Abstract
The human immunodeficiency virus type 1 (HIV-1) RNA genome is flanked by a repeated sequence (R) that is required for HIV-1 replication. The first 57 nucleotides of R form a stable stem-loop structure called the transactivation response element (TAR) that can interact with the virally encoded transcription activator protein, Tat, to promote high levels of gene expression. Recently, we demonstrated that TAR is also important for efficient HIV-1 reverse transcription, since HIV-1 mutated in the upper stem-loop of TAR showed a reduced ability both to initiate and to complete reverse transcription. We have analyzed a series of HIV-1 mutant viruses to better defined the structural or sequence elements required for natural endogenous reverse transcription and packaging of virion RNA. Our results indicate that the requirement for TAR in reverse transcription is conformation dependent, since mutants with mutations that alter the upper stem-loop orientation are defective for reverse transcription initiation and have minor defects in RNA packaging. In contrast, TAR mutations that allowed the formation of alternative upper stem-loop structure greatly reduced RNA packaging but did not affect reverse transcription efficiency. These results are consistent with direct involvement of the upper stem-loop structure in packaging of genomic RNA and suggest that the TAR RNA stem-loop from nucleotide +18 to +42 interacts with other components of the reverse transcription initiation complex to promote efficient reverse transcription.
The human immunodeficiency virus type 1 (HIV-1) RNA genome can form numerous RNA structures, many of which regulate virus replication. Viral RNA structures are required for many processes, including transcription by RNA polymerase II, polyadenylation of viral mRNA, transport of singly spliced and unspliced viral mRNA, viral genomic RNA dimerization and packaging, and reverse transcription. For example, the TAR element, which comprises the first 57 nucleotides (nt) of the viral transcript, can form a stable stem-loop structure. The Tat protein and cellular kinases, which are recruited by Tat, can bind to the TAR stem-loop to enable efficient transcription by RNA polymerase II (for reviews, see references 12a and 24a). Another example is the HIV-1 RNA packaging signal, which includes a series of RNA stem-loop structures (designated SL1, SL3, and SL4) which flank the 5′ major splice donor site (3, 9, 19, 27) and bind to the nucleocapsid domain of the pr55 Gag protein to promote packaging of unspliced HIV-1 transcripts into virions (2, 5, 12, 29). Another regulatory viral RNA structure is formed by sequences flanking the primer binding site in conjunction with cellular tRNA3Lys. Interactions between an A-rich loop upstream of the virus primer binding site and the anticodon loop of tRNA3Lys are reported to be required for the formation of an efficient reverse transcription initiation complex (21, 23, 24).
It has long been appreciated that a single viral RNA structure can have effects on multiple steps in the replication cycle. The TAR element, for example, has also been shown to reduce translation of viral mRNA both in Xenopus oocyte microinjection assays and in cell-free translation systems (6, 31). Genetic analysis of TAR showed that maintenance of the TAR stem-loop structure and the primary sequence of the loop were required for inhibition of HIV-1 translation. Mutations that disrupted the TAR lower stem increased translation efficiency, while compensatory mutations that restored stem base pairing also restored TAR-mediated inhibition of translation. More recently, TAR has been implicated in the encapsidation of virus genomic RNA (8, 11, 20, 28). Viruses lacking the TAR element or carrying mutations that disrupted the lower portion of the TAR stem structure reduced RNA encapsidation, and some, but not all, of these mutations reduced reverse transcription efficiency. As with translation, compensatory mutations that restored TAR base pairing also restored RNA encapsidation, indicating that TAR structure, but not primary sequence, is required for efficient RNA encapsidation. How TAR acts in packaging is not known, but evidence that distinct pools of genomic RNA are packaged into virions makes it plausible that TAR-mediated inhibition of translation may contribute to the selection of nontranslated genomic RNA by Gag in the packaging process (26). In a previous study, we showed that mutations within the upper TAR stem-loop structure greatly reduced reverse transcription efficiency, but no defect in RNA encapsidation was observed (18). In the present study, we constructed further HIV-1 TAR mutations and determined their effects on viral reverse transcription and RNA packaging. Our results suggest that TAR's role in reverse transcription is dependent on the conformation of the upper stem-loop structure, from nucleotides +18 to +42, and that this region of TAR also supports RNA encapsidation.
MATERIALS AND METHODS
Plasmids and sequences.
All HIV-1 sequence notations take the transcription start site (GGG of TAR) as +1. The HIV-1 infectious molecular construct pBRDH2-neo and procedures used to introduce TAR mutations into this construct have been previously described (15). Briefly, a plasmid containing HIV-1 sequences from position −160 to +988 was mutagenized using the QuickChange mutagenesis system and oligonucleotides that included the desired mutations in both DNA strands. The mutations were confirmed by DNA sequence determination using the Sequenase system (Amersham Pharmacia Biotec) and ligated into pBRDH2-neo. The runoff RNA used as an internal control in quantitative reverse transcriptase PCR (RT-PCR) analysis was synthesized from a pGEM3z (Promega Australia Pty. Ltd.) construct containing HIV-1 sequences from position −22 to +553 with an internal deletion from +80 to +151. The construct was linearized with EcoRI and used as the template for in vitro transcription by SP6 RNA polymerase. The runoff RNA used for quantitative RNA protection assays was synthesized from a pGem4z (Promega Australia Pty. Ltd.) construct containing HIV-1 sequences from position +113 to +553. This construct was linearized with EcoRI and used as the template for in vitro synthesis of radiolabeled RNA riboprobe using [32P]CTP (400 Ci/mmol) (ICN Pharmaceuticals) and T7 RNA polymerase (Promega Australia Pty. Ltd.). All in vitro transcripts were purified from 5% polyacrylamide gels before use.
