Abstract
Sindbis virus contains two membrane glycoproteins, E1 and E2, which are organized into 80 trimers of heterodimers (spikes). These trimers form a precise T=4 icosahedral protein lattice on the surface of the virus. Very little is known about the organization of the E1 and E2 glycoproteins within the spike trimer. To gain a better understanding of how the proteins E1 and E2 are arranged in the virus membrane, we have used the techniques of limited proteolysis and amino acid chemical modification in combination with mass spectrometry. We have determined that at neutral pH the E1 protein regions that are accessible to proteases include domains 1–21 (region encompassing amino acids 1 to 21), 161–176, and 212–220, while the E2 regions that are accessible include domains 31–84, 134–148, 158–186, 231–260, 299–314, and 324–337. When Sindbis virus is exposed to low pH, E2 amino acid domains 99–102 and 262–309 became exposed while other domains became inaccessible. Many new E1 regions became accessible after exposure to low pH, including region 86–91, which is in the putative fusion domain of E1 of Semliki Forest virus (SFV) (M. C. Kielian et al., J. Cell Biol. 134:863–872, 1996). E1 273–287 and region 145–158 were also exposed at low pH. These data support a model for the structure of the alphavirus spike in which the E1 glycoproteins are centrally located as trimers which are surrounded and protected by the E2 glycoprotein. These data improve our understanding of the structure of the virus membrane and have implications for understanding the protein conformational changes which accompany the process of virus-cell membrane fusion.
Sindbis virus, the prototype of the Alphavirus subgroup of the Togaviridae, has a well-defined T=4 icosahedral structure composed of three structural proteins E1, E2, and C (3, 10, 32, 49). The capsid protein combines with the virus's positive-sense 42S RNA genome in the infected cell cytoplasm and is enveloped in a host-derived membrane bilayer which contains the two virus structural glycoproteins E1 and E2 (7, 8, 10, 47). The E1 and E2 glycoproteins form trimers of heterodimers in the endoplasmic reticulum of the infected cell. The trimers are exported to the plasma membrane, where interaction with nucleocapsids followed by lateral associations between 80 of the trimers produces the icosahedral lattice of spike complexes on the surface of the virus (3, 32, 36, 47). These protein-protein associations are responsible for the precise icosahedral structure of the virus membrane and are stable even in the presence of a nonionic detergent (36). The virus nucleocapsid is situated within the virus membrane and is composed of 240 copies of the capsid protein organized into a T=4 icosahedron matching, precisely, the geometry of the glycoproteins in the virus membrane. The inner icosahedral protein shell is connected to the outer membrane protein shell through specific interactions between the capsid protein and the endodomain of glycoprotein E2 (22, 23).
It has been difficult to study the configuration of the structural glycoproteins in the Sindbis virus membrane bilayer. No crystal structure exists for the membrane glycoproteins, and although the technology has greatly improved, it is unlikely that electron cryomicroscopy will produce images at a resolution sufficient to ascertain protein configurations at atomic resolution. It is important to understand how these proteins are arranged in the lipid bilayer, how they contact one another, which regions of these proteins are exposed on the surface of the spike complex, and which are buried within this structure. This information is important in evaluating domains which are critical for maintaining the structural integrity of the virus and determining the protein domains responsible for virus host interactions, penetration, membrane fusion, and antibody response. Furthermore, Sindbis virus is currently being considered as a vector for human genetic engineering and gene therapy (52). Knowing how its two structural proteins are organized can help produce a more precisely targeted vector and may help prevent unwanted immune reactions.
The precise configuration of the E1 and E2 glycoproteins in the spike complex remains unclear. Enzymatic radiolabeling of exposed tyrosines has indicated that while E1 and E2 have similar numbers of tyrosines in their ectodomains, the tyrosines in E2 are more accessible for labeling than those of E1. Chemical cross-linking studies conducted on the Sindbis virus spike complex in the mature virion have shown that E1-E1 associations stabilize the spike trimer and that the E2 members of the trimer are located on the periphery of the spike, where they form E2-E2 interactions around the fivefold and sixfold axis (3). Electron cryomicroscopy imaging of a PE2-containing Sindbis virus mutant suggested that the E3 region of PE2 lies at the periphery of the spike complex (33). Electron cryomicroscopy of virus complexed with anti-E2 Fab monoclonal antibody fragments showed the Fab fragment to bind on the spike periphery (45). All of these data support an organization of the spike which has E1 centrally located and protected by E2. Others studies have resulted in a model in which the E2 glycoprotein occupies the central portion of the spike and prevents contact between the E1 proteins. This conclusion is based on the observation that multimers of E1 are not detected in detergent-lysed Semliki Forest virus (SFV) until the virus is exposed to low pH. Upon exposure to acid conditions, trimers of E1 can be recovered (39, 50, 51). A morphological study of SFV conducted by electron cryomicroscopy claimed to visualize a process in which the E1 and E2 proteins changed positions in the spike. In this structural reorganization, the E2 protein, which separates the peripheral E1 proteins, moves from the center of the spike to the periphery. This process was called “swiveling” (16).
Several domains of the spike complex have been located on the surface by the technique of escape mutation mapping using monoclonal antibody probes. These studies have shown that (i) the major viral antigenic determinant is on the E2 protein in the region from amino acids 173 to 220 (region 173–220) and (ii) E2 region 186–212 is located on the surface of the spike complex. Also, a charge escape mutation in E1 132 blocked the monoclonal antibody to region 186–212, suggesting that it may be located near this region (46). Finally, it was shown that antibodies responsible for virus neutralization recognized multiple epitopes on E2 and one epitope on E1 (40).
Conformational changes occur in the alphavirus spike as the process of attachment and membrane fusion takes place. The process of virus membrane-cell membrane fusion can be induced by transient exposure of Sindbis virus-cell complexes to acid pH followed by return to neutral pH (1, 12, 13, 25). We have proposed that this in vitro low-pH-mediated fusion is a two-step event. The initial conformational change required for fusion occurs at acid pH; however, return to neutral-pH conditions is required to complete a second step in the fusion process (12). We have proposed that the return to neutrality is required to establish conditions which allow the reduction of disulfide bridges which stabilize the alphavirus structure (1, 4). E1 region 75–98 is the putative fusion domain, and mutations in this domain in SFV (a related alphavirus) abolish fusion activity (19, 20). There are five cysteine residues between the amino terminus of E1 and the putative fusogenic domain. Evidence suggests that these cysteines are involved in disulfide bridges which would produce a complex three-dimensional structure. It is unlikely that this complex structure could penetrate the host cell membrane to position the fusion domains such that a hydrophobic channel could be created. We have proposed that these disulfides must be reduced before the interaction of the fusogenic domain with the target (host cell) membrane (1).
The precise conformational changes produced in the glycoprotein spike by low-pH exposure are unknown. Reported changes include a decrease in sedimentation velocity (13), a change in sensitivity to proteolytic enzymes (4, 13), an increase in hydrophobicity (30), an increase in the radii of the spike complex (48), and a possible swiveling of the E1 glycoprotein from the periphery to the center of the spike complex (16). A recent study using monoclonal antibody probes determined that upon low-pH exposure, a new epitope is exposed and that a mutation in E1 157 will abolish the binding of the antibody to this epitope (2).
