Abstract
Vaccinia virus complement control protein (VCP) has been shown to possess the ability to inhibit both classical and alternative complement pathway activation. The newly found ability of this protein to bind to heparin has been shown in previous studies to result in uptake by mast cells, possibly promoting tissue persistence. It has also been shown to reduce chemotactic migration of leukocytes by blocking chemokine binding. In addition, this study shows that VCP—through its ability to bind to glycosaminoglycans (heparin-like molecules) on the surface of human endothelial cells—is able to block antibody binding to surface major histocompatibility complex class I molecules. Since heparin binding is critical for many functions of this protein, we have attempted to characterize the molecular basis for this interaction. Segments of this protein, generated by genetic engineering of the DNA encoding VCP into the Pichia pastoris expression system, were used to localize the regions with heparin binding activity. These regions were then analyzed to more specifically define their properties for binding. It was found that the number of putative binding sites (K/R-X-K/R), the overall positive charge, and the percentage of positively charged amino acids within the protein were responsible for this interaction.
Poxviruses are complex double-stranded DNA viruses capable of causing disease in a wide variety of animals, including humans (22). Much of the success of this family of viruses is due to the ability to encode several proteins that can subvert the host's immune response. Poxviruses produce two major families of proteins involved in evasion of host defense: cell-associated cytokine response-modifying proteins (serpin-related proteins) (14) and secreted proteins termed virokines (13). The secreted protein family is made up of several cytokine-chemokine-binding proteins (termed viroreceptors) (29), a neurovirulence factor (10), and a complement control protein (13). We have been most interested in studying the many properties of the vaccinia virus complement control protein (VCP). VCP, the first identified soluble microbial protein to have complement binding activity, is composed of four short consensus repeats (SCR), making it and other homologous proteins members of the complement control protein (RCA) superfamily (13). VCP exhibits functional similarity to factor H (fH), membrane cofactor protein, decay-accelerating factor, and complement receptor one and is structurally similar to human complement 4b binding protein. Biologically active VCP has been shown to inhibit the classical pathway of complement activation through its ability to bind C3 and C4 and act as a cofactor for factor I cleavage of C3b and C4b (11, 19). It has also been shown to inhibit the alternative pathway (to a lesser extent) by using the same mechanism of action, resulting in the cleavage of C3b into iC3b, thereby preventing the formation of the alternative pathway C3 convertase (24). It has been shown that VCP can prevent antibody-mediated virus neutralization (8). Also, by blocking complement activation at multiple sites, there is a large reduction of C3a, C4a, and C5a proinflammatory chemotactic factors, resulting in reduced cellular influx and inflammation. Support for this comes from in vivo experiments using BALB/c mice injected in the footpad with cowpox virus (CPV) expressing or lacking the VCP homolog in CPV, termed the inflammation modulatory protein (IMP) (21). Results showed a significantly greater influx of inflammatory cells into the tissue when infected with CPV lacking IMP than that found in IMP-expressing CPV-infected footpads. Another experiment, using BALB/c congenically matched C5-sufficient and C5-deficient mice injected in the footpad with CPV, showed that C5-deficient mice exhibited a significantly greater swelling response that persisted longer and also showed signs of hemorrhage, edema, and ulceration (20). In addition, the presence of IMP drastically diminished the inflammatory response elicited by CPV (7, 12).
The most recently identified property of the multifunctional VCP is its ability to bind heparin. It has been shown in previous studies, as well as this one, to exhibit lysozyme-like heparin binding activity. Because of this activity, VCP can be taken up by mast cells and possibly persist in the tissue for extended periods of time, helping to preserve the viral habitat (9). It has also been shown to reduce chemotactic migration of leukocytes in the presence and absence of the chemokine MIP-1α (23). This suggests that VCP can bind to heparin-like molecules lining the surface of endothelial cells, blocking chemokine binding and thereby blocking the chemotactic signal.
In this paper, we have attempted to further characterize the biological significance of VCP's ability to bind heparin. Using flow cytometry, the amount of specific antibody binding to human endothelial cells—in the presence and absence of VCP—was measured. It was found that VCP was able to inhibit antibody binding to major histocompatibility complex class I molecules on human endothelial cells. This suggests that VCP can interfere with molecular interactions with infected cells and could prevent antibody-dependent cell-mediated cytotoxicity as well as other cytotoxic cell interactions with target cells. The ability of VCP to bind heparin-like molecules suggests that it plays many roles and therefore may have a variety of applications. It is for these reasons that we have been interested in obtaining a better understanding of the molecular basis for the VCP-heparin interaction. Through examination of several recombinant VCP (rVCP) fragments, it has now been determined that the percentage of positively charged amino acids, overall charge, and the number of putative heparin binding sites are all important factors governing the heparin binding ability of VCP.
MATERIALS AND METHODS
Flow microfluorimetric analysis.
