless of whether PKR was expressed ectopically by transient transfection or endogenously after IFN induction.
FIG. 5.
Poly(I)-poly(C)-induced phosphorylation of eIF2α is not affected by induction of the HCV proteins. (A) UHCV-11 cells were seeded at a density of 106/10-cm-diameter petri dish. After 24 h, they were washed three times with PBS to remove tetracycline and then incubated in culture medium alone (control [C]) or containing IFN at 500 U/ml. After 18 h of incubation, the cells were washed twice in phosphate-free, serum-free medium and further incubated in 2.5 ml of this medium supplemented with 100 μg of poly(I)-poly(C) (PL Biochemicals), either alone (IC) or mixed with 25 μg of Lipofectin (Gibco BRL) (lipo+IC). [32P]orthophosphate (Amersham) was then added (750 μCi/dish), and an incubation was carried out for 90 min at 37°C. The medium was removed, the cells were washed twice and scraped off in PBS, and the cell pellets were recovered by centrifugation and lysed with 600 μl of buffer I supplemented with 10 mM β-glycerophosphate, 10 mM NaF, 10 mM p-nitrophenyl-phosphate, and 300 μM Na3VO4 as phosphatase inhibitors. After centrifugation at 12,000 × g, 5 μl of each of the crude extracts was subjected to SDS–12.5% PAGE and the rest was immunoprecipitated with anti-PKR antibodies as described in Materials and Methods. After incubation at 4°C for 18 h, the beads were washed three times with buffer I and the proteins were separated by SDS–12.5% PAGE. IPLab Gel-based quantification gave estimates of 1.12-fold [treatment with poly(I)-poly(C) alone] and 3.2-fold [treatment with Lipofectin plus poly(I)-poly(C)] for the increases in intensity of the phosphorylated bands in the IFN lane compared with the control lanes. (B) UHCV-11 cells were treated with poly(I)-poly(C) either alone or in the presence of Lipofectin (Lipo) as for the experiment shown in A, except that all dishes of cells had been previously treated with IFN and induced (Tet−) or not induced (Tet+) for expression of the HCV proteins. The proteins of the cell extracts were separated by SDS–12.5% PAGE and analyzed by immunoblotting. Use of antibodies directed specifically against a phosphorylated eIF2α peptide (eIF2α-P) demonstrated that eIF2α phosphorylation had occurred, whereas antibodies directed against total eIF2 (eIF2α) revealed the total levels of eIF2α. In this experiment, eIF2α was found to migrate as two bands in the gel. Both bands were specific since they were recognized by the two different anti-eIF2α antibodies.
DISCUSSION
Resistance to the antiviral action of IFN is one major characteristic of HCV of genotype 1. To study the mechanisms of resistance of HCV to IFN, it is necessary to characterize the interactions between the viral and cellular components. However, a cell culture system for HCV infection is not yet available. The human osteosarcoma-derived UHCV-11 cell line, which is regulated by tetracycline for the expression of all HCV structural and nonstructural proteins, has recently been described and provides a good approach for this study (34).
By using two different types of viruses, the picornavirus EMCV and the rhabdovirus VSV, we first showed that UHCV-11 cells developed an antiviral state after treatment with IFN. The VSV yields were found to be inhibited by 1.8 to 2.7 logs (data not shown), while the EMCV yields were inhibited by 3.8 to 5 logs depending on the MOI (Table 1; see Δlog values corresponding to expression of polyprotein). To determine whether expression of the HCV proteins interfered with IFN action, the growth of each virus in cells treated with IFN and induced to express the HCV polyprotein was compared to its growth in IFN-treated cells in which expression of the polyprotein was repressed. We observed that expression of HCV proteins had no effect on VSV growth (data not shown), whereas it increased the growth efficiency of EMCV. The most significant EMCV rescue (90-fold) was observed when the cells were infected at a low MOI (0.01).