To further elucidate the structure of the alphavirus spike, we have used the techniques of limited proteolysis and selective surface chemical modification in combination with mass spectrometry (MS) to determine which regions of the E1 and E2 proteins are exposed on the virus surface. We have also used this technology to determine how protein conformations change when the virus is exposed to low pH. This technique has been successfully used to determine viral protein dynamics in several non-membrane-containing viruses including flock house virus and rhinovirus (5, 6, 24, 43). This technique is based on the principle that sequences of amino acids that are exposed on the surface of the spike complex will be digested by small concentrations of proteases whereas those buried in the spike complex will be inaccessible. The peptides produced are separated from the remaining virus and identified by MS-based sequencing. This technique is fast and accurate and needs only femtomole amounts of peptide to be accurately identified. This procedure can identify all regions accessible on the surface at the same time, in the same experiment, without the necessity to modify the amino acid sequence or bind anything to the virus that could change its conformation.
MATERIALS AND METHODS
Materials.
All chemicals were HPLC (high-pressure liquid chromatography) or ultra grade and purchased from Sigma Chemical. 125I was purchased from New England Nuclear. Sequencing-grade modified porcine trypsin was purchased from Promega and stored at −80°C until use. Peptides for MALDI-TOF (matrix-assisted laser desorption–ionization time of flight) MS mass calibration, iodination, and trypsin controls were purchased from Sigma. Immobilized pepsin was purchased from Pierce. Iodogen (1,3,4,6-tetrachloro-3α,6α-diphenylglycoluril) tubes and micro BCA (bicinchoninic acid) protein assays were purchased from Pierce. Amicon 50-kDa-cutoff membrane tubes were purchased from Millipore. C18 Zip-Tips were purchased from Millipore. Poros R3 medium was obtained from PE Applied Biosystems.
Virus and cells.
BHK-21 cells were cultured at 37°C in Gibco minimal essential medium supplemented with 10% fetal bovine serum, 5% tryptose phosphate broth, 2 mM l-glutamine, and 50 μM gentamicin in 75-cm2 flasks. Heat-resistant Sindbis virus (SVHR) was originally provided by E. R. Pfefferkorn and was passaged in BHK-21 cells as previously described (35).
Virus purification.
Medium from infected cells was harvested after an 18-h infection. Cellular debris was removed by centrifugation at 5,000 rpm at room temperature for 15 min in a clinical centrifuge. The virus was then purified twice by gradient centrifugation. The first gradient purification was through a 15 to 35% potassium tartrate step gradient in phosphate-buffered saline (PBS-D). The virus band was collected and then purified through a second 10 to 40% linear potassium tartrate gradient in PBS-D. Both centrifugations were done in a Beckman SW28 rotor at 24,000 rpm for 16 h at 4°C. The purified virus band was collected and concentrated by pelleting at 24,000 rpm in a Beckman SW28 rotor through a 20% sucrose cushion at 4°C. The virus was stored at 4°C under sterile conditions for no longer than 14 days. Protein content of the virus pellet was assayed by a micro BCA assay and checked for purity on a mini-sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel that was subsequently stained using Coomassie brilliant blue R250. The virus was examined by negative-stain electron microscopy to ensure that the virus was intact.
Predigestion of Pierce immobilized pepsin.
To remove any free pepsin and to reduce autodigest products, the immobilized pepsin was allowed to autodigest at 37°C for 45 min; then 200 μl of Pierce immobilized pepsin was placed at 37°C. After 45 min, the immobilized pepsin was washed in 1 ml of PBS-D (pH 4.5) three times using a 0.2-mm syringe filter. The immobilized pepsin was then centrifuged at 1,500 rpm, and the excess PBS-D was discarded. The washed pepsin was then placed on ice until use.
Limited virus digestion and peptide purification.
Virus was diluted to 5 μg in 50 μl of reaction buffer with PBS-D at either pH 7.6 or pH 4.5. Proteases were added to the virus suspension: either 2 μl of Pierce sequencing-grade modified trypsin at a concentration of 0.52 mg/ml or 2 μl of washed immobilized pepsin. The trypsin reaction was allowed to proceed for various lengths of time at 37°C and then stopped by adding 4 μl of 100 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) in 0.1 N HCl. The pepsin digest was carried out at 37°C in an Eppendorf shaker at 1,050 rpm. Removing the immobilized pepsin from the virus stopped the reaction. The digested peptides and immobilized pepsin were removed from the remaining virus by centrifugation through an Amicon 50-kDa-cutoff membrane tube. The peptides were stored at −80°C until MS analysis could be conducted.
MS and MS/MS analysis for trypsin and pepsin digest products.
Liquid chromatography (LC)/MS analyses of aliquots of each digest of Sindbis virus were done using an Ultimate capillary LC system (LC Packings, San Francisco, Calif.; equipped with a Famos autosampler) coupled to a quadrupole time-of-flight (Q-TOF) mass spectrometer (Micromass, Manchester, United Kingdom) with a Z-spray ion source. Aliquots of each digest were preconcentrated and desalted onto a guard column (300 μm [inside diameter {i.d.}] by 1 mm; LC Packings) packed with Pepmap C18 material using the Famos autosampler. After the preconcentration step, the guard column was switched in-line with the analytical capillary column. Peptides were then separated using a 75-μm-i.d. by 15-cm capillary column packed with 3 μm of Pepmap C18 material. Mobile phase A consisted of 0.1% formic acid in a 2% acetonitrile solution, while mobile phase B consisted of 0.1% formic acid in an 80-20% acetonitrile-water solution. Peptides were eluted from the column into the microelectrospray ion source of the Q-TOF mass spectrometer using a gradient of 5% to 50% B in 30 min. The outlet of the capillary column was coupled to a platinum-coated 360-μm-outside-diameter- by 20-μm-i.d. fused silica spray tip (10-μm tip i.d. outlet; New Objective, Inc., Cambridge, Mass.) which made electrical contact through the Picotip holder (New Objective) in the Z-spray ion source. MS survey scans were acquired at a rate of 2 per s from m/z (mass-to-charge ratio) 400 to 2,000. The instrument was operated in a data-dependent MS-to-tandem MS (MS/MS) switching mode where peptide ions detected in MS survey scans triggered a switch to MS/MS for obtaining peptide product ion spectra. To identify peptides contained in the digest, uninterpreted peptide product ion spectra were searched against a protein database containing the sequences of bovine trypsin, porcine pepsin, and Sindbis virus capsid, E1, and E2 proteins using the SEQUEST program (14). Alternatively, product ion spectra were searched against the nonredundant protein database using the Mascot search program (Matrix Sciences, Ltd., London, United Kingdom). Searches were done both with and without the protease specificities option turned on.
Iodination of intact virus.