Cell lines, viruses, and infections.
All cells were incubated at 37°C in a humidified 5% CO2 atmosphere. The method by which stable cell lines were generated is described in detail elsewhere (15). Briefly, 293 cells were transfected with the mutated pBRDH2-neo construct, allowed to recover for 48 h, and then serially diluted in Iscove's modified Dulbecco minimal essential medium (IMDM) supplemented with 1% penicillin-streptomycin, 1 mM Glutamax, 5% newborn calf serum, and 2% fetal bovine serum (complete IMDM) plus 800 μg of G418 sulfate (Life Technologies) per ml. Individual cell foci were isolated and expanded, and supernatant from each culture was assayed for HIV-1 p24 antigen (Ag). Cell lines that produced p24 Ag were further characterized.
Virus stocks were produced and assayed as previously described (32). Briefly, 293 cell lines stably transfected with either wild-type or TAR mutant virus were grown in 100-mm-diameter tissue culture dishes in complete IMDM supplemented with 500 μg of G418 sulfate per ml. When the cells reached 50% confluency, the supernatant was replaced with complete IMDM lacking G418 sulfate, and the plates were incubated for a further 18 to 24 h. The medium was collected, filtered through a 0.45-mm-pore-size PES membrane, and stored in aliquots at 80°C. The virus stocks were assayed for HIV-1 p24 Ag using a commercial enzyme-linked immunosorbent assay (NEN Life Science Products) and for RT activity using the RT Detect Assay (Roche Diagnostics Australia Pty. Ltd.).
To infect Jurkat cells, filtered culture supernatant containing 90 mU of RT activity was adjusted to 10 ml with cell-conditioned culture medium and incubated with 2 × 106 cells for 2 h with gentle rotation. Mock infections were carried out with wild-type virus inactivated by incubation at 60°C for 20 min. The infected Jurkat cells were washed three times with culture medium to remove residual virus and incubated in RPMI 1640 medium supplemented with 1% penicillin-streptomycin, 1 mM Glutamax, and 10% fetal bovine serum. The cells were passaged twice weekly and assayed for p24 Ag by enzyme-linked immunosorbent assay as described above.
PCR and RT-PCR assays.
Chromosomal DNA was isolated from 293 cell lines using DNAzol (Life Technologies). The long terminal repeats (LTRs) of TAR mutant viruses were amplified by 30 cycles of PCR (95, 55, and 72°C for 1 min each) using purified chromosomal DNA (500 ng) as a template, 1.25 U of Platinum Taq DNA polymerase (Life Technologies), and 50 ng of each of the following oligonucleotide pairs: 5′ LTR, −436/−415 (5′-CCC AAA CAA GAC AAG AGA TTG A, sense) and +242/+219 (5′-CCT GCG TCG AGA GAG CTC CTC TGG, antisense); 3′ LTR, +8605/+8625 (5′-GCA GCT TTA GAT ATT AGC CAC, sense) and +9282/+9258 (5′-CTG CTA GAG ATT TTT CCA CAC TGA C, antisense). The 678- and 677-bp PCR products amplified from the 5′ and 3′ LTRs, respectively, were ligated into pGemTeasy (Promega) and analyzed by DNA sequencing.
Quantitative RT-PCR was carried out on virion-associated RNA isolated from pelleted virus particles. DNase I-treated stocks of wild-type or TAR mutant virus were subjected to centrifugation through a 20% sucrose cushion at 120,000 × g for 90 min at 4°C and resuspended in serum-free IMDM. The viral suspensions were assayed for p24 Ag and RT activity as described above. Equal amounts of p24 Ag (100 ng) were solubilized with Trizol (Life Technologies) according to the manufacturer's recommendations, and 5 ng of internally deleted HIV-1 internal control RNA was added to monitor RNA recovery and in vitro reverse transcription efficiency. Nucleic acids were precipitated overnight and recovered by centrifugation at 15,000 × g at 28°C for 1 h. The visible pellets were rinsed with 70% ethanol and dissolved in 30 μl of water. Virion RNA was reverse transcribed using C-Therm DNA polymerase (Roche Diagnostics Australia Pty. Ltd.) according to the manufacturer's instructions in duplicate reaction mixtures containing 10 μl of viral RNA and 50 ng of either first-strand primer B (+412/+387; 5′-GAC TGC GAA TCG TTC TAG CTC CCT GC, antisense) or first-strand primer A (+288/+267; 5′-CAG TCG CCG CCC CTC GCC TCT TG, antisense). The RT reaction mixtures were serially diluted and assayed for the internal control RNA by 27 cycles of PCR as described above with oligonucleotide primers complementary to the polylinker region of pGem3z (5′-GGG AGA CAA GCT TGC ATG CCT G, sense) and to HIV-1 sequences from position +65 to +46 (5′-AAG CAG TGG GTT CCC TAG TTA G, antisense). The HIV-1-specific primer was radiolabeled using T4 polynucleotide kinase and [γ-32P]ATP. The reaction mixtures were normalized according to internal control cDNA concentrations and assayed for HIV-1 cDNA by 30 cycles of PCR as described above with a radiolabeled HIV-1-specific oligonucleotide, primer +96/+118 (5′-CAA GTA GTG TGT GCC CGT CTG TT, sense), and an unlabeled primer, +182/+158 (5′-CTG CTA GAG ATT TTT CCA CAC TGA C, antisense). The PCR products were then separated on 5% polyacrylamide (19:1 acrylamide/bisacrylamide ratio) gels and quantified by PhosphorImager (Molecular Dynamics) analysis using PCR standard curves generated from plasmids containing the target sequences. All PCRs were performed in the linear range of the assay.