Human umbilical cord vascular endothelial cells (HUVECs) were obtained from the American Type Culture Collection (Manassas, Va.) at passage 13. Monolayer cultures were maintained by using F12K Ham's media supplemented with 10% fetal bovine serum (Sigma), 30 μg of endothelial cell growth supplement (Sigma) per ml, and 100 μg of heparin (Sigma) per ml at 37°C in humidified air containing 5% CO2. Cells were cultured to approximately 80% confluency in 75-cm2 vented flasks (Falcon, Lincoln Park, N.J.) coated with 1.5% gelatin (Sigma) in phosphate-buffered saline. Cells were trypsinized (0.25% trypsin, 1 mM EDTA; Sigma), and volumes at 4 × 105 cells/ml were placed in six-well flat-bottom culture plates (3.5-cm-diameter wells; Falcon) coated with 1.5% gelatin and incubated for 24 h in F12K media without growth factor (15). In order to determine the ability of rVCP, rVCP SCR (3, 4), and heparin binding protein (HBP) to bind HUVECs, duplicate wells (2 ml each) were trypsinized and washed with fluorescent treponemal antibody (FTA) hemagglutination buffer (Becton Dickinson, San Jose, Calif.) and stained for 30 min on ice with either 20 μg of fluorescein isothiocyanate (FITC)-labeled rVCP, 30 μg of FITC-labeled rVCP SCR (2, 3), or 30 μg of FITC-labeled HBP. For analysis of antibody interaction with cell surface class I HLA-ABC molecules, triplicate wells (2 ml each) were trypsinized and washed with FTA hemagglutination buffer and stained for 30 min on ice with 0.25 μg of phycoerythrin-conjugated mouse anti-human HLA-ABC monoclonal antibody (MAb) (Caltag, Burlingame, Calif.) or a mouse immunoglobulin G2a (IgG2a) MAb (an isotype-match negative control) in the presence or absence of 2 or 5 μg of VCP that was 90 to 98% pure from natural infection, prepared as described before (11). After incubation, cells were washed three times in FTA buffer and then fixed in Hank's balanced salt solution containing 2% paraformaldehyde. Before being stained, cell cultures were assessed for viability by trypan blue dye exclusion, and in all cases, the cells were found to be >95% viable. The percentage of positively stained cells was determined by using a flow cytometer (Becton Dickinson FACScan) equipped with a single 15-mW argon laser tuned to 488 nm. Forward and 90° angle light scatter and integrated log phycoerythrin and FITC fluorescence signals were collected and analyzed. Variability between duplicate samples was less than 10%. To compensate for any background fluorescence, the control threshold was set at less than 1% binding of control MAbs. Data were acquired from the analysis of >3,000 events. A single homogenous cell population was indicated as detected by forward and 90° light scatter.
Cloning of VCP.
Fragments of VCP were expressed in Pichia pastoris cells by using the secretory expression vector pPIC9. The fragments corresponded to amino acids 18 to 146 [rVCP SCR (1,2)], 82 to 204 [rVCP SCR (2,3)], 145 to 263 [rVCP SCR (3,4)], and 18 to 262 [rVCP SCR (1-4)]. Genomic DNA from vaccinia virus was used as template for the amplification of the DNA fragments encoding the above protein fragments by PCR. In all cases except for rVCP SCR (3,4), the oligonucleotides used introduced a 5′ EcoRI site and a 3′ NotI site, which were used for cloning the fragments into the expression vector. For rVCP SCR (3,4), cloning was carried out as described earlier (30).
The selection of clones and expression of rVCP SCR (3,4) have been described previously (30). For rVCP SCR (1,2), rVCP SCR (2,3), and rVCP SCR (1-4), clones transformed with the expression vectors described above were selected on the basis of their ability to grow on histidine-deficient medium. Small-scale expression screening was performed by inoculating 5 ml of buffered minimal glycerol (BMG) and growing it at 30°C until the optical density at 600 nm (OD600) reached between 6 and 10. The cells were harvested and resuspended in 2 ml (for KM71 cells) or 15 ml (for GS115 cells) of buffered minimal methanol (BMM). Cells were grown with vigorous shaking for 4 to 5 days with daily addition of methanol to reach a concentration of 0.5%. Media from these inductions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the highest level expressers were selected. For rVCP SCR (1,2), (2,3), and (1-4), the KM71 cell line proved to produce the most protein. For large-scale growth of rVCP SCR (1,2) and rVCP SCR (1-4), 100 ml of BMG was inoculated with 5 ml of an overnight culture and grown at 30°C with vigorous shaking overnight. This culture was then used to inoculate several liters of BMG. The cultures were grown for 48 h until the OD reached approximately 20. The cells were spun down and resuspended in the same volume of BMM and grown for 4 to 5 days with vigorous shaking. Cells were fed methanol to a concentration of 1% every 24 h. For rVCP SCR (2,3), BMG media were inoculated with an overnight culture and grown at 30°C with vigorous shaking until an OD of approximately 6 had been attained. The pellet was then harvested by centrifugation, resuspended in BMM media (40% of the total BMG volume), and grown for 5 days with daily addition of methanol to a concentration of 0.5%.