Sindbis virus was iodinated with 125I or 127I using Pierce iodotubes. Iodination with 125I was carried out with 1 mCi of 125I per reaction sample. The iodination reaction was stopped with the addition of saturated (0.453 mg/ml, 25°C) tyrosine. Excess 125I was removed by gel filtration using Sephadex G-25. Radiolabeled virus was stored at 4°C until use. Virus was iodinated with 127I using 50 μM K125I in PBS-D (pH 7.4). The reaction was stopped by the addition of excess tyrosine and stored at −80°C until use. The virus was denatured by the addition of 1% BME and heated at 99°C for 25 min. The denatured virus was then digested with trypsin for 18 h at 37°C; the peptides were not removed from the remaining virus after the digest was completed.
Digestion with protease V8.
The digestion conditions and virus concentration were the same as for the trypsin digest of iodinated and noniodinated virus. V8 was added at 4 μg of V8 per 100 μl of SVHR. The iodinated and noniodinated SVHR was denatured and reduced by heating at 99°C in the presence of 2-mercaptoethanol (BME) (same conditions as for trypsin digest). The digestion was allowed to proceed 16 h at 37°C. Digestion was stopped by freezing the sample at −80°C.
MALDI-MS of iodinated virus peptides.
Viral tryptic peptides iodinated with 127I were analyzed using a Bruker Proflex linear TOF mass spectrometer equipped with a MALDI source, delayed extraction, and a 1-GHz digitizer. The peptides were desalted with either C18 Zip-Tips or homemade micro-desalting columns using Poros R3 medium; 1.5 μl of desalted peptides were combined with 1.5 μl of saturated α-cyano-4-hydroxycinnamic acid in 50% acetonitrile with 0.1% trifluoroacetic acid. The above mixture was applied to the MALDI target using the dried droplet method. Mass calibration was obtained first through an external calibration and then through an internal calibration using diagnostic capsid tryptic masses. The mass spectrometer used in this study typically has a mass accuracy of 0.05 to 0.1% if calibrated internally. Iodinated peptides were compared to noniodinated peptide controls, and differences in the spectra were used to identify the iodinated masses. Comparing these shifted masses to masses computed from a theoretical trypsin digest of the iodinated Sindbis virus structural proteins then identified the peptides with the modified tyrosines. Cases where masses obtained by MALDI-MS coincidentally match more than one possible Sindbis virus peptide are given less weight than masses that have only one possible Sindbis virus peptide match.
SDS-polyacrylamide gel electrophoresis of iodinated and noniodinated virus proteins.
Iodinated and noniodinated viral proteins were separated on a 12.5% polyacrylamide gel. The gel was dried and visualized by autoradiography (29). Nonradiolabeled viral proteins were identified by silver staining as previously described (42).
RESULTS
Protein domains exposed on the surface of the virus spike complex at neutral pH.
To determine which domains of E1 and E2 are accessible to proteases in an aqueous neutral-pH environment, we performed a number of limited proteolysis experiments on intact Sindbis virus. Limited proteolysis when combined with MS can be used to probe for higher-order structure of proteins and protein complexes (for a review, see reference 21). Freshly prepared Sindbis virus was treated with trypsin or pepsin as described in Materials and Methods. The peptides released from the virus by protease treatment were separated from the partially digested virus and identified by MS as described in Materials and Methods. The released peptides were separated by nanoscale capillary LC which was coupled through a microelectrospray interface to a Q-TOF mass spectrometer. Peptides detected by the mass spectrometer were subjected to MS/MS in order to generate product ion spectra. MS/MS is a technique by which a selected precursor ion is fragmented, and the resulting fragment ions are mass analyzed to yield a product ion (fragment ion) spectrum. Peptide primary sequence can be derived from product ion spectra (31, 37), and computer algorithms can rapidly search protein databases with uninterpreted product ion spectra for matching peptide sequences (14).
A representative product ion spectrum of E2 peptide 31–44 and corresponding sequence is shown in Fig. 1. The total numbers of peptides recovered from trypsin digests of 1, 5, and 15 min are shown in Fig. 2. No Sindbis virus glycoprotein fragments were seen in the 1-min digest. Figure 2 shows the peptides released from E1, E2, and capsid. Peptides released from the virus at neutral pH are primarily from E2. The peptides released from E2 are in amino acid regions 31–84, 134–148, 158–186, 231–260, 299–314, and 324–337. This constitutes 39% of the amino acids of E2 on the ectodomain of the virus. By contrast, few E1 peptides were released. The peptides released from E1 are in amino acid regions 1–21, 161–176, and 212–220. This constitutes only 13% of the available amino acids on the ectodomain of the virus. Identification of these regions does not exclude other protein domains from being exposed on the surface, especially regions containing a high number of disulfide bonds. Because the SEQUEST and Mascot algorithms used for database searching do not allow for identification of disulfide-linked peptides, peptide molecular weights from spectra not matched to Sindbis virus peptides with SEQUEST or Mascot were searched against a protein database using a disulfide scanning program (available at Rockefeller University [http://www.proteometrics.com]). This program models every possible disulfide linkage in a protein of interest, calculates the expected molecular weights of all disulfide-linked peptide combinations, and compares these expected molecular weights to the input molecular weights. Using this program, we did not find any masses that matched disulfide-bonded peptides. This indicates that if these regions were on the surface, their conformation might be constrained by the disulfide bonds in such a way as to prevent protease cleavage sites from being exposed. The MS proteolytic cleavages indicate that many more E2 than E1 regions are accessible at neutral pH. These data are in agreement with previously published radioiodination studies which showed E2 to be more exposed on the virus surface than E1 (41, 44). These data are also consistent with the monoclonal antibody studies described above (28, 46). These data support the structural model of E2 occupying the outer surface of the spike complex and shielding a centrally located E1.
FIG. 1.
Representative product ion spectra of a peptide released from the Sindbis virus spike complex. Sindbis virus was digested with trypsin for 15 min as described in Materials and Methods. The released peptides were removed from the remaining virus, separated by capillary HPLC, and analyzed by LC/MS with data-dependent MS/MS switching. This peptide was matched to E2 31-44 using the Mascot search engine.
FIG. 2.
Tryptic peptides released from the Sindbis virus spike complex at neutral pH. Virus was digested at 1, 5, and 15 min, and the released peptides were removed from the remaining virus, separated by capillary HPLC, and analyzed by LC/MS with data-dependent MS/MS switching. The x axis indicates the amino acid number from the amino to carboxy terminus. No Sindbis virus peptides were found in the 1-min digests.
As indicated in Fig. 2, capsid fragments were found in the 5- and 15-min samples along with E1 and E2 fragments. As capsid is located in the interior of the virus, it should not be accessible to protease. It is unlikely that these capsid fragments are transiently exposed on the surface of the virus, as has been indicated in similar studies of flock house and rhinoviruses (5, 24). It is also highly unlikely that fragments from the capsid protein could cross a lipid bilayer or that trypsin could cross the membrane bilayer to digest capsid. The fragments that are found constitute almost the entire capsid protein. The origin of these fragments is best explained by the possibility that either some virus has been destroyed during purification or fragments of capsid proteins from the debris of infected cells adhere to the virus as it is purified. Capsid protein has a large number of hydrophobic residues and is likely very “sticky.” To determine if a subpopulation of the virus particles were damaged during virus purification and were releasing the capsid protein, we iodinated the whole purified virus with 125I. Previous studies have shown that capsid protein is not iodinated if the virus is intact (41, 44). Figure 3 shows iodinated Sindbis virus proteins from intact (used in the MS analysis described above) or lysed virus separated on a reducing SDS-polyacrylamide gel. The capsid protein is clearly iodinated in the lysed virus, but its iodinated form is absent in the nonlysed virus preparation. E2 is more heavily labeled than E1 in the sample of intact virus. E1 and E2 acquire similar amounts of label when virus is lysed before iodination. The glycoprotein E1 contains 16 tyrosine residues in its ectodomain, while E2 contains 20. These results are consistent with previously published radioiodination studies of Sindbis virus (41, 44). These data suggest that the capsid fragments seen in the MS analysis are not the result of degrading capsid protein in a population of broken virus particles; rather, these fragments are fortuitously bound to the surface of the particles.