RNA protection assay.
RNA protection assays were performed using an RPAII kit (Ambion) according to the manufacturer's instructions. Briefly, DNase I-treated sucrose cushion-purified virus containing 100 ng of p24 Ag was solubilized along with 10 μg of purified yeast RNA and a radiolabeled RNA riboprobe (Promega Australia Pty. Ltd.) using Trizol reagent (Life Technologies). The RNA mixtures were incubated in the hybridization solution provided with the kit at 46°C for 16 h and digested with a combination of RNase T1 and RNase A. The digested RNAs were precipitated with ethanol and centrifuged at 12,000 × g at 4°C, and the pellets were resuspended in the RNA loading dye provided. The digested RNAs were separated on denaturing (7.3 M urea) 5% polyacrylamide gels and visualized and quantitated by PhosphorImager (Molecular Dynamics) analysis.
RT assays.
Wild-type and TAR mutant virus stocks were assayed for total RT activity on a synthetic homopolymer template in the presence of detergent using the RT Detect Assay (Roche Diagnostics) according to the manufacturer's instructions. Aliquots of virus (typically 1.0 mU of RT activity) were supplemented with 10 mM MgCl2 and incubated for 30 min at 37°C with 100 U of DNase I in a final volume of 200 μl of IMDM. Reverse transcription was terminated in half of the DNase I-treated virus stock by the addition of 150 μl of stop solution (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 20 mg of sheared salmon sperm DNA per ml, and 50 mg of proteinase K per ml) followed by incubation at 37°C for 10 min and then boiling for 10 min. The remaining 100 μl was supplemented with 200 μM deoxynucleoside triphosphates and incubated at 37°C for 90 min, and then the reaction was terminated as described above. The stopped reaction mixtures were centrifuged briefly at 14,000 × g, and 10 μl of each was assayed for negative-strand strong-stop DNA by 34 cycles of PCR (65°C for 2 min and 93°C for 1 min) using the HIV-1-specific oligonucleotide primers +96/+118 (radiolabeled) and +158/+182 in the presence of 3.5 mM MgCl2 to compensate for EDTA present in the stop mix. PCR standard curves were generated by amplifying serial dilutions of an HIV-1 proviral plasmid, and all PCRs were performed in the linear range of the assay.
RESULTS
Isolation and characterization of cell lines making wild-type and TAR mutant HIV-1.
In a previous study we identified HIV-1 viruses with mutated TAR RNA sequences which supported transactivation by Tat and high levels of gene expression but did not support efficient virus replication (18). Our original analysis showed that these viruses were defective for reverse transcription. However, two recent reports have shown that mutations of the HIV-1 lower stem-loop resulted in decreased packaging of virus genomic RNA and supported packaging of HIV-1 spliced RNA transcripts (8, 11). Our original study found either minor or no packaging defects but did not exclude the possibility that packaging of spliced RNA transcripts was responsible for the observed reverse transcription defects.
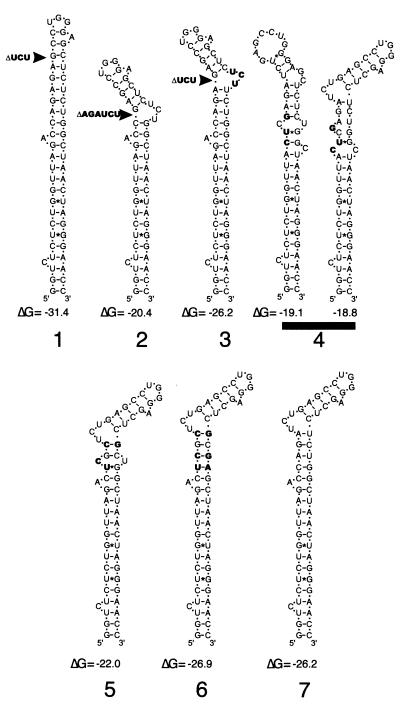
The following mutations were introduced into the TAR RNA upper stem-loop structure in the HIV-1 proviral construct pBRDH2-neo: a six-base deletion of nt +19 to +24 (TAR2), a three-base deletion of nt +22 to +24 in combination with a compensatory UCU insertion (TAR3), and a three-base substitution at nt +15, +16, and +18 (TAR4) (Fig. 1). Each mutated proviral DNA was transfected into 293 cells, and stable cell lines producing each TAR mutant virus were selected and characterized as previously described (15). In addition, three stable cell lines which have been described elsewhere (15) were used to produce viruses carrying the following TAR RNA mutations: a three-base deletion from nt +22 to +24 (TAR1); a four-base substitution at nt +18, +19, +21 and +39 (TAR5); and a six-base substitution mutation, nt +18, +19, +21/+39, +41, and +42, that maintained TAR RNA structure (TAR6) (Fig. 1). Supernatant from each cell line was subjected to ultracentrifugation through a 20% sucrose cushion. The virus pellets were resuspended in serum-free RPMI 1640 medium, and their p24 Ag contents and total RT activities were measured. More than 90% of the p24 Ag present in culture supernatants was recovered from the pellets, and RT activity was detected only in the virus pellets (data not shown). The ratio of p24 Ag concentration to total RT activity was calculated for each virus stock, and the p24/total RT ratios observed in TAR mutant and wild-type virus stocks fell within the same range (Table 1).
