Abstract
A herpes simplex virus type 1 (HSV-1) OriS analogue in which the A+T sequence linking the box I and II elements was replaced by two single-stranded oligo(dT)s is unwound by the UL9 protein-ICP8 complex. Unwinding of wild-type OriS by the UL9 protein-ICP8 complex was also observed under conditions which destabilize the A+T sequence. These experiments support a model for the unwinding of OriS in which destabilization of the A+T sequence can generate a single-stranded DNA binding site for ICP8, which then associates with the UL9 protein bound to boxes I and II to promote the bidirectional unwinding of OriS.
Herpes simplex virus type 1 (HSV-1) encodes a 94-kDa origin binding protein, the product of the UL9 gene, which is essential for HSV-1 DNA replication (5, 10). The origin binding protein (UL9 protein) is a homodimer that binds the two inverted pentanucleotide repeats, boxes I and II of the HSV-1 origin of replication, OriS. Boxes I and II are linked by an A+T-rich sequence of 18 nucleotide residues (5). In addition to its origin binding activity, the UL9 protein possesses DNA-dependent ATPase and 3′-5′ helicase activities (3, 6). Despite its helicase activity, we have been unable to detect the unwinding of duplex DNA containing OriS (9).
Previous studies have shown that a complex of the UL9 protein and the HSV-1-encoded single-stranded DNA binding protein, ICP8, can efficiently unwind a duplex box I if it possesses a 3′ single-stranded tail at least 18 nucleotides in length, positioned downstream of box I (8). These findings suggested a model for the unwinding of OriS in which a complex of the UL9 protein bound to boxes I and II and ICP8 bound to single-stranded DNA generated at the A+T-rich linker, possibly as a consequence of transcription (4), unwinds the origin of DNA replication to provide access to the replication machinery, thereby permitting the initiation of DNA replication.
To test this model, we have investigated the ability of the UL9 protein-ICP8 complex to unwind an OriS analogue in which the A+T-rich linker has been replaced by two single strands each consisting of 18 deoxythymidylate residues. We have found this OriS analogue to be unwound by the complex. In addition, we have found that at temperatures that would be expected to destabilize the A+T-rich linker, some unwinding of wild-type OriS can be observed. OriS bearing mutations in boxes I and II which inhibit binding of UL9 protein could not be unwound under these conditions. These findings support the model in which destabilization of the A+T-rich linker to provide a binding site for ICP8 represents the initial event in the UL9 protein-promoted unwinding of OriS.
The structure of the duplex oligonucleotides used are shown in Fig. 1. The single-stranded oligonucleotides were purchased from Operon Technologies, Inc. They were purified prior to use by 16% polyacrylamide electrophoresis under denaturing conditions. The concentration of single-stranded oligonucleotides was determined spectrophotometrically assuming that 1 absorbance unit at 260 nm is equivalent to 30 μg/ml. For double-stranded DNA 1 absorbance unit is equivalent to 50 μg/ml. Oligonucleotides were labeled with [γ-32P]ATP, using T4 polynucleotide kinase according to a protocol supplied by the U.S. Biochemical Corp. Unincorporated nucleotide was removed by Sephadex G-50 spin column chromatography (Quickspin DNA; Boehringer Mannheim). Duplex oligonucleotides were formed by briefly mixing equimolar amounts of the two complementary single-stranded oligonucleotides in 50 mM Tris buffer, pH 8.0, containing 0.1 M NaCl at 90°C for 5 min and then allowing the mixture to cool slowly to room temperature overnight. The annealed DNA was passed through a Sephadex G-50 spin column to remove single-stranded DNA.
FIG. 1.
Helicase substrates used. nt, nucleotides.
To form the box I substrate, the 44-mer 5′-CGCGAAGCG TTCGCACTTCGTCCCGCCTTCCTGCGCCTTCCTGT-3′ was annealed to the 36-mer 5′-CATGCTCGCAGCGGGACGAAGTGCGAACGCTTCGCG-3′ to generate a 26-bp box I duplex with an 18-nucleotide 3′ and a 10-nucleotide 5′ single-stranded tail.
To form the duplex OriS, 5′-CGCGAAGCGTTCGCACTT CGTCCCAATATATATATATTATTAGGGCGAAGTGCG AGCACTGGC-3′ was annealed to 5′-GCCAGTGCTCGCA CTTCGCCCTAATAATATATATATATTGGGACGAAGT GCGAACGCTTCGCG-3′. The mutant OriS was generated by annealing 5′-CGCGAAGCGGGCGCACTTCGTCCCAATA TATATATATTATTAGGGCGAAGTGCGCCCACTGGC- 3′ to 5′-GCCAGTGGGCGCACTTCGCCCTAATAATA TATATATATTGGGACGAAGTGCGCGCTCCTCGCG-3′. OriST18 was generated by annealing 5′-CGCGAAGCGTTCG CACTTCGTCCCTTTTTTTTTTTTTTTTTTGGGCGAAGT GCGAGCACTGGC-3′ to 5′-GCCAGTGCTCGCACTTCGC CCTTTTTTTTTTTTTTTTTTGGGACGAAGTGCGAACG CTTCGCG-3′.