FIG. 3.
Purified Sindbis virus is intact, and capsid protein is not iodinated unless it is lysed (+) with 0.1% SDS prior to iodination. In the nonlysed viral sample (−), no capsid is seen, and E2 is iodinated to a greater extent than E1, which is consistent with previous studies.
Protein domains exposed on the surface of the virus spike complex at low pH.
Having identified the amino acid sequences accessible to proteases at neutral pH, we next conducted experiments to determine how the spike proteins change conformation when the virus is exposed to low pH. When Sindbis virus is exposed to low pH, a conformational change takes place that is the first step in a two-step process which can lead to the fusion of the virus membrane with a target membrane (12). The second step in membrane fusion requires a return to neutral-pH conditions. We have presented evidence that this step is necessary to allow reshuffling of disulfide bridges which maintain the structure of the virus membrane (1, 13, 25). Fuller et al. (16) have indicated that the conformational change occurring at low pH involves a dramatic reorganization of the virus membrane in which the E1 and E2 proteins change their protein-protein contacts and their physical positions in the protein lattice, a process referred to as swiveling. Swiveling results from the breaking of the heterodimer of E1 and E2, allowing the E1 proteins to move from the periphery of the spike to the center, forming E1 homotrimers (16). These homotrimers then initiate the fusion reaction which in SFV involves the putative fusion domain E1 75–98 (20). This low-pH conformational change renders Sindbis virus noninfectious while largely preserving the overall virus structure (4, 13). If reducing agents are then added to low-pH-treated virus, the virus will disassemble, releasing its internal, capsid protein-complexed, RNA genome (4).
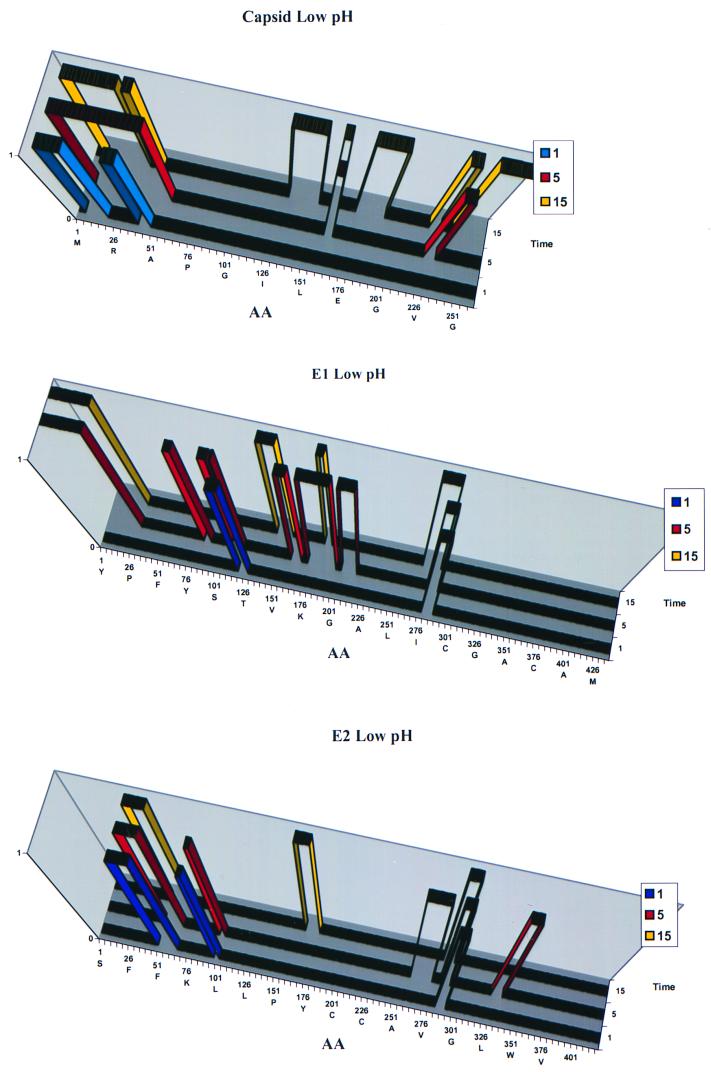
To determine the protein rearrangements that take place when Sindbis virus is exposed to a low-pH environment, Sindbis virus was exposed to pH 4.5 and then digested with pepsin for various periods of time. Pepsin is a quasi-specific protease that cleaves at the carboxyl side of hydrophobic, preferentially aromatic, amino acids. Pepsin has an optimum pH of 2.0 but is capable of digesting proteins at pH 5.0 or less. To control the digestion process, pepsin immobilized on agarose beads was used as described in Materials and Methods. The beads, with associated enzyme, were removed from the virus by filtration. The Sindbis virus peptides released from pepsin digests of acid-treated virus at 1, 5, and 15 min are shown in Fig. 4. Capsid fragments are again seen in the pepsin digests of low-pH-treated virus.
FIG. 4.
Peptides released from the Sindbis virus spike complex at low pH. Sindbis virus was digested with immobilized pepsin at pH 4.5 for 1, 5, and 15 min. The reaction was stopped, and the released peptides were removed from the remaining virus, separated by capillary HPLC, and analyzed by LC/MS with data-dependent MS/MS switching. The x axis indicates the amino acid number from the amino to carboxy terminus.
The conformational changes which occur at low pH involve the exposure of many E1 regions not detected at neutral pH. Some of the E1 peptides released from the surface of low-pH-treated Sindbis virus include regions 86–91, 110–119, and 273–287; only two new regions of E2, 99–102 and 262–309, were exposed, and many E2 regions that were accessible to trypsin at neutral pH were inaccessible to pepsin at pH 4.5 (see Fig. 7). Some of these E2 peptides seen at neutral pH but not at pH 4.5 include regions 67-84, 158-167, and 310-314. Low-pH treatment appears to expose regions of E1 to the environment while causing domains of E2 to be protected.
FIG. 7.
Amino acid regions of E1 and E2 accessible to proteases at pH 7.4 and 4.5.
Chemical modification of tyrosines on the surface of the spike complex at neutral pH.