FIG. 1.
RNA stem-loop structures and stem energies of wild-type and mutated TAR RNA sequences. TAR mutations are shown as follows: 1, 3-nt deletion from position +22 to +24 (TAR1); 2, 6-nt deletion from position +19 to +24 (TAR2); 3, combined 3-nt deletion from position +22 to +24 and compensatory 3-nt insertion of UCU at position +39 (TAR3); 4, three nucleotide substitutions at positions +15, +16, and +18 (TAR4); 5, four nucleotide substitutions at positions +18, +19, +21, and +39 (TAR5); and 6, six nucleotide substitutions, at nt +18, +19, +21, +39, +41, and +42, that maintained the stem-loop structure (TAR6). RNA stem-loop structure energies (ΔG), shown in kilocalories per mole, were predicted using M-fold version 3.0 (copyright 1996 by M. Zuker; http://mfold.burnet.edu.au).
TABLE 1.
Analysis of partially purified virusa
| Virusb | p24 Ag (ng/ml)c | RT activity (nU/μl)d | p24 Ag/RTe | Virus replicationf (%) |
|---|---|---|---|---|
| TAR1 | 115 | 3.0 | 38.3 | Negative |
| TAR2 | 92 | 2.1 | 43.8 | Negative |
| TAR3 | 103 | 2.4 | 42.9 | Negative |
| TAR4 | 257 | 7.8 | 32.9 | 3 |
| TAR5 | 1,212 | 26.9 | 45.0 | 5 |
| TAR6 | 261 | 8.2 | 31.7 | 100 |
| Wild type | 714 | 15.8 | 45.2 | 100 |
The results are representative of those from three independent experiments.
The TAR RNA sequence is shown in Fig. 1.
Concentration of p24 Ag measured in virus pelleted through a 20% sucrose cushion.
RT activity of virus pelleted through a 20% sucrose cushion.
Absolute value of p24 Ag concentration divided by RT activity.
Peak p24 Ag concentration in culture supernatant relative to the wild-type level.
Jurkat cells were infected with equivalent amounts of each virus for 2 h, after which the virus was removed. Infected and mock-infected cells were cultured for 3 weeks, and the culture supernatant was sampled twice weekly. The peak amount of p24 Ag produced by each infection was used as a measure of the replication potential of each virus and was expressed relative to the wild-type level (Table 1). TAR6 virus, which had an intact TAR-like RNA stem-loop structure, replicated at wild-type levels as previously observed. In contrast, the TAR1, TAR2, and TAR3 mutants failed to replicate at all, while TAR4 and TAR5 replicated at very low levels. These results are consistent with previous reports on the effect of TAR mutations on virus replication kinetics (15, 25). The previously observed capacity of these TAR structures to support activation of HIV-1 gene expression by Tat correlated with both the replication kinetics observed in Jurkat cells and the levels of virus made by stably transfected 293 cells (4, 13, 14, 16).
An intact upper TAR structure is required for natural endogenous reverse transcription.
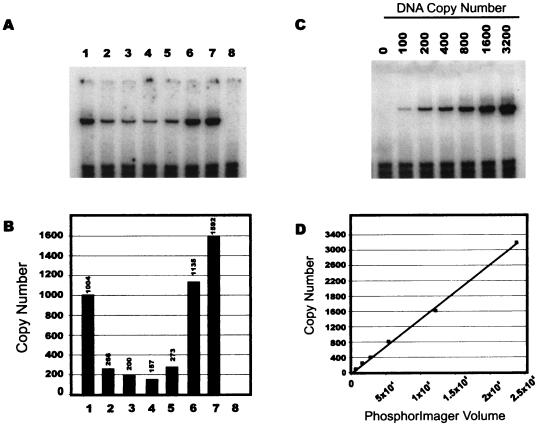
We used PCR to measure the amounts of negative strong-stop DNA synthesized by each of these viruses in natural endogenous reverse transcription (NERT) assays (32, 35), and we expressed the NERT capacity of each virus relative to that of the wild type. Consistent with previous results in a cell infection assay system, the TAR1 and TAR6 mutants produced 63 and 71%, respectively, of wild-type amounts of negative strong-stop DNA in NERT assays (Fig. 2). In contrast, the TAR2, TAR3, TAR4, and TAR5 mutants displayed only 10 to 17% of wild-type NERT capacity (Fig. 2). It is therefore apparent that deletion of sequences from position +22 to +24 had only minor effects on reverse transcription, while transposition of the bulge (TAR3, nt +37 to +38) resulted in defective reverse transcription. These NERT results confirmed that the stem structure, but not the primary sequence, of TAR from position +18 to +21 and +39 to +42 is required for optimal HIV-1 reverse transcription initiation. TAR5, which has a symmetrical internal loop at +19/+41, was also defective for negative strong-stop DNA synthesis in NERT assays. Finally, the TAR4 mutation, which, according to stem energy calculations (36), favors an alternate structure over a less stable TAR-like RNA conformation, also reduced negative strong-stop DNA synthesis relative to that of the wild type.
FIG. 2.