Purification of rVCP SCR (3,4) has been described previously (30). For purification of rVCP SCR (1,2), the medium was concentrated down to a small volume by using a combination of a Millipore Prep/Scale-TFF cartridge (3-kDa molecular mass cutoff) and an Amicon stirred cell (with 3-kDa molecular mass cutoff). The sample was exchanged into 50 mM Tris-HCl (pH 9.0) by using a Pharmacia PD-10 column and loaded onto a Pharmacia Mono-Q column equilibrated in the same buffer. The protein was then eluted with an NaCl gradient of 0 to 100% over 20 min. Fractions containing rVCP SCR (1,2) were collected and concentrated by using Amicon stirred-cell ultrafiltration and then loaded onto a Brownlee Aquapore C4 reversed-phase column, and the protein was eluted with an acetonitrile gradient. Fractions containing pure rVCP SCR (1,2) were collected and lyophilized. For purification of rVCP SCR (2,3), the medium was concentrated to approximately 50 ml by using an Amicon concentrator (3-kDa cutoff) before being centrifuged at 20,000 × g for 1 h, and the pellet was discarded. The sample was exchanged into 5 mM sodium acetate (pH 4.0) by using a PD-10 column and applied to a Mono-S cation-exchange column (Amersham Pharmacia, Uppsala, Sweden) equilibrated in the same buffer. Protein was eluted with an NaCl gradient, and fractions corresponding to pure rVCP SCR (2,3) were pooled and lyophilized. For purification of rVCP SCR (1-4), the medium was concentrated to a small volume by the same means as rVCP SCR (1,2). The protein was exchanged into 20 mM phosphate (pH 6) by using a PD-10 column and dried.
Heparin binding ability.
In order to establish a basis for comparison, 10 μg each of bovine serum albumin (BSA), HBP (described earlier [5]), lysozyme, and MIP-1α were pooled, dissolved in 1 ml of ultrapure water, and passed through a 1-ml HiTRAP heparin column, and the unbound material was collected. The bound proteins were washed with 1 ml of ultrapure water and then eluted with increasing sodium chloride concentrations ranging from 250 mM to 4.0 M. Next, 20 μg of purified rVCP SCR (1,2), rVCP SCR (2,3), rVCP SCR (3,4), and rVCP or 10 μg of wild-type VCP were each dissolved in 1 ml of ultrapure water and passed through separate 1-ml HiTRAP heparin columns, and the unbound materials were collected. After the column was washed with 1 ml of ultrapure water, the proteins were eluted with sodium chloride concentrations ranging from 250 mM to 2.5 M. The fractions were then separated by using SDS-PAGE and silver stained. Densitometric readings were taken by using the AlphaImager 2000 software system. Finally, 0.5 ml of the fraction containing the protein was concentrated by ultrafiltration and its activity was tested by using the hemolysis assay.
Sequencing of the VCP homolog in MPV.
Sequencing of the VCP homolog in monkeypox virus (MPV) was done as described previously (29).
Hemolysis assay.
The biological activity of the proteins was determined by using the hemolysis assay, which was described earlier (12). In order to more specifically quantitate their activity, each protein was tested as a series of dilutions ranging from 0.5 to 5.0 μg.
Model of VCP.
The structures of the two unknown VCP modules (VCP SCR 1 and VCP SCR 2) were modeled by homology with the four known VCP module structures, VCP SCR 3, VCP SCR 4, fH SCR 15, and fH SCR 16, by using the program MODELLER (25). Five models of each module were created. The model which was closest to the average structure (as measured by root-mean-square deviation) was used in the construction of the VCP SCR (1-4) model. Construction of VCP SCR (1-4) was undertaken by using the molecular visualization package InsightII (Msi Inc., San Diego, Calif.). This was achieved by simply bonding the two modeled VCP structures to the nuclear magnetic resonance-derived structure of the VCP SCR (3,4) module pair (30). No attempt was made to accurately model the junctions between modules 1 and 2 and modules 2 and 3. Rather, torsion angles in the two residues linking the module pairs were set to yield a relatively extended conformation.