Chemical modification of amino acids and the identification of the protein domains containing those modified amino acids by MS have proven to be a powerful tool for the determination of protein conformation. Using iodine activated by immobilized Iodogen, we have modified tyrosines on the surface of the Sindbis virus spike complex (described in Materials and Methods). Activated iodine can also modify histidines if a high concentration of iodine is used and/or the reaction is done at pH 9.0 (38). We used a very small concentration of activated iodine, 50 μm KI at pH 7.4. At these conditions, no histidine is iodinated and only the surfaces of cells and viruses are iodinated (26). Iodine has a mass of 126.9 Da, and the sequence of the virus glycoproteins is precisely known. It is therefore possible to precisely identify those tyrosines which are exposed on the surface of the spike complex. Purified Sindbis virus was iodinated with 127I using the lodogen procedure (described in Materials and Methods). The reaction was quenched with an equal volume of cold tyrosine, and the iodinated virus was denatured by adding BME to a final concentration of 1% and heating the mixture at 99°C for 25 min. The denatured virus was then proteolytically digested with trypsin or V8 protease. The resulting digests of iodinated virus were compared to noniodinated control digests by MALDI-TOF MS. In these experiments, aliquots of each digest mixture were analyzed without any separation of the resulting peptides. Peptides containing iodotyrosines were identified by identifying masses in the iodinated sample that had shifted by multiples of 126 Da from the noniodinated controls.
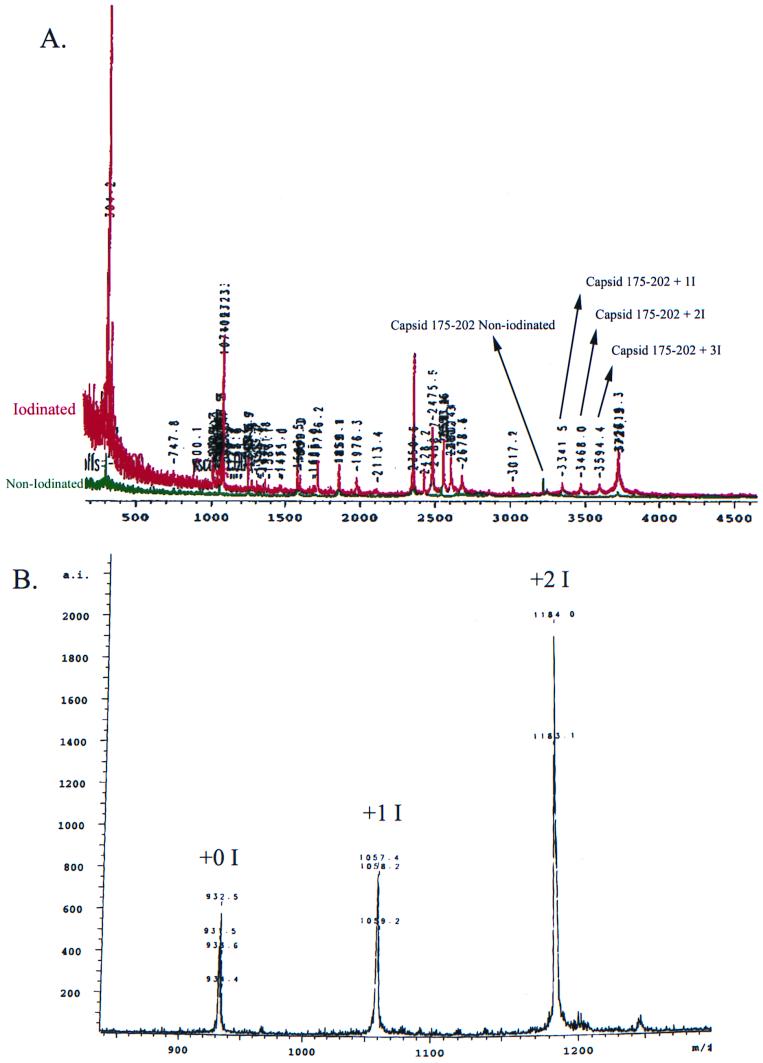
A sample MALDI-TOF spectrum is shown in Fig. 5. Figure 5A is an overlay of two spectra, one from iodinated and one from noniodinated virus; Fig. 5B shows a similar profile from iodinated angiotensin III that was used as a control. Using capsid fragments as internal calibrants, accurate masses were obtained by MALDI-TOF MS. All of the iodinated peptides found by this analysis are shown in Table 1. Masses that can coincidently match more than one Sindbis virus tryptic peptide are given less confidence than masses that can be assigned to only one possible peptide. Masses that have an ambiguous assignment, for reasons described in Materials and Methods, and masses that have only one possible match are shown. MALDI-TOF MS analysis identified 17 Sindbis virus spike peptides digested with trypsin and V8 protease having iodine-modified tyrosines. Of these 17 peptides, 5 are located on E1, 8 are located on E2, 2 are located on capsid, and 2 are located on sequences identified as PE2 (a small amount of PE2 is incorporated naturally into virions [34]). The E1 peptides that are iodinated are in amino acid region 1–30, which contains tyrosines at positions 1, 15, and 24. No peptides were generated that separated Y1 from Y15; thus, we were unable to determine if both tyrosines 1 and 15 were iodinated. It is, however, likely that both are labeled, as peptide 14 (Table 1) indicates that Y24 is iodinated as well.
FIG. 5.
Representative MALDI-TOF spectrum comparing iodinated and noniodinated Sindbis virus peptides. (A) Comparison of a tryptic digest of Sindbis virus iodinated with 127I (red) to a noniodinated Sindbis digest control (green). (B) Angiotensin III control iodinated with 127I.
TABLE 1.