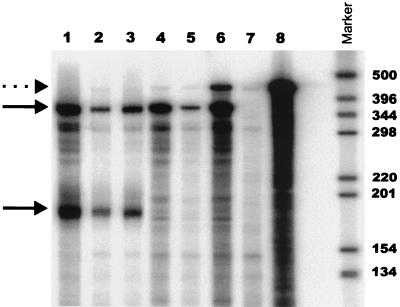
Representative NERT assay of wild-type and TAR mutant viruses. (A) negative strong-stop DNA detected by PCR in NERT reactions with TAR1, TAR2, TAR3, TAR4, TAR5, TAR5, TAR6, or wild-type virus (lanes 1 to 7, respectively) or mock supernatant (lane 8). (B) PhosphorImager analysis of the PCR shown in panel A; DNA copy number is indicated. (C) PCR standard curve generated using an HIV-1 proviral plasmid. (D) PhosphorImager analysis of the gel shown in panel C; r2 = 0.996.
An intact upper TAR structure is required for efficient packaging of HIV-1 genomic RNA.
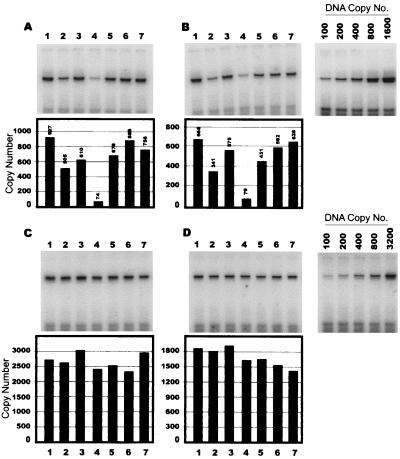
A previous analysis of HIV-1 TAR mutants (18) failed to detect significant defects in RNA packaging, but that study did not eliminate the possibility that defects in RNA packaging resulted in the encapsidation of spliced HIV-1 RNA species. Therefore, we performed both RT-PCR and RNA protection assays to quantitate and characterize virion RNA content in viruses with mutated TAR sequences. The RT-PCR strategy was similar to a previously described strategy (Fig. 3). Briefly, an in vitro-transcribed HIV-1 RNA with an internal 71-nt deletion was used as an internal control in the RT-PCR assay. Partially purified virus containing 100 ng of p24 Ag was lysed in Trizol solution, and 5 ng of synthetic internal control RNA was added. First-strand cDNA synthesis was performed in separate reactions using primer A, which hybridized to both spliced and unspliced RNA, and primer B, which hybridized only to unspliced RNA. The cDNA products were analyzed by PCR using primers specific for HIV-1 or internal control sequences. The PCR products were separated by polyacrylamide gel electrophoresis and then visualized and quantitated with a Molecular Dynamics PhosphorImager. As shown in Fig. 4, similar levels of HIV-1 genomic RNA were measured by RT-PCR in the TAR1, TAR5, TAR6, and wild-type viruses. RNA packaging in the TAR2 and TAR4 viruses was reduced to approximately 55% and approximately 10%, respectively, of the wild-type level. The overall reduction in copy numbers detected in first-strand reactions containing primer B, compared to primer A, was due to the lower efficiency of primer B in first-strand synthesis and was consistent between HIV-1-specific and internal control primers.
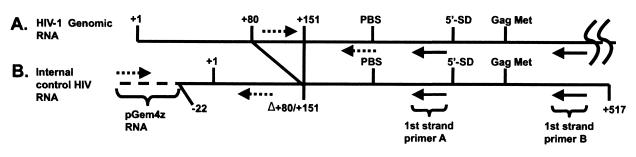
FIG. 3.
RT-PCR strategy. HIV-1 genomic RNA (A) was isolated from virus containing 100 ng of p24 Ag along with added synthetic internal control RNA (B) containing plasmid sequence (dotted line) and HIV-1 sequences from position −22 to +517, except for an internal deletion from +80 to +151. The isolated RNA was reverse transcribed in separate reactions using the first-strand primers shown (solid arrows). Primer A can anneal to either spliced or unspliced HIV-1 RNA, while primer B can anneal to unspliced but not to spliced HIV-1 RNA; both primers can anneal to internal control RNA. HIV-1 and internal control cDNAs were detected by PCR using the primers shown (dotted arrows). PBS, primer binding site.
FIG. 4.
RT-PCR analysis of virion RNA levels. Virion RNA was isolated from partially purified TAR1, TAR2, TAR3, TAR4, TAR5, TAR6, and wild-type virus (lanes 1 to 7 respectively), to which internal control RNA had been added. The isolated RNAs were reverse transcribed in either the presence or absence (not shown) of C-Therm DNA polymerase and first-strand primer A (A and C) or first-strand primer B (B and D). PCR to detect HIV-1 cDNA (A and B) or internal control cDNA (C and D) was performed using the primers shown in Fig. 3. The PCR products were separated by polyacrylamide gel electrophoresis and analyzed with a Molecular Dynamics PhosphorImager. The copy numbers shown for HIV-1 cDNA (A and B) are normalized to internal control cDNA copy numbers (C and D). The correlation coefficients (r2) of standard curves generated with plasmid DNA were 0.992 for HIV-1-specific primers and 0.997 for internal control-specific primers (data not shown). The data shown are representative of those from three similar replicate experiments using different virus stocks.