To measure the unwinding of the duplex oligonucleotides, reaction mixtures (25 μl) containing 50 mM HEPES-KOH (pH 8.15), 10 mM NaCl, 10 MgCl2, 2.0 mM dithiothreitol, 10% (vol/vol) glycerol, 10 μg of bovine serum albumin, 2 pmol of box I substrate, 1.0 pmol of OriS, 1.8 pmol of OriST18 1.8 pmol of mutant OriS, 1.6 pmol ICP8 (8), and 1.6 pmol of UL9 protein (8) were incubated on ice for 5 min. ATP (50 μM) was added, and the reaction mixtures were incubated at 37°C for 60 min. The reactions were terminated by the addition of 6.5 μl of stop solution (100 mM EDTA, 1% sodium dodecyl sulfate, 20 μg of proteinase K) for an additional 10 min. at 37°C, followed by electrophoresis through a 15% polyacrylamide gel at 10 V/cm. The gel was then dried on Whatman DE81 paper at 80°C under vacuum and quantitated with a PhosphorImager (Molecular Dynamics) or exposed to Kodak XAR X-ray film.
Unwinding of OriST18.
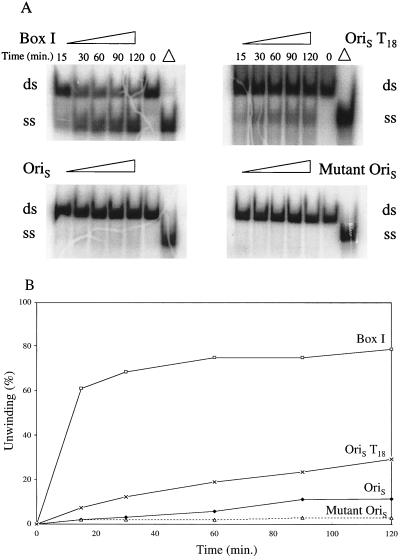
Earlier studies with the box I substrate had led to a model in which destabilization of the A+T sequence linking boxes I and II to provide a binding site for ICP8 was the first step in the unwinding of OriS by the UL9 protein (8). Unwinding of OriS is required to provide an entry site for the replication machinery needed to initiate DNA replication. In vivo, destabilization could result from transcription from promoter sites flanking OriS (4). To test this notion, we generated an OriS duplex, OriST18, in which the A+T region was replaced by two unpaired single strands, each containing 18 thymidylate residues, a structure analogous to OriS in which the A+T sequence had been unwound (Fig. 1). As shown in Fig. 2, in the presence of UL9 protein and ICP8, substantial (20%) unwinding of this structure occurred in 60 min; at 120 min, 30% if the OriST18 was unwound. Under the same conditions, the extent of unwinding of the wild-type OriS duplex was 5 and 8%, respectively. The extent of unwinding of the wild-type OriS, though small, is nevertheless significant, since less than 2% of the mutant OriS was unwound under these conditions. This mutant contained base changes in both boxes I and II and did not bind the UL9 protein (7).
FIG. 2.
Unwinding of OriS, mutant OriS, OriST18, and box I substrate by UL9 protein and ICP8. Individual reaction mixtures containing 1.0 pmol of OriS, 1.8 pmol of mutant OriS, 1.6 pmol of OriST18, or 2.0 pmol of box I substrate, 1.6 pmol of UL9 protein, and 1.6 pmol of ICP8 were prepared as described in the text. They were incubated at 37°C for the times indicated. The products were electrophoresed through a 15% polyacrylamide gel. (A) Autoradiogram; (B) quantitation of products with a PhosphorImager. Percent unwinding is the ratio of the intensities of the single-stranded DNA product and the double-stranded DNA substrate. Δ, double-stranded DNA substrate heated at 100°C for 5 min and then chilled on ice. ss, single stranded; ds, double stranded.
Unwinding of OriS.