Iodinated tyrosines on the surface of the E1 and E2 glycoproteins at neutral pH
| Confidence level | Peptide no. | Enzyme used | Protein | Amino acids | Tyrosine(s) | No. of modifying iodines | Avg M+Hb
|
Avg M+H+I expected | Error (%) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Observed | Expected | |||||||||
| Low | 1 | V8 | E1 | 1–20 | 1 and 15 | 1 | 2,101.23 | 1,978.29 | 2,104.09 | 0.14 |
| 2 | Trypsin | E2 | 150–172 | 155 and 165 | 3 | 3,099.4 | 2,723 | 3,100.4 | 0.03 | |
| 3 | Trypsin | E2 | 160–190 | 165, 176 and 179 | 2 | 3,796.54 | 3,545.95 | 3,797.55 | 0.03 | |
| 4 | Trypsin | PE2 | 33–55 | 46 | 2 | 2,889.08 | 2,640.94 | 2,892.54 | 0.12 | |
| 5 | Trypsin | E1 | 1–16 | 1 and 15 | 4 | 2,359.62 | 1,858.09 | 2,361.29 | 0.07 | |
| High | 6 | Trypsin | E1 | 1–21 | 1 and 15 | 1 | 2,551.5 | 2,427 | 2,552.6 | 0.04 |
| 6 | Trypsin | E1 | 1–21 | 1 and 15 | 2 | 2,678.4 | 2,427 | 2,678.6 | 0.01 | |
| 7 | Trypsin | E1 | 1–16 | 1 and 15 | 2 | 2,111.43 | 1,858.09 | 2,109.69 | 0.08 | |
| 8 | Trypsin | E2 | 45–63 | 53 | 1 | 2,072.26 | 1,945.04 | 2,070.84 | 0.07 | |
| 9 | Trypsin | E2 | 149–157 | 155 | 2 | 1,346.28 | 1,095 | 1,346.6 | 0.02 | |
| 10 | Trypsin | E2 | 187–195 | 188 | 1 | 1,074.07 | 947.11 | 1,072.91 | 0.11 | |
| 11 | Trypsin | E2 | 138–148 | 140 | 1 | 1,408.66 | 1,283.46 | 1,409.26 | 0.04 | |
| 12 | Trypsin | Capsid | 175–202 | 189 and 198 | 1 | 3,340.51 | 3,216.4 | 3,342.2 | 0.05 | |
| 12 | Trypsin | Capsid | 175–202 | 189 and 198 | 2 | 3,466.62 | 3,216.4 | 3,468 | 0.04 | |
| 12 | Trypsin | Capsid | 175–202 | 189 and 198 | 3 | 3,592.29 | 3,216.4 | 3,593.8 | 0.04 | |
| 12 | Trypsin | Capsid | 175–202 | 189 and 198 | 4 | 3,717.78 | 3,216.4 | 3,719.6 | 0.05 | |
| 13 | Trypsin | PE2 | 29–64 | 46 | 2 | 4,282.19 | 4,029.47 | 4,281.07 | 0.03 | |
| 14 | V8 | E1 | 21–30 | 24 | 2 | 1,354.23 | 1,104.24 | 1,355.84 | 0.12 | |
| 15 | V8 | Capsid | 134–163 | 162 | 2 | 3,580.08 | 3,326.96 | 3,578.56 | 0.04 | |
| 16 | V8 | E2 | 328–347 | 328 and 339 | 1 | 2,412.77 | 2,289.46 | 2,415.26 | 0.10 | |
| 16 | V8 | E2 | 328–347 | 328 and 339 | 3 | 2,664.77 | 2,289.46 | 2,666.86 | 0.08 | |
| 16 | V8 | E2 | 328–347 | 328 and 339 | 4 | 2,790.41 | 2,289.46 | 2,792.66 | 0.08 | |
| 16 | V8 | E2 | 325–347 | 328 and 339 | 3 | 2,967.33 | 2,588.78 | 2,966.18 | 0.04 | |
| 17a | V8 | E2 | 325–347 | 328 and 339 | 4 | 3,092.97 | 2,588.78 | 3,091.98 | 0.03 | |
| V8 | E2 | 325–347 | 328 and 339 | 5 | 3,218 | 2,588.78 | 3,217.78 | 0.01 | ||
| V8 | E2 | 325–347 | 328 and 339 | 6 | 3,343.6 | 2,588.78 | 3,343.58 | 0.00 | ||
It is not clear if six iodines can modify this peptide. This peptide contains one histidine which may be iodinated. No other peptides can be matched to these masses.
M+H, protonated molecular ion.
The E2 iodinated peptides are in regions 45–63, 138–190, and 325–347. Peptide 9 (Table 1) matches E2 regions 45–63, which contains an iodine-modified tyrosine at position 53. E2 region 138–190 contains six tyrosines at positions 140, 155, 165, 176, 179, and 188. While peptides 2 and 3 (Table 1) match E2 iodinated peptides, peptide 2 can also be matched to the noniodinated peptide E1 51–79 and peptide 3 can be matched to the noniodinated peptide capsid 4–35. The data presented in Table 1 indicate that tyrosines 140, 155, 165, and 188 are iodinated. Tyrosines 176 and 179 may be iodinated; however, we are uncertain of this because no iodinated fragments containing only these tyrosines were found. Peptide 3 matches E2 160–190 with three added iodines. It is likely that either Y176 or Y179 is iodinated; the addition of three iodines to Y165 seems unlikely, as the chemistry of iodine modification indicates that tyrosines have two preferred sites that can be iodinated (11). E2 region 325–347 contains two tyrosines, at positions 328 and 339. Peptides 16 and 17 (Table 1) indicate that both of these tyrosines are iodinated. Peptide 17 matches E2 325–347 with four, five and six modifying iodines. The addition of three iodines per tyrosine molecule seems unlikely, as the angiotensin control did not indicate any tri-iodinated species. Although unlikely, no other Sindbis virus tryptic peptides could be matched to these masses.
Ions corresponding to iodinated capsid and PE2 peptides were also found. Peptide 4 (Table 1) matches PE2 33–55 and indicates that Y46 may be iodinated. Peptide 4 can also be matched to the noniodinated peptide E2 45–70, which could be a product of an incomplete digest. Peptide 13 (Table 1) matches PE2 29–64 and also indicates that Y46 is iodinated. Some PE2 can be found in Sindbis virus (34), and PE2 peptides were found in the trypsin limited digest experiments (data not shown). Peptides 12 and 15 (Table 1) match capsid 175–202 and 134–163, respectively, and indicate that Y162, Y189, and Y198 are iodinated. The presence of iodinated capsid fragments is consistent with the limit digest studies, and their presence is explained above. In addition, previous studies have shown that capsid Y180 is the only exposed tyrosine residue on the surface of intact Sindbis nucleocapsids (10). The fact that we find iodinated tyrosines other then Y180 provides additional evidence that the capsid peptides found in this study were capsid fragments released during a lytic infection and which purified with the intact virus.
It is unclear why E2 fragments containing tyrosines at positions 53, 64, and 66 were not found to be iodinated. The limit digest data suggested that the tyrosines were included in the peptides released from the spike complex within 15 min. This may be due to MALDI selectivity in ionizing certain peptides (9) or our ability to control the extent of the trypsin digestion. To determine the extent to which iodinated tyrosines were being released from the spike complex, Sindbis virus was iodinated with 125I as described in Materials and Methods. Samples of iodinated and nonlabeled virus were digested for 1, 5, 15, and 90 min. The virus digests labeled with 125I were separated on an SDS-polyacrylamide gel and visualized by autoradiography. The nonradiolabeled digests were also separated on an SDS-polyacrylamide gel and visualized by silver staining. These results are shown in Fig. 6. Within 15 min, the proteins labeled with 125I appear to be completely digested, while the nonlabeled proteins appear to be relatively intact even at 90 min. This result could suggest that the tyrosines modified by 125I are almost completely digested from the surface of the proteins, while the majority of the unlabeled proteins remain largely intact. It is more likely, however, that the superficial position of the iodine-labeled tyrosines results in their being rapidly cleared from the surface, and because they are very radioactive they easily overexpose the gel, giving the impression that the iodinated residues are more sensitive to proteolysis. In reality the presence of modifications on the tyrosines probably reduces susceptibility to proteolysis through steric hindrance.
FIG. 6.
Tyrosines are digested from the viral spike surface, while the spike proteins remain largely intact. Sindbis virus was iodinated with 125I. The iodinated virus was then digested with trypsin for 1, 5, 15, and 90 min, and the resulting mixture was separated on SDS-polyacrylamide gels. Radiolabeled peptides were visualized by autoradiography and compared to the nonradiolabeled viral proteins, which were visualized by silver staining.