RNase protection assays were performed on virion RNA and cellular RNA from the corresponding 293 stable cell lines, using a 550-bp antisense RNA probe that spanned the 5′ major splice donor site. Full-length HIV-1 genomic RNA overlaps with 363 nt of the probe, while spliced HIV-1 RNA overlaps with only 189 bp. Virion RNA isolated from partially purified virus containing 100 ng of p24 Ag, or 10 μg of total cellular RNA isolated from stable 293 cell lines, was incubated with the radiolabeled antisense RNA probe. As shown in Fig. 5, both genomic and spliced HIV-1 RNA species were detected in cell lines producing TAR5, TAR4, and wild-type viruses. The ratios of full-length to spliced RNA were 0.82, 0.51, and 0.83 in TAR5, TAR4, and wild-type virus, respectively. These ratios are similar to the RNA ratios in wild-type and TAR mutant viruses reported by Clever et al. (8). Virion RNA from TAR5, TAR4, and wild-type virus protected an RNA fragment indicative of full-length genomic RNA, but little if any spliced RNA was detected. In this and other similar RNase protection assays, the RNA packaging efficiency of the TAR5 mutant ranged from 50 to 60% of the wild-type level, compared to the value of 80% obtained from the RT-PCR assay. The RNA packaging efficiency of TAR4 ranged from 8 to 11% of the wild-type level, consistent with the RT-PCR assay results.
FIG. 5.
RNA protection assay of cellular and virion RNAs. Total cellular RNA was isolated from stable 293 cell lines producing TAR4 (lane 2), TAR5 (lane 1), and wild-type (lane 3) HIV-1. Ten micrograms of total cellular RNA or virion RNA isolated from TAR4 (lane 5), TAR5 (lane 4), and wild-type (lane 6) virus containing 100 ng of p24 Ag was annealed to excess radiolabeled RNA probe. The same RNA probe was also incubated with 10 μg of yeast RNA (lanes 7 and 8). The annealed RNA mixtures were digested with RNase T1 and RNase A. Intact probe is shown (lane 8 and dotted arrow). Protected fragments corresponding to unspliced (top arrow, 363 nt) or spliced (bottom arrow, 189 nt) HIV-1 RNA are indicated.
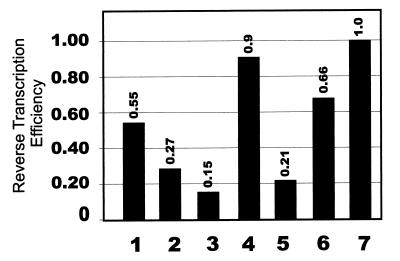
The relative reverse transcription efficiency of each TAR RNA mutant was calculated as the percentage of wild-type NERT activity divided by the percentage of wild-type RNA packaging level (Fig. 6). This analysis revealed that the NERT defect observed in the TAR4 virus could be attributed almost entirely to decreased RNA packaging, while the TAR1 and TAR6 viruses both had relative reverse transcription efficiencies which were reduced by less than onefold, which was consistent with previous results (18, 20). The TAR2, TAR3, and TAR5 viruses were all defective for reverse transcription; the TAR3 mutant had a sevenfold defect compared to wild-type virus. The reverse transcription defects associated with TAR5 were lower in NERT assays than previously reported for cell infection assays (13) and were partly due to reduced packaging efficiency.
FIG. 6.
Relative reverse transcription efficiencies of wild-type and TAR mutant viruses. The reverse transcription efficiency of each virus was calculated as the percentage of wild-type negative strong-stop DNA synthesis measured by NERT assay divided by the percentage of wild-type virion RNA detected by RT-PCR (values were averaged over two different first-strand primers). Relative reverse transcription efficiencies of TAR1, TAR2, TAR3, TAR4, TAR5, TAR6, and wild-type viruses (lanes 1 to 7, respectively) are shown.
DISCUSSION
In this study we confirmed that TAR is required for efficient HIV-1 reverse transcription. The upper stem-loop from position +18 to +42 is required for this function of TAR, but reverse transcription is not strictly dependent on the bulge sequences from position +22 to +24, since deletion of these sequences had no effect. However, reverse transcription was down regulated when the bulge was either transposed to the opposite side of the stem or shifted by an internal deletion, indicating that the role of TAR in reverse transcription is dependent on its secondary structure. The mechanism by which TAR exerts its effect remains unknown, but it may involve the binding of viral or cellular proteins that facilitate reverse transcription. Alternatively, TAR may form alternative RNA structures that stabilize the reverse transcription complex. The TAR RNA upper stem-loop structure is the binding site for the HIV-1 Tat protein and cellular protein complexes that contain the RNA polymerase II large subunit, Trp-185, and Tat-cyclin-T1 (33, 34). The binding events involving these proteins share a common dependence upon single-stranded regions of the loop and.or bulge regions of TAR. The binding of protein to RNA is commonly accompanied by a conformational change in RNA structure. For example, crystal structure analysis of the anticodon loops of tRNAs complexed with the appropriate aminoacyl-tRNA synthetases showed that protein interactions facilitate unstacking of anticodon sequences, allowing them to bind in separate recognition pockets in the RNA structure (10). In TAR RNA, the three-nucleotide bulge causes a distortion in the RNA duplex which makes the major groove accessible, and binding of ligands to this region results in the formation of a triple base pair between U23 and A27-U38 that restores TAR RNA to an A-form-like duplex (1, 7, 30). It is intriguing that the TAR bulge deletion mutation, an A-form RNA duplex structure, was the only bulge mutation that supported high levels of reverse transcription. It is possible that Tat or another TAR binding protein may induce a similar conformational change in TAR structure and that this conformational change favors efficient reverse transcription. In a separate study, Tat was shown to be required for efficient early reverse transcription in the absence of obvious biochemical defects in the virions (17, 32), but numerous attempts to detect Tat in purified HIV-1 have been unsuccessful. Studies are under way in our laboratory to determine whether other viral proteins such as RT and Ncp7 specifically bind to the TAR element.