RESULTS
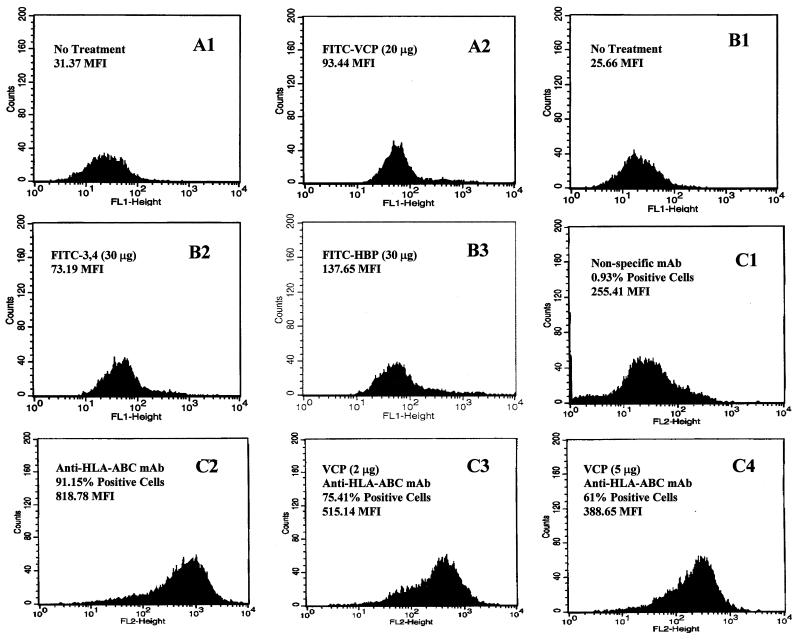
In order to determine the biological consequences of heparin binding by VCP, we tested whether VCP was able to block functional molecular interactions with human vascular endothelial cells. First, the ability of rVCP, rVCP SCR (3,4), and HBP to bind the surface of HUVECs was tested by using flow cytometric analysis. As shown in Fig. 1A and B, all three proteins possess the ability to bind strongly to HUVECs. A 3.0-, 2.9-, or 5.4-fold increase in fluorescent intensity over that of the control was observed when 20 μg of FITC-labeled rVCP, 30 μg of FITC-labeled rVCP SCR (3,4), or 30 μg of FITC-labeled HBP, respectively, was added to the suspension of HUVECs. Next, using flow cytometric analysis of HUVECs treated with mouse MAbs to human major histocompatibility complex class I molecules in the presence and absence of VCP, VCP was shown to be capable of modulating antibody binding in a dose-dependent manner. As shown in Fig. 1C, addition of 5 μl (2 μg) of VCP to antibody-treated HUVECs reduced antibody binding from 91.5 to 75.4%. An additional 15 μl (5 μg total) of VCP reduced antibody binding from 91.5 to 61.0%, suggesting that it blocks in a dose-dependent manner.
FIG. 1.
Representative histograms showing that VCP is able to reduce mouse anti-human HLA class I antibody binding to HUVECs. The results of flow microflourimetric analysis are as follows. (A1) No treatment control. (A2) Binding of FITC-labeled rVCP to HUVECs. (B1) No treatment control. (B2) Binding of FITC-labeled rVCP SCR (3,4) to HUVECs. (B3) Binding of FITC-labeled HBP to HUVECs. (C1) Nonspecific mouse MAb (isotype-matched mouse IgG2a MAb) binding to HUVECs (negative control). (C2) Mouse monoclonal anti-human HLA-ABC antibody binding to HUVECs (positive control). (C3) Mouse monoclonal anti-human HLA-ABC antibody binding to HUVECs in the presence of 5 μl (2 μg) of VCP. (C4) Mouse anti-human HLA-ABC antibody binding to HUVECs in the presence of 20 μl (5 μg) of VCP.
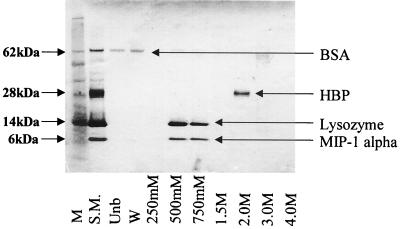
To calibrate the HiTRAP heparin column, several proteins having different binding affinities for heparin were passed through the column and eluted with increasing sodium chloride concentrations. SDS-PAGE analysis of the eluted fractions containing these proteins of various heparin-binding abilities can be seen in Fig. 2. Analysis suggests that BSA does not bind heparin, since it was present only in the unbound and wash fractions. Lysozyme and MIP-1α, which bound heparin with moderate and equal affinity, were contained within the 500 and 750 mM sodium chloride fractions. HBP, showing the highest affinity for heparin, was eluted by a sodium chloride concentration of 2.0 M.
FIG. 2.
PAGE analysis of the heparin binding activity of BSA, HBP, lysozyme, and MIP-1α. In order to calibrate the HiTRAP heparin column, various proteins (BSA, HBP, lysozyme, and MIP-1α) with differing affinities for heparin were passed through the column and eluted with sodium chloride concentrations ranging from 250 mM to 4.0 M. Lanes: M, molecular mass marker; S.M., starting material; Unb, unbound fraction; W, wash.