DISCUSSION
The stable and precise icosahedral structure of Sindbis virus makes it an excellent model for studies on the structure of virus membranes. We have used limited proteolysis in combination with MS to determine which amino acids are accessible on the surface of the glycoprotein spike complex and how the conformation of these protein domains change when the virus is exposed to low pH. This MS-based mapping technique has determined the conformational dynamics of two non-membrane-containing viruses, flock house virus and rhinovirus (5, 24). The MS-based mapping not only matched the crystal structure for each virus but also identified protein domains of the viruses that dynamically “breathe.” These dynamic conformational changes were not seen or predicted from the viral crystal structures. MS-based mapping is, therefore, able to assay flexible conformational changes in viral proteins that were previously unknown. The study presented here is similar to these previous studies with one major exception: Sindbis virus is a membrane-containing virus, and no crystal structure exists for any of its membrane components.
The data presented above indicate that the configuration of E1 and E2 in the spike complex at neutral pH is most likely one in which E1 occupies the center of the structure and is almost entirely shielded by E2 (Fig. 7). The E2 protein domains found on the surface of the spike are consistent with the observation that E2 173–220 is the major antigenic site on E2 (46). Strauss and coworkers (46) also identified E2 domain 186–212 as exposed at neutral pH. We found that regions 158–186 and 231–260 are located on the surface of the spike. Our experiments may have missed the region between E2 amino acids 186 and 231 because there is a carbohydrate binding site at E2 position 196. Any peptides containing carbohydrate would be missed in our study because of the unknown mass additions and heterogeneity of the carbohydrate. In addition, this region may not have been cleaved efficiently due to steric hindrance caused by the carbohydrate moiety. The three domains of E1 that are accessible and not shielded by E2 are 1–21, 161–176, and 212–220. E1 contains a number of cysteines between amino acids 49 and 114. It is possible that these domains are on the surface of the spike complex, even though these peptides were not detected. It is possible that disulfides constrain the structure in this region to such an extent that trypsin was not able to access potential cleavage sites. E1 and E2 both contain two carbohydrate binding sites, E1 139 and 245 and E2 196 and 318, and as indicated above, peptides containing this modification would have escaped detection. The technique of tyrosine modification by iodination before protease digestion can overcome some of these limitations although iodinated tyrosines located in the same peptide as a carbohydrate will still be missed.
Exposure of virus to low pH produced dramatic changes in the outcome of these experiments (Fig. 7). The amount of E1 protein accessible to protease increased from 13% to 35% of the total protein in the E1 ectodomain. The domains of E1 exposed by low pH included regions 86–91 and 110–119, which are near to and overlap the putative fusion domain E1 75–98 described for SFV (20). E1 region 145–158 also became accessible to protease after exposure to low pH. A recent study showed that a charge mutation in E1 157 will block the binding of an E1 low-pH-specific monoclonal antibody (2), indicating that this amino acid, and the region around it, are not exposed in a neutral pH environment. Our data are consistent with these findings and further indicate that region 145–158 is exposed at low pH while E1 region 161–170 is exposed at both neutral and low pH. Region 160–175 is also conserved among the alphaviruses and has a conserved hydrophobic region from amino acids 161 to 166. E1 region 273–287 was also exposed during low-pH exposure. This domain is conserved among the alphaviruses and contains a number of hydrophobic amino acids between E1 276 and E1 287. It is interesting that region E1 161–166 is exposed at both neutral and low pH whereas region 273–287 is exposed only at low pH.
Little is known about the fusion mechanism of Sindbis virus. It is possible that Sindbis virus has a novel fusion motor, a contention supported by its dissimilarity to other membrane-containing viruses. Many viruses such as influenza virus and human immunodeficiency virus employ hydrophobic coiled-coil domains located at the amino termini of glycoproteins in their fusion motors. The sequence of protein around the putative fusion domain in the alphavirus SFV does not contain a predicted coiled-coil region. The E1 glycoproteins containing the putative fusion domain also have 74 disulfide cross-bridged amino acids between the amino-terminal ends of the protein and the fusion domain. Clearly this globular structure cannot be forced through the membrane of a host cell to allow integration of the fusion domain. Thus, the alphaviruses may not follow the influenza virus paradigm regarding the fusion of membranes by virus glycoproteins. The data presented above also indicate that other conserved hydrophobic regions of the E1 protein, probably region 273–287 and possibly region 161–166, may play a part in this fusion process.
Our data indicate that the E2 protein actually becomes less exposed at low pH. This would seem to contradict previous studies that have shown that upon low-pH exposure E2 becomes more accessible to trypsin (3, 13). An explanation for this disparity is that we are identifying only the most superficial domains of the spike complex. Studies indicating an increase in E2 sensitivity at low-pH conditions typically used a large amount of enzyme for a considerable period of time. It is probable that under these conditions, additional conformational changes take place in the E2 protein as a result of the protease digestion itself and that these conformational changes then lead to the complete digestion of E2.
The data presented above support the contention that the organization of the alphavirus spike is one in which the E1 glycoprotein is centrally situated and exists as E1 trimers. The E1 trimers are surrounded by, and protected from the environment by, E2. This model is in agreement with numerous other studies utilizing chemical cross-linkers, antibody binding, electron microscopy, and genetics. It greatly simplifies proposed mechanisms for the interaction of virus glycoproteins with host cells during attachment and penetration. In this model, the virus glycoproteins are not required to change position in the envelope, a difficult process considering the multiple protein interactions which hold the two nested icosahedra together (see Introduction). The observation that E1 trimers are recovered after detergent lysis of virus exposed to low pH (the basis for proposing the alternative model) is likely the result of conformational changes occurring low pH and stabilizing the hydrophobic domains in the centrally located E1 trimer. The stabilized E1 trimers can then be recovered after detergent lysis. Furthermore, the morphological studies involving electron cryomicroscopy which purported to visualize the proposed change in position of E1 and E2 (the second observation leading to the alternative model) compared images of low-pH- and neutral-pH-exposed virus at greatly different resolutions (16), and the conformational changes reported may be due to differences in the resolution of the data sets.
ACKNOWLEDGMENTS
This research was supported in part by grants from The Foundation for Research, Carson City Nevada, and the National Institutes of Health (grant AI42775 to D.T.B.).