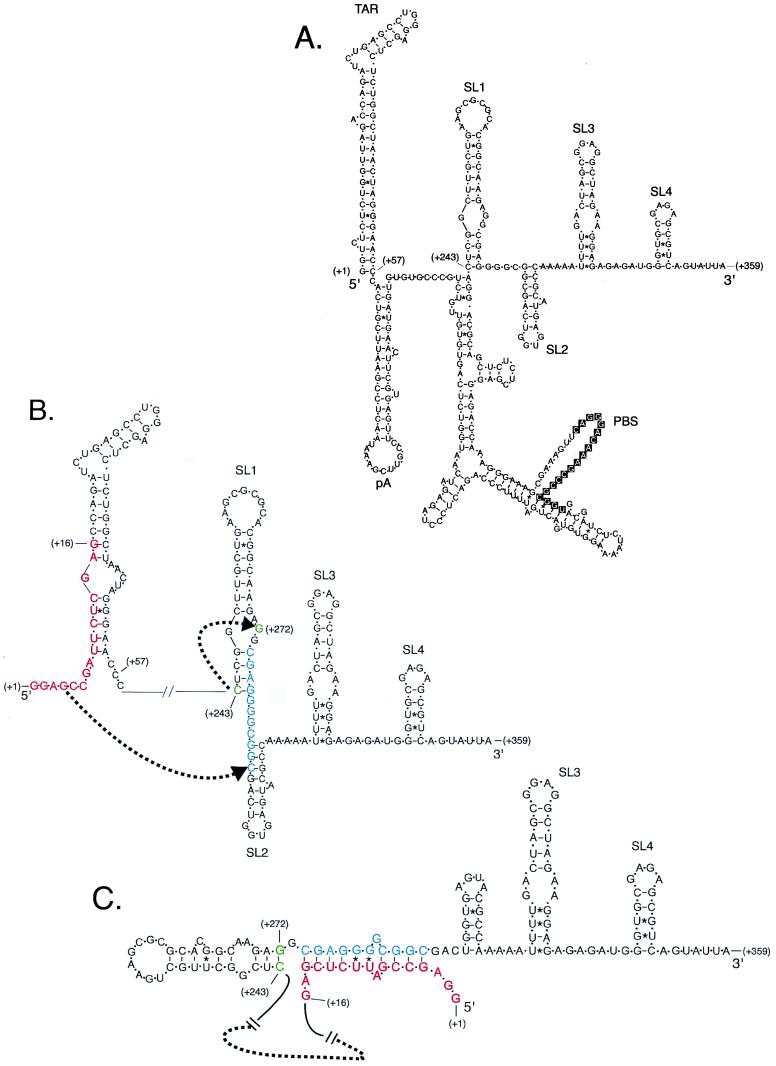
The multiple functions associated with TAR include transcription, translation, genomic RNA packaging, and reverse transcription. Some of these functions are at least partially controlled by complex RNA structures. For example, packaging depends on the so-called psi sequences, which include the stem-loop structures SL1, SL2, and SL4 (Fig. 7A) (3, 9, 19, 27), and reverse transcription requires RNA interactions between the primer binding site and flanking sequences that bind the anticodon loop of tRNA3Lys (22, 36, 37). Some earlier studies of TAR mutations used gross clustered mutations that greatly altered TAR structure. For example, a mutation called Xho+10 destabilized TAR from position +2 to +16 and reduced packaging efficiency by more than 90% (11). However, these large mutations may allow alternative Watson-Crick base pairing with nearby sequences, and this may interfere with other natural RNA structures. Computer analysis indicates that Xho+10 sequences have a strong potential to base pair with nearby sequences within SL1, which could cause marked changes in the RNA structures of SL1 and SL2 (Fig. 7B and C). Our results confirm that TAR plays a role in packaging, since viruses with altered TAR structures have packaging defects. In this study and others, viruses that formed non-TAR-like structures, or which destabilized the lower stem structure, were associated with packaging defects of 90% or greater (8, 11, 20, 28). Mutations in TAR that destabilized portions of the lower stem without disturbing other structural elements greatly reduced RNA packaging, and second-site mutations that restored the stem base pairing also restored RNA packaging. These observations indicate that structure rather than primary RNA sequence is important for this function of TAR. The seminal observations that distinct pools of genomic RNA traffic to either translation or packaging processes (26), that TAR RNA inhibits translation (6, 31), and that the TAR lower stem region contributes to an efficient packaging signal (8, 11, 20) all suggest that specific features of the TAR lower stem are one determinant of RNA packaging and translation pathways that is codependent upon other RNA structures or RNA binding proteins.
FIG. 7.