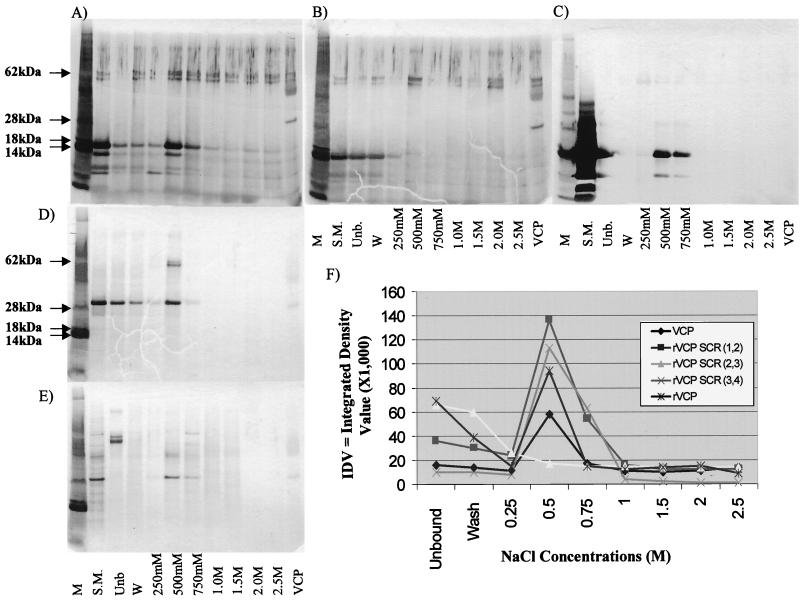
In order to better characterize the heparin binding ability of the VCP molecule, VCP, rVCP, and various recombinant segments of VCP were passed through HiTRAP heparin columns and analyzed by using SDS-PAGE, shown in Fig. 3. The results suggest that the full-length natural and recombinant proteins bind heparin with an affinity close to or equal to that of lysozyme and MIP-1α; both VCP and rVCP were concentrated primarily in the 500 and 750 mM sodium chloride fractions. PAGE analysis of the various rVCP segments revealed even more interesting data. rVCP SCR (1,2) and rVCP SCR (3,4) bound heparin with the same strength as the full-length protein, eluting once again at 500 and 750 mM. rVCP SCR (2,3), on the other hand, did not bind heparin at all and was found primarily in the unbound and wash fractions. The activity of the purified proteins was then tested by using the hemolysis assay. The results indicate that only the full-length protein inhibits lysis of sensitized sheep red blood cells—anywhere from 60 to 90% inhibition. The rVCP segments showed no inhibition of lysis, suggesting that the whole protein is needed to block complement activation. However, the naturally truncated VCP homolog produced by MPV (Fig. 4 and 5), which lacks almost the entire fourth SCR, has been shown to inhibit hemolysis of sensitized sheep red blood cells, thus indicating that it blocks the classical pathway of complement activation.
FIG. 3.
PAGE analysis of the heparin binding activity of VCP and rVCPs. VCP and various rVCPs were passed through separate HiTRAP heparin columns and eluted with sodium chloride concentrations ranging from 250 mM to 2.5 M. The fractions were collected, run on SDS-PAGE gels, and silver stained, and the band densities were measured. The results are shown as follows. (A) rVCP SCR (1,2). (B) rVCP SCR (2,3). (C) rVCP SCR (3,4). (D) rVCP. (E) VCP from the natural infection process. Lanes (A to E): M, molecular mass marker; S.M., starting material; Unb, unbound fraction; W, wash; VCP, VCP from natural infection. (F) Densitometric scanning of the gels (A to E).
FIG. 4.
Sequence alignment, including termini of rVCP constructs and putative heparin binding sites. Multiple alignment of the four SCR from orthopoxviruses VAC-COP (vaccinia virus, Copenhagen strain) (6), VAC-WR (vaccinia virus, Western Reserve strain) (10), CPV-GRI (cowpox virus, Russian isolate from human patient) (28), CPV-BRI (cowpox virus, Brighton strain) (20), VAR-BSH (variola virus, Bangladesh strain) (17), VAR-IND (variola major virus, India strain) (27), VAR-GAR (alastrim variola minor virus, Garcia strain) (18), and MPV-ZAI (MPV isolated from a human patient from Zaire in 1996). The putative heparin binding sites (K/R-X-K/R) are marked with solid bars, arrows indicate the termini of the rVCP constructs, and the cysteines are highlighted.
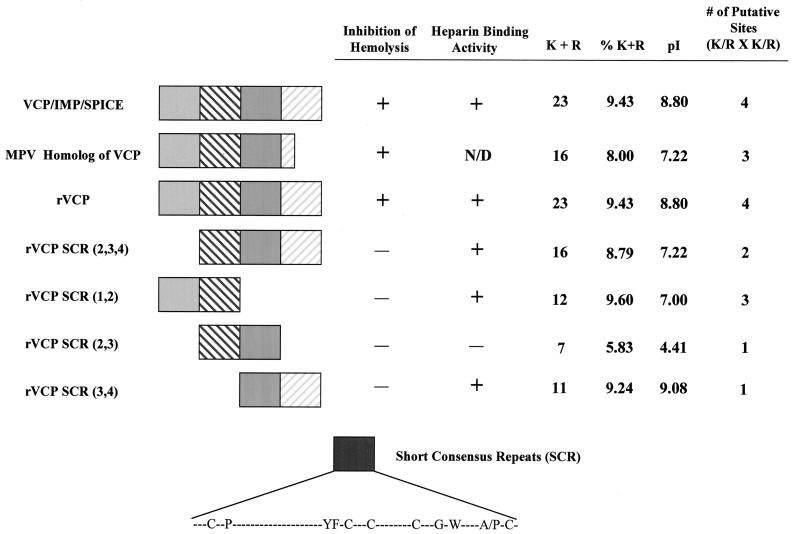
FIG. 5.
Structure-function summary table of VCP, VCP homologs, and rVCPs. VCP/IMP/SPICE, the MPV homolog of VCP, rVCP, and four recombinant segments of VCP are shown along with whether they are able to inhibit hemolysis of sensitized sheep red blood cells and/or bind heparin. Also listed are the number of positively charged amino acids (K + R) found in the protein, the percentage of positively charged amino acids (%K + R) making up the protein, the pI of the protein, and the number of putative heparin binding sites found on the surface of the protein.