REFERENCES
- 1.Abell B A, Brown D T. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J Virol. 1993;67:5496–5501. doi: 10.1128/jvi.67.9.5496-5501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn A, Klimjack M R, Chatterjee P K, Kielian M. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J Virol. 1999;73:10029–10039. doi: 10.1128/jvi.73.12.10029-10039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony R P, Brown D T. Protein-protein interactions in an alphavirus membrane. J Virol. 1991;65:1187–1194. doi: 10.1128/jvi.65.3.1187-1194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony R P, Paredes A M, Brown D T. Disulfide bonds are essential for the stability of the Sindbis virus envelope. Virology. 1992;190:330–336. doi: 10.1016/0042-6822(92)91219-k. [DOI] [PubMed] [Google Scholar]
- 5.Bothner B, Dong X F, Bibbs L, Johnson J E, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- 6.Bothner B, Schneemann A, Marshall D, Reddy V, Johnson J E, Siuzdak G. Crystallographically identical virus capsids display different properties in solution. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 7.Brown D T, Waite M R F, Pfefferkorn E R. Morphology and morphogenesis of Sindbis virus as seen with freeze-etching techniques. J Virol. 1972;10:534–536. doi: 10.1128/jvi.10.3.524-536.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi H K, Tong L, Minor W, Dumas P, Boege U, Rossmann M G, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S L, Chait B T. Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal Chem. 1996;68:31–37. doi: 10.1021/ac9507956. [DOI] [PubMed] [Google Scholar]
- 10.Coombs K, Brown D T. Topological organization of Sindbis virus capsid protein in isolated nucleocapsids. Virus Res. 1987;7:131–149. doi: 10.1016/0168-1702(87)90075-x. [DOI] [PubMed] [Google Scholar]
- 11.Creighton T E. Proteins. 2nd ed. Philadelphia, Pa: W. H. Freeman and Company; 1993. [Google Scholar]
- 12.Edwards J, Brown D T. Sindbis virus-mediated cell fusion from without is a two-step event. J Gen Virol. 1986;67:377–380. doi: 10.1099/0022-1317-67-2-377. [DOI] [PubMed] [Google Scholar]
- 13.Edwards J, Mann E, Brown D T. Conformational changes in Sindbis virus envelope proteins accompanying exposure to low pH. J Virol. 1983;45:1090–1097. doi: 10.1128/jvi.45.3.1090-1097.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eng J K, McCormack A L, Yates J R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 15.Ferlenghi I, Gowen B, de Haas F, Mancini E J, Garoff H, Sjoberg M, Fuller S D. The first step: activation of the Semliki Forest virus spike protein precursor causes a localized conformational change in the trimeric. J Mol Biol. 1998;283:71–81. doi: 10.1006/jmbi.1998.2066. [DOI] [PubMed] [Google Scholar]
- 16.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 17.Glomb-Reinmund S, Kielian M. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology. 1998;248:372–381. doi: 10.1006/viro.1998.9275. [DOI] [PubMed] [Google Scholar]
- 18.Kielian M, Helenius A. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol. 1985;101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kielian M C, Keranen S, Kaariainen L, Helenius A. Membrane fusion mutants of Semliki Forest virus. J Cell Biol. 1984;98:139–145. doi: 10.1083/jcb.98.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriwacki R, Siuzdak G. Combined use of proteases and mass spectrometry in structural biology. J Biomol Tech. 1998;9:5–15. [Google Scholar]
- 22.Lee H, Ricker P D, Brown D T. The configuration of Sindbis virus envelope proteins is stabilized by the nucleocapsid protein. Virology. 1994;204:471–474. doi: 10.1006/viro.1994.1557. [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Owen K E, Choi H K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J K, Bothner B, Smith T J, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci USA. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann E, Edwards J, Brown D T. Polycaryocyte formation mediated by Sindbis virus glycoproteins. J Virol. 1983;45:1083–1089. doi: 10.1128/jvi.45.3.1083-1089.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markwell M A, Fox C F. Surface-specific iodination of membrane proteins of viruses and eucaryotic cells using 1,3,4,6-tetrachloro-3alpha,6alpha-diphenylglycoluril. Biochemistry. 1978;17:4807–4817. doi: 10.1021/bi00615a031. [DOI] [PubMed] [Google Scholar]
- 27.Meyer W J, Gidwitz S, Ayers V K, Schoepp R J, Johnston R E. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J Virol. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer W J, Johnston R E. Structural rearrangement of infecting Sindbis virions at the cell surface: mapping of newly accessible epitopes. J Virol. 1993;67:5117–5125. doi: 10.1128/jvi.67.9.5117-5125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulvey M, Brown D T. Formation and rearrangement of disulfide bonds during maturation of the Sindbis virus E1 glycoprotein. J Virol. 1994;68:805–812. doi: 10.1128/jvi.68.2.805-812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omar A, Koblet H. Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology. 1988;166:17–23. doi: 10.1016/0042-6822(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 31.Papayannopoulos I A. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom Rev. 1995;14:49–73. [Google Scholar]
- 32.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paredes A M, Heidner H, Thuman-Commike P, Prasad B V, Johnston R E, Chiu W. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J Virol. 1998;72:1534–1541. doi: 10.1128/jvi.72.2.1534-1541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Presely J F, Brown D T. The proteolytic cleavage of PE2 to envelope glycoprotein E2 is not strictly required for the maturation of Sindbis virus. J Virol. 1989;63:1975–1980. doi: 10.1128/jvi.63.5.1975-1980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renz D, Brown D T. Characteristics of Sindbis virus temperature-sensitive mutants in cultured BHK-21 and Aedes albopictus (mosquito) cells. J Virol. 1976;19:775–781. doi: 10.1128/jvi.19.3.775-781.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice C M, Strauss J H. Association of Sindbis virion glycoproteins and their precursors. J Mol Biol. 1982;154:325–348. doi: 10.1016/0022-2836(82)90067-5. [DOI] [PubMed] [Google Scholar]
- 37.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 38.Salacinski P R, McLean C, Sykes J E, Clement-Jones V V, Lowry P J. Iodination of proteins, glycoproteins, and peptides using a solid-phase oxidizing agent, 1,3,4,6-tetrachloro-3 alpha,6 alpha-diphenyl glycoluril (Iodogen) Anal Biochem. 1981;117:136–146. doi: 10.1016/0003-2697(81)90703-x. [DOI] [PubMed] [Google Scholar]
- 39.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki forest virus. II. Cleavage-dependant reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmaljohn A L, Kokubun K M, Cole G A. Protective monoclonal antibodies define maturational and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology. 1983;130:144–154. doi: 10.1016/0042-6822(83)90124-1. [DOI] [PubMed] [Google Scholar]
- 41.Sefton B M, Wickus G G, Burge B W. Enzymatic iodination of Sindbis virus proteins. J Virol. 1973;11:730–735. doi: 10.1128/jvi.11.5.730-735.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 43.Siuzdak G. Probing viruses with mass spectrometry. J Mass Spectrom. 1998;33:203–211. doi: 10.1002/(SICI)1096-9888(199803)33:3<203::AID-JMS653>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 44.Smith J F, Brown D T. Envelopments of Sindbis virus: synthesis and organization of proteins in cells infected with wild type and maturation-defective mutants. J Virol. 1977;22:662–678. doi: 10.1128/jvi.22.3.662-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith T J, Cheng R H, Olson N H, Peterson P, Chase E, Kuhn R J, Baker T S. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc Natl Acad Sci USA. 1995;92:10648–10652. doi: 10.1073/pnas.92.23.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauss E G, Stec D S, Schmaljohn A L, Strauss J H. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J Virol. 1991;65:4654–4664. doi: 10.1128/jvi.65.9.4654-4664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stubbs M J, Miller A, Sizer P J, Stephenson J R, Crooks A J. X-ray solution scattering of Sindbis virus. Changes in conformation induced at low pH. J Mol Biol. 1991;221:39–42. doi: 10.1016/0022-2836(91)80200-e. [DOI] [PubMed] [Google Scholar]
- 49.VonBonsdorff C H, Harrison S C. Sindbis virus glycoproteins form a regular icosahedral surface lattice. J Virol. 1978;28:578. doi: 10.1128/jvi.16.1.141-145.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahlberg J M, Garoff H. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]