Computer model of potential HIV-1 RNA secondary structures in the presences of stem-destabilizing clustered mutations. (A) HIV-1 RNA secondary structure predicted from the wild-type HIV-1 HXB2 sequence from position +1 to +359 using M-fold version 3.0. Stem-loop structures corresponding to TAR, pA, SL1, SL2, SL3, and SL4 as well as the primer binding site (PBS) are indicated. (B) Predicted secondary structure of TAR RNA (positions +1 to +57) showing the clustered mutation Xho+10 (10) indicated by red letters. HIV-1 nt +274 to +284 (shown in blue) can base pair to sequences in Xho+10. A dotted arrow indicates a shift by cytidine +243 (green) to base pair with guanine +272 (green). (C) The new secondary structure maintains TAR and pA stem-loop structures (not shown), SL3, and SL4 but eliminates SL1 and SL2. The free energy of this structure is −40.2 kcal/mol compared to the structure shown in panel B, from position +243 to +359, with free energy of −33.9 kcal/mol calculated by M-fold version 3.0 using standardized parameters (36).
Our results provide evidence that the TAR upper stem structure is also important for RNA packaging, since the TAR4 mutant, which has an altered upper stem-loop structure but a conserved lower stem mutation, was defective for RNA packaging. However, not all of the TAR mutations fit this model. For example, a TAR deletion from position +34 to +37 would be expected to form an alternative RNA structure, yet genomic RNA carrying this deletion was efficiently packaged into virions (20). At least a partial explanation for this may lie in the different methods which have been used to generate virus in different studies. Several studies produced virus by transient expression from proviral plasmids, and in some cases these plasmids carried a simian virus 40 origin of replication. Transfection of these plasmids into either COS or 293T cells, both of which make simian virus 40 large T antigen, results in amplification of the plasmid and synthesis of large quantities of viral transcripts. In contrast, the present study used isolated cell lines with stably integrated proviral constructs, resulting in levels of virus production more nearly approximating those in a natural infection. The impact of this technical difference is not known and requires further analysis.
Further study is also required to identify the specific features of TAR that support reverse transcription and genomic RNA packaging. Attempts to identify potential cofactors of reverse transcription are in progress in our laboratory.
ACKNOWLEDGMENTS
We thank William B. Lott for critical reading of the manuscript.
This work was supported by grants from the National Centre for HIV-1 Virology Research (to D.H.).
Footnotes
Publication number 106 from Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin F, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, Jeang K T. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991;19:6169–6176. doi: 10.1093/nar/19.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 6.Braddock M, Powell R, Blanchard A D, Kingsman A J, Kingsman S M. HIV-1 TAR RNA-binding proteins control TAT activation of translation in Xenopus oocytes. FASEB J. 1993;7:214–222. doi: 10.1096/fasebj.7.1.8422967. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky A S, Erlacher H A, Williamson J R. NMR evidence for a base triple in the HIV-2 TAR C-G.C+ mutant-argininamide complex. Nucleic Acids Res. 1998;26:1991–1995. doi: 10.1093/nar/26.8.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever J L, Eckstein D A, Parslow T G. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article] [PubMed] [Google Scholar]
- 11.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Holland E C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 14.Garcia J A, Harrich D, Soultanakis E, Wu F, Mitsuyasu R, Gaynor R B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989;8:765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrich D, Hsu C, Race E, Gaynor R B. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J Virol. 1994;68:5899–5910. doi: 10.1128/jvi.68.9.5899-5910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrich D, Mavankal G, Mette-Snider A, Gaynor R B. Human immunodeficiency virus type 1 TAR element revertant viruses define RNA structures required for efficient viral gene expression and replication. J Virol. 1995;69:4906–4913. doi: 10.1128/jvi.69.8.4906-4913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrich D, Ulich C, Garcia-Martinez L F, Gaynor R B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrich D, Ulich C, Gaynor R B. A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4017–4027. doi: 10.1128/jvi.70.6.4017-4027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison G P, Lever A M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helga-Maria C, Hammarskjold M L, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 22.Isel C, Keith G, Ehresmann B, Ehresmann C, Marquet R. Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res. 1998;26:1198–1204. doi: 10.1093/nar/26.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isel C, Lanchy J M, Le Grice S F, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 24.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 24a.Jeang K T, Xiao H, Rich E A. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 25.Klaver B, Berkhout B. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 1994;13:2650–2659. doi: 10.1002/j.1460-2075.1994.tb06555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin J G, Rosenak M J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. . (Erratum, 71:858, 1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon D T, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puglisi J D, Tan R, Calnan B J, Frankel A D, Williamson J R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 31.SenGupta D N, Berkhout B, Gatignol A, Zhou A M, Silverman R H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulich C, Dunne A, Parry E, Hooker C W, Gaynor R B, Harrich D. Functional domains of Tat required for efficient human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:2499–2508. doi: 10.1128/jvi.73.3.2499-2508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 34.Wu-Baer F, Sigman D, Gaynor R B. Specific binding of RNA polymerase II to the human immunodeficiency virus trans-activating region RNA is regulated by cellular cofactors and Tat. Proc Natl Acad Sci USA. 1995;92:7153–7157. doi: 10.1073/pnas.92.16.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Dornadula G, Pomerantz R J. Natural endogenous reverse transcription of HIV type 1. AIDS Res Hum Retroviruses. 1998;14(Suppl. 1):S93–S95. [PubMed] [Google Scholar]
- 36.Zhang Z, Kang S M, Li Y, Morrow C D. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA. 1998;4:394–406. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Kang S M, Morrow C D. Genetic evidence of the interaction between tRNA(Lys,3) and U5 facilitating efficient initiation of reverse transcription by human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1998;14:979–988. doi: 10.1089/aid.1998.14.979. [DOI] [PubMed] [Google Scholar]
- 38.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]