To more precisely identify the molecular basis behind the VCP-heparin interaction, the amino acid sequences were analyzed by using the MacVector software system. Putative heparin binding sites (K/R-X-K/R) were first identified—shown in Fig. 4 along with the sequence alignment of different pox virus VCP homologs—and predicted to be located on the surface of the protein. The amino acid sequences were then scanned to determine the total number of positive amino acids, the percentages of these positively charged amino acids, and the overall pI of the protein, and the results are summarized in Fig. 4. All of the proteins shown to bind heparin had an overall pI of greater than 7.0 and, more importantly, were made up of more than 9% positively charged amino acids. The results also suggest that SCR 1 and SCR 4 contribute the most to this interaction. Although the results show there is no simple answer to how VCP binds heparin, it is clear that the overall positive charge, the number of putative binding sites, and the percentage of positive amino acids making up the protein have a significant effect on its ability to interact with and bind heparin.
DISCUSSION
The ability of VCP to bind heparan sulfate proteoglycan molecules adds a new dimension to its role in modulating the host immune response. In previous studies, the ability of VCP to bind heparin was found to permit uptake by mast cells, possibly allowing for tissue persistence over extended periods of time. In addition, earlier studies have shown that binding to heparin-like molecules on the surface of endothelial cells can block chemokine adherence, resulting in reduced chemotactic signaling. The next step was to determine if VCP bound to the endothelial cell surface could interfere with antibody binding. In this study, flow cytometry was used to demonstrate that VCP is able to bind the surface of endothelial cells, interfering with antibody binding. More importantly, this study demonstrates that addition of VCP significantly decreases the amount of antibody able to attach to HUVECs in a dose-dependent manner. It is postulated that VCP, binding to heparin-like molecules on the surface of endothelial cells, prevents antibody adherence. This binding may therefore block antibody attachment through steric interference and block both molecular and cellular interactions, which are dependent on target cell adhesion molecules binding to cytotoxic cells (F. Al-Mohanna, R. Parhar, and G. J. Kotwal, submitted for publication). Furthermore, this novel mechanism can protect virus-infected cells or the antigen-presenting cell from destruction by a host defense mechanism. Interestingly, it was estimated that the amount of VCP required for inhibition of roughly 5 × 105 cells is approximately the same as that produced by 2 × 105 infected cells, and therefore each infected cell can protect itself and one or two surrounding cells. Thus, VCP enables orthopoxviruses, all of which encode homologs of VCP, to evade the immune system by multiple mechanisms, e.g., by blocking complement pathways, by blocking antibody-dependent cytotoxic cell activity, or by interfering with attachment of cytotoxic T cells to virus-infected cells. In addition, as explained below, VCP can inhibit alternative pathway activation in a manner similar to that of fH, another heparin binding protein.
Due to the apparent importance heparin binding has in many of the biological activities exhibited by VCP, we became interested in better characterizing the molecular reasons for this interaction. Recombinant segments of VCP, each representing two of the four SCRs, were generated by genetic engineering vaccinia virus DNA encoding VCP into the P. pastoris expression system. The ability of these recombinant segments to bind heparin was then tested. It was determined that recombinant proteins rVCP SCR (1,2) and rVCP SCR (3,4) retained the same binding affinity as the full-length native protein while recombinant protein rVCP SCR (2,3) retained no heparin binding ability at all. In order to explain this, the amino acid sequences of the recombinant protein segments were more closely examined. It was determined that the number and, more importantly, the percentage of positively charged amino acids making up the protein contributed greatly to its ability to interact with heparin. Also, the overall charge and the number of putative binding sites (K/R-X-K/R) making up the protein appear to play a role in heparin binding. For the most part, the ability of VCP to bind heparin lies in its first and fourth SCR. These two SCR possess within them the highest percentage of positive amino acids and the greatest number of putative binding sites and have a very strong overall positive charge (data not shown).
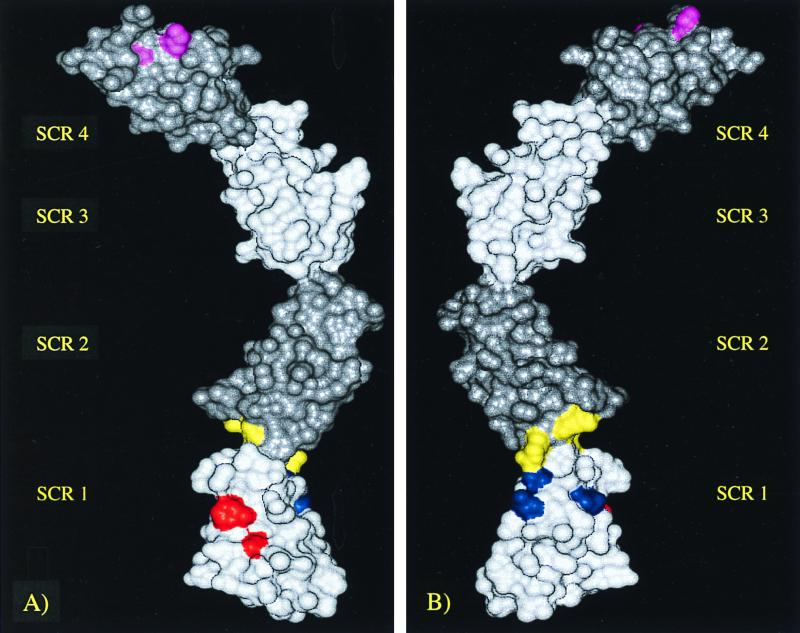
In order to better visualize the positioning of heparin binding sites in the VCP molecule, a model was developed by using four known VCP module structures: VCP SCR 3, VCP SCR 4, fH SCR 15 and fH SCR 16 (Fig. 6). As can be seen in this model, heparin binding exists primarily at the ends (SCR 1 and 4). The middle of the molecule therefore does not contribute to the binding [hence, VCP SCR (2,3) showed no ability to bind].