In a recent study, HeLa and osteosarcoma cells engineered to express the nonstructural protein NS5A of either genotype 1a or 1b HCV under the control of a tetracycline-dependent promoter were found to provoke a slight rescue of the growth of VSV and to differentially rescue EMCV. Only clones expressing NS5A 1b rescued EMCV (37). Another study showed that NS5A from a different HCV-1b subtype can rescue EMCV but only minimally rescues VSV (40). The UHCV-11 cell line, which is inducible for the expression of the HCV polyprotein of genotype 1a, was also found to have a very limited rescue effect (if any) on VSV. However, in contrast with the data presented for the NS5A 1a-expressing clones, the use of the UHCV-11 cell line, which expresses not only NS5A 1a but also all of the remaining HCV 1a proteins, allowed us to show a rescue of the growth of EMCV. This indicates that HCV proteins other than NS5A may be involved in the mechanism of resistance to IFN, either independently or in combination with NS5A.
IFN treatment of cells results in the establishment of an antiviral state after de novo induction of cellular proteins. Only a few of the induced genes have been shown to be involved in antiviral mechanisms. For instance, the human MxA protein can induce resistance to influenza virus, VSV (36), Semliki Forest virus (27), measles virus (39), and Thogoto virus (12). PKR and 2-5A synthetase were both demonstrated to be involved in resistance to EMCV but not VSV (8, 33). MxA, PKR, and the 2-5A synthetase pathway are therefore considered to be the principal components used by IFNs to mount an antiviral state. However, alternative antiviral pathways exist, as shown recently by the ability of IFN to provoke an antiviral state against EMCV or VSV in cultured embryonic fibroblasts obtained from mice deficient in Mx1, RNase L, and PKR (46). For example, the human guanylate binding protein GBP-1 was recently found to mediate antiviral resistance to both EMCV and VSV (1).
Expression of the HCV polyprotein in UHCV-11 cells was recently shown to block IFN-mediated signal transduction through the Jak-STAT pathway, possibly by inhibiting the binding of Stat1 to their DNA response element (17). Our data, however, show that IFN can normally induce the synthesis of PKR and the p100 2-5A synthetase in UHCV-11 cells, in spite of the fact that the expression of both genes is dependent on Stat1. This apparent contradiction can be explained by the fact that in our experiments, IFN treatment of UHCV-11 cells was performed a few hours before the HCV proteins were synthesized. This provided a window of time sufficient for induction of IFN-stimulated genes before IFN signaling could be inhibited. Since PKR and 2-5A synthetase have long half-lives (6.6 and 8 h, respectively [22, 23]), their ability to act as antiviral proteins does not depend on continuous induction. The HCV-mediated inhibition of Stat1 may indicate, therefore, that some other IFN-induced genes encoding short-half-life proteins play a critical role in HCV-mediated virus rescue.
What is the importance of NS5A with respect to the antiviral action of IFN? This protein has been the subject of great interest and controversy. Studies conducted on Japanese patients have related the IFN responsiveness of HCV patients to their ISDRs (11). However, studies conducted on European and North American patients did not show a correlation between their ISDRs and patterns of response to IFN (19, 25, 41, 45).
We studied the impact of expression of HCV proteins on PKR, a key component of the antiviral and antiproliferative effects of IFN. This study allowed the examination of the potential importance of the PKR-NS5A interaction in the context of expression of all viral proteins. Coprecipitation experiments failed to show a significant interaction between PKR and NS5A. Indeed, confocal-microscopy analysis did not show striking colocalization of PKR and NS5A; PKR localized throughout the cytoplasm, whereas NS5A localized predominantly around the nuclei, probably at the level of the ER.
In a functional assay of PKR performed in UHCV-11 cells, we found that expression of the HCV proteins did not reverse the action of PKR, in contrast with the action of the PKR inhibitor TRBP. Furthermore, expression of HCV proteins neither prevented the poly(I)-poly(C) activation of IFN-induced endogenous PKR of UHCV-11 cells nor inhibited the subsequent phosphorylation of its substrate, eIF2α.
