High-resolution mapping of the mouse axonal connectome reveals new foundational wiring principles of the mammalian brain.
Abstract
Fine-grained descriptions of brain connectivity are required to understand how neural information is processed and relayed across spatial scales. Previous investigations of the mouse brain connectome have used discrete anatomical parcellations, limiting spatial resolution and potentially concealing network attributes critical to connectome organization. Here, we provide a voxel-level description of the network and hierarchical structure of the directed mouse connectome, unconstrained by regional partitioning. We report a number of previously unappreciated organizational principles in the mammalian brain, including a directional