FIG. 6.
VCP model showing the heparin binding sites. Front (A) and back (B) views of the modeled structure of VCP SCR (1-4) showing the heparin binding sites (differentially colored). In order to differentiate the extents of the individual modules, they are shaded appropriately.
It is clear that the ability to bind polyanions, such as heparin, is crucial for the function of many immune-regulating proteins. fH, for example, binds to the polyanion sialic acid on the surface of mammalian cells, preventing activation of the alternative pathway. Like VCP, fH is made up of SCR, two of which have been shown to have heparin binding abilities (1). It is postulated that these heparin-binding domains are responsible for binding sialic acid, thereby preventing alternative pathway activation. The heparin binding regions of many other proteins exhibiting heparin binding activity have been mapped (2, 3, 16, 26, 31). In most cases (including ours), binding is the result of dense regions of positively charged amino acids. These arginine-rich and/or lysine-rich regions vary in sequence and length among heparin binding proteins. Interestingly, the heparin binding sequence used by VCP and its homologs is short and relatively conserved.
In summary, by examining the heparin-binding ability of VCP, we have gained a better understanding of the molecular basis by which this interaction occurs. In addition, the data presented here further enhance our understanding of this truly unique, yet compact, multifunctional viral protein. It resembles in structure and function the giant complex human complement regulatory proteins human C4b binding protein and complement receptor 1, respectively. Not only is VCP a miniature version of these large molecules, it has retained all the complement modulating functions and, as we have described here, added a number of additional functions to its repertoire due to its ability to bind heparin-like molecules. This better understanding may someday help in the exploitation of VCP's possible therapeutic properties in the treatment of inflammatory conditions, such as Alzheimer's disease (4), restenosis, accidental spinal cord injury, and xenograft transplant rejections.
ACKNOWLEDGMENTS
The HBP was kindly provided by Hans Flodgaard from Novo Nordisk, Copenhagen, Denmark (5). We would also like to thank Sam Welhausen of the J. Graham Brown Cancer Center for the use of his FACS facilities.
Partial support for this work was provided by the Kentucky Spinal Cord and Head Injury Research Trust (KSCHIRT).
REFERENCES
- 1.Blackmore T K, Hellwage J, Sadlon T A, Higgs N, Zipfel P F, Ward H M, Gordon D L. Identification of the second heparin-binding domain in human complement factor H. J Immunol. 1998;160:3342–3348. [PubMed] [Google Scholar]
- 2.Cardin A D, Demeter D A, Weintraub H J, Jackson R L. Molecular design and modeling of protein-heparin interactions. Methods Enzymol. 1991;203:556–583. doi: 10.1016/0076-6879(91)03030-k. [DOI] [PubMed] [Google Scholar]
- 3.Cardin A D, Weintraub H J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 4.Daly J, Kotwal G J. Pro-inflammatory complement activation by the A beta peptide of Alzheimer's disease is biologically significant and can be blocked by vaccinia virus complement control protein. Neurobiol Aging. 1998;19:619–627. doi: 10.1016/s0197-4580(98)00100-6. [DOI] [PubMed] [Google Scholar]
- 5.Flodgaard H, Ostergaard E, Bayne S, Svendsen A, Thomsen J, Engels M, Wollmer A. Covalent structure of two novel neutrophile leucocyte-derived proteins of porcine and human origin. Neutrophile elastase homologues with strong monocyte and fibroblast chemotactic activities. Eur J Biochem. 1991;197:535–547. doi: 10.1111/j.1432-1033.1991.tb15942.x. [DOI] [PubMed] [Google Scholar]
- 6.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–263. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 7.Howard J, Justus D E, Totmenin A V, Shchelkunov S, Kotwal G J. Molecular mimicry of the inflammation modulatory proteins (IMPs) of poxviruses: evasion of the inflammatory response to preserve viral habitat. J Leukoc Biol. 1998;64:68–71. doi: 10.1002/jlb.64.1.68. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs S N, Kotwal G J, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci USA. 1992;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotwal G J, Reynolds D, Keeling K, Howard J, Justus D E. Vaccinia virus complement control protein is a virokine with lysozyme-like heparin-binding activity: possible implications in prolonged evasion of host immune response. In: Talwar G P, Nath I, Ganguly N K, Rao K V S, editors. 10th International Congress of Immunology. Bologna, Italy: Monduzzi Editore; 1998. pp. 315–320. [Google Scholar]