In conclusion, we have shown that concomitant expression of the HCV nonstructural and structural proteins can alter the antiviral mechanisms of action of IFN. However, this HCV-mediated inhibition could not be attributed to an inhibition of PKR's ability to regulate protein synthesis through eIF2α phosphorylation. A number of studies have shown that NS5A has the ability to interact with and to inhibit PKR. We stress here that the impact of NS5A on PKR activity may differ when NS5A is expressed individually or in the context of the other HCV proteins. We showed by confocal microscopy that PKR and NS5A are not colocalized in UHCV-11 cells induced to express the HCV proteins. This situation may prevent NS5A from interacting efficiently with PKR in order to prevent the antiviral action of PKR against EMCV. It is possible that during the course of HCV infection, NS5A plays a role in inhibiting PKR locally at the site of HCV protein synthesis. NS5A might, however, participate in the blockage of IFN's antiviral action through other mechanisms, such as the recently reported interaction with the Ras-associated Grb-2 protein (42). In view of the results presented here, it will certainly be necessary to reconsider all types of interactions between any particular HCV protein and its cellular partner(s) in the context of expression of the other HCV proteins.
ACKNOWLEDGMENTS
This work was supported in part by a grant (Sub990014DR18; CNRS) from the Ministère de la Recherche et de l'Enseignement, by a grant from the Agence National de la Recherche contre le SIDA (to A.G.), and by a grant from the Programme Hospitalier de Recherche Clinique de Picardie 1997. D.M. and H.E.B. were supported by grant Mo 799/1-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Anderson S L, Carton J M, Lou J, Xing L, Rubin B Y. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 2.Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. doi: 10.1074/jbc.270.29.17423. [DOI] [PubMed] [Google Scholar]
- 3.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K-T. Oncogenic potential of TAR RNA-binding protein TRBP and its regulatory interaction with protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besse S, Rebouillat D, Marie I, Puvion-Dutilleul F, Hovanessian A G. Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp Cell Res. 1998;239:379–392. doi: 10.1006/excr.1997.3908. [DOI] [PubMed] [Google Scholar]
- 5.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 6.Chong K, Feng L, Donahue T F, Friesen J D, Meurs E, Hovanessian A G, Williams B R G. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens M J, Androulla E. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 8.Coccia E M, Romeo G, Nissim A, Marziali G, Albertini R, Affabris E, Battistini A, Fiorucci G, Orsatti R, Rossi G B, Chebath J. A full-length murine 2-5A synthetase cDNA transfected in NIH-3T3 cells impairs EMCV but not VSV replication. Virology. 1990;179:228–233. doi: 10.1016/0042-6822(90)90292-y. [DOI] [PubMed] [Google Scholar]
- 9.Duh E J, Maury W J, Folks T M, Fauci A S, Rabson A B. Tumor necrosis factor α activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373–1381. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 12.Frese M, Kochs G, Meier-Dieter U, Siebler J, Haller O. Human MxA protein inhibits tick-borne Thogoto virus but not Dhori virus. J Virol. 1995;69:3904–3909. doi: 10.1128/jvi.69.6.3904-3909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale M, Jr, Blakely C M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale M J, Jr, Korth M J, Tang N M, Tan S L, Hopkins D, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 15.Gatignol A, Buckler-White A, Berkhout B, Jeang K-T. Characterization of human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 16.Gatignol A, Duarte M, Daviet L, Chang Y N, Jeang K T. Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors. Gene Expr. 1996;5:217–228. [PMC free article] [PubMed] [Google Scholar]
- 17.Heim M H, Moradpour D, Blum H E. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469–8475. doi: 10.1128/jvi.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershey J W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 19.Hofgärtner W T, Polyak S J, Sullivan D G, Carithers R L, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 20.Hovanessian A G. Interferon-induced dsRNA-activated protein kinase (PKR): antiproliferative, antiviral and antitumoral functions. Semin Virol. 1993;4:237–245. [Google Scholar]
- 21.Hovanessian A G, Galabru J. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur J Biochem. 1987;167:467–473. doi: 10.1111/j.1432-1033.1987.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 22.Hovanessian A G, Galabru J, Meurs E, Buffet-Janvresse C, Svab J, Robert N. Rapid decrease in the levels of the double-stranded RNA-dependent protein kinase during virus infections. Virology. 1987;159:126–136. doi: 10.1016/0042-6822(87)90355-2. [DOI] [PubMed] [Google Scholar]
- 23.Hovanessian A G, Svab J, Marie I, Robert N, Chamaret S, Laurent A G. Characterization of 69- and 100-kDa forms of 2-5A-synthetase from interferon-treated human cells. J Biol Chem. 1988;263:4945–4959. [PubMed] [Google Scholar]
- 24.Jeffrey I W, Kadereit S, Meurs E F, Metzger T, Bachmann M, Schwemmle M, Hovanessian A G, Clemens M J. Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp Cell Res. 1995;218:17–27. doi: 10.1006/excr.1995.1126. [DOI] [PubMed] [Google Scholar]
- 25.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209–2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 26.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 27.Landis H, Simon-Jödicke A, Klöti A, Di Paolo C, Schnorr J-J, Schneider-Schaulies S, Hefti H P, Pavlovic J. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J Virol. 1998;72:1516–1522. doi: 10.1128/jvi.72.2.1516-1522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to interferon induced 68,000 Mr protein and their use for the detection of double-stranded RNA dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Major M E, Feinstone S M. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 30.Marie I, Blanco J, Rebouillat D, Hovanessian A G. 69-kDa and 100-kDa isoforms of interferon-induced (2′-5′) oligoadenylate synthetase exhibit differential catalytic parameters. Eur J Biochem. 1997;248:558–566. doi: 10.1111/j.1432-1033.1997.t01-1-00558.x. [DOI] [PubMed] [Google Scholar]
- 31.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meurs E F, McMillan N, Williams B R, Hovanessian A G, Southern P J. Human PKR transfected into murine cells stimulates expression of genes under control of the HIV1 or HTLV-I LTR. Virology. 1995;214:653–659. doi: 10.1006/viro.1995.0080. . (Erratum, 247:125, 1998.) [DOI] [PubMed] [Google Scholar]
- 33.Meurs E F, Watanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R G, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moradpour D, Kary P, Rice C M, Blum H E. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology. 1998;28:192–201. doi: 10.1002/hep.510280125. [DOI] [PubMed] [Google Scholar]
- 35.Patel R C, Stanton P, Sen G C. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 36.Pavlovic J, Zürcher T, Haller O, Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polyak S J, Paschal D M, McArdle S, Gale M J, Jr, Moradpour D, Gretch D R. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262–1271. doi: 10.1002/hep.510290438. [DOI] [PubMed] [Google Scholar]
- 38.Roy S, Agy M, Hovanessian A G, Sonenberg N, Katze M G. The integrity of the stem structure of human immunodeficiency virus type 1 Tat-responsive sequence RNA is required for interaction with the interferon-induced 68,000-Mr protein kinase. J Virol. 1991;65:632–640. doi: 10.1128/jvi.65.2.632-640.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnorr J-J, Schneider-Schaulies S, Simon-Jödicke A, Pavlovic J, Horisberger M A, ter Meulen V. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J Virol. 1993;67:4760–4768. doi: 10.1128/jvi.67.8.4760-4768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Fujii M, Wang F, Itoh M, Hotta H. The NS5A protein of hepatitis C virus partially inhibits the antiviral activity of interferon. J Gen Virol. 1999;80:879–886. doi: 10.1099/0022-1317-80-4-879. [DOI] [PubMed] [Google Scholar]
- 41.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 42.Tan S L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a SRC homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M C. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 44.Tong C Y, Gilmore I T, Hart C A. HCV-associated liver cancer. Lancet. 1995;345:1058–1059. doi: 10.1016/s0140-6736(95)90804-8. [DOI] [PubMed] [Google Scholar]
- 45.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 46.Zhou A, Paranjape J, Der S, Williams B, Silverman R. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]