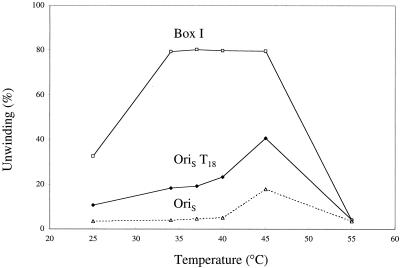
In another approach, unwinding of wild-type OriS was measured under conditions which would be expected to destabilize preferentially the A+T region, i.e., elevated temperature. As shown in Fig. 3, increasing the temperature of the reaction from 40 to 45°C resulted in an increase in the extent of unwinding from 5 to 18%. Previous experiments had demonstrated that the nonspecific helicase activity of the UL9 protein increased linearly from 33 to 45°C, then decreased at 55°C (2), where denaturation of the enzyme occurs (data not shown). The greater-than-threefold increase in the extent of unwinding of OriS with a 5°C increase in the temperature of incubation is consistent with the destabilization of the A+T sequence linking boxes I and II to provide a single-stranded DNA binding site for ICP8. In contrast to OriS, the difference in the extent of unwinding of the OriST18 analogue between 40°C and 45°C was only 1.6-fold, an increase to be expected from the Q10 of the enzyme (2). The box I substrate, which is efficiently unwound over a broad range of temperatures, was maximally unwound (>80%) at 35°C.
FIG. 3.
Effect of temperature on unwinding of OriS, OriST18, and box I substrate by UL9 protein and ICP8. Reaction mixtures were prepared as described in the text and incubated for 60 min at the temperatures indicated. The products were electrophoresed through a 15% polyacrylamide gel and quantitated with a PhosphorImager.
Effect of the UL9 protein/ICP8 ratio.
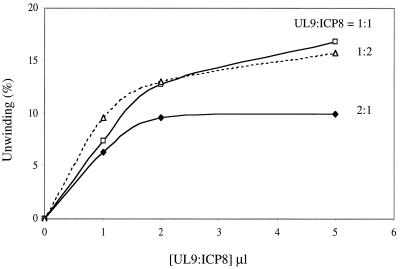
ICP8 presumably serves two functions in the unwinding of OriS: (i) as a component of the UL9 protein-ICP8 complex which constitutes the active helicase (8) and (ii) as a single-stranded DNA binding protein that binds the single strands generated as a consequence of helicase action, thereby preventing their reannealing (1, 11). We therefore investigated the effect of varying the concentration of ICP8 relative to the UL9 protein on the extent of unwinding of the OriS analogue OriST18. As shown in Fig. 4, in accord with a dual function for ICP8, the maximal extent of unwinding was significantly reduced at a UL9 protein to ICP8 ratio of 2:1.
FIG. 4.
Effect of the UL9 protein/ICP8 ratio on unwinding of OriST18. Reaction mixtures were prepared as described in the text. Increasing amounts of enzyme solution containing the indicated UL9 protein/ICP8 ratios were added, and the reaction mixtures were incubated for 60 min. The products were electrophoresed through a 15% polyacrylamide gel and quantitated with a PhosphorImager.
It is also possible that an excess of UL9 protein may have a dominant negative effect on unwinding.
Acknowledgments
This work was supported by NIH research grant AI26538.
REFERENCES
- 1.Bayliss G J, Marsden H S, Hay J. Herpes simplex virus proteins: DNA-binding proteins in infected cells and in the virus structure. Virology. 1975;68:124–134. doi: 10.1016/0042-6822(75)90154-3. [DOI] [PubMed] [Google Scholar]
- 2.Boehmer P E, Dodson M S, Lehman I R. The herpes simplex virus type 1 origin binding protein: DNA helicase activity. J Biol Chem. 1993;268:1220–1225. [PubMed] [Google Scholar]
- 3.Bruckner R C, Crute J J, Dodson M S, Lehman I R. The herpes simplex virus 1 origin binding protein: a DNA helicase. J Biol Chem. 1991;266:2669–2674. [PubMed] [Google Scholar]
- 4.Dabrowski C E, Schaffer P A. Herpes simplex virus type 1 origin-specific binding protein: oriS-binding properties and effects of cellular proteins. J Virol. 1991;65:3140–3150. doi: 10.1128/jvi.65.6.3140-3150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elias P, Lehman I R. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc Natl Acad Sci USA. 1988;85:2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fierer D S, Challberg M D. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J Virol. 1992;66:3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez T R, Dutch R E, Lehman I R, Gustafsson C, Elias P. Mutations in a herpes simplex virus type 1 origin that inhibit interaction with origin-binding protein also inhibit DNA replication. J Virol. 1991;65:1649–1652. doi: 10.1128/jvi.65.3.1649-1652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S S-K, Lehman I R. Unwinding of the box I element of a herpes simplex virus type I origin by a complex of the virus origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc Natl Acad Sci USA. 1997;94:2838–2842. doi: 10.1073/pnas.94.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehman I R, Boehmer P E. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 10.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinn J P, McGeoch D J. DNA sequence of the region of the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985;13:8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]