- 10.Kotwal G J, Hugin A W, Moss B. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology. 1989;171:579–587. doi: 10.1016/0042-6822(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 11.Kotwal G J, Isaacs S N, McKenzie R, Frank M M, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 12.Kotwal G J, Miller C G, Justus D E. The inflammation modulatory protein (IMP) of cowpox virus drastically diminishes the tissue damage by down-regulating cellular infiltration resulting from complement activation. Mol Cell Biochem. 1998;185:39–46. doi: 10.1023/a:1006844624825. [DOI] [PubMed] [Google Scholar]
- 13.Kotwal G J, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 14.Kotwal G J, Moss B. Vaccinia virus encodes two proteins that are structurally related to members of the plasma serine protease inhibitor superfamily. J Virol. 1989;63:600–606. doi: 10.1128/jvi.63.2.600-606.1989. . (Erratum, 64:966, 1990.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lian R H, Kotwal G J, Wellhausen S R, Hunt L A, Justus D E. IFN-gamma-induced MHC class II gene expression is suppressed in endothelial cells by dextran sulfate. J Immunol. 1996;157:864–873. [PubMed] [Google Scholar]
- 16.Marino M, Friedlander J A, McCluskey R T, Andrews D. Identification of a heparin-binding region of rat thyroglobulin involved in megalin binding. J Biol Chem. 1999;274:30377–30386. doi: 10.1074/jbc.274.43.30377. [DOI] [PubMed] [Google Scholar]
- 17.Massung R F, Liu L I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 18.Massung R F, Loparev V N, Knight J C, Totmenin A V, Chizhikov V E, Parsons J M, Safronov P F, Gutorov V V, Shchelkunov S N, Esposito J J. Terminal region sequence variations in variola virus DNA. Virology. 1996;221:291–300. doi: 10.1006/viro.1996.0378. [DOI] [PubMed] [Google Scholar]
- 19.McKenzie R, Kotwal G J, Moss B, Hammer C H, Frank M M. Regulation of complement activity by vaccinia virus complement-control protein. J Infect Dis. 1992;166:1245–1250. doi: 10.1093/infdis/166.6.1245. [DOI] [PubMed] [Google Scholar]
- 20.Miller C G, Justus D E, Jayaraman S, Kotwal G J. Severe and prolonged inflammatory response to localized cowpox virus infection in footpads of C5-deficient mice: investigation of the role of host complement in poxvirus pathogenesis. Cell Immunol. 1995;162:326–332. doi: 10.1006/cimm.1995.1086. [DOI] [PubMed] [Google Scholar]
- 21.Miller C G, Shchelkunov S N, Kotwal G J. The cowpox virus-encoded homolog of the vaccinia virus complement control protein is an inflammation modulatory protein. Virology. 1997;229:126–133. doi: 10.1006/viro.1996.8396. [DOI] [PubMed] [Google Scholar]
- 22.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 1163–1197. [Google Scholar]
- 23.Reynolds D, Keeling K, Molestina R, Srisatajluk R, Butterfield J H, Ehringer W, Justus D E, Kotwal G J. Heparin binding activity of vaccinia virus complement control protein confers additional properties of uptake by mast cells and attachment to endothelial cells. In: Jameel S, Villareal L, editors. Advances in animal virology. New Delhi, India: Oxford and IBN Publishing; 1999. [Google Scholar]
- 24.Sahu A, Isaacs S N, Soulika A M, Lambris J D. Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J Immunol. 1998;160:5596–5604. [PubMed] [Google Scholar]
- 25.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 26.Sendak R A, Bensadoun A. Identification of a heparin-binding domain in the distal carboxyl-terminal region of lipoprotein lipase by site-directed mutagenesis. J Lipid Res. 1998;39:1310–1315. [PubMed] [Google Scholar]
- 27.Shchelkunov S N, Resenchuk S M, Totmenin A V, Blinov V M, Marennikova S S, Sandakhchiev L S. Comparison of the genetic maps of variola and vaccinia viruses. FEBS Lett. 1993;327:321–324. doi: 10.1016/0014-5793(93)81013-p. [DOI] [PubMed] [Google Scholar]
- 28.Shchelkunov S N, Safronov P F, Totmenin A V, Petrov N A, Ryazankina O I, Gutorov V V, Kotwal G J. The genomic sequence analysis of the left and right species-specific terminal region of a cowpox virus strain reveals unique sequences and a cluster of intact ORFs for immunomodulatory and host range proteins. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- 29.Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 30.Wiles A P, Shaw G, Bright J, Perczel A, Campbell I D, Barlow P N. NMR studies of a viral protein that mimics the regulators of complement activation. J Mol Biol. 1997;272:253–265. doi: 10.1006/jmbi.1997.1241. [DOI] [PubMed] [Google Scholar]
- 31.Wong P, Hampton B, Szylobryt E, Gallagher A M, Jaye M, Burgess W H. Analysis of putative heparin-binding domains of fibroblast growth factor-1. Using site-directed mutagenesis and peptide analogues. J Biol Chem. 1995;270:25805–25811. doi: 10.1074/jbc.270.43.25805. [DOI] [PubMed] [Google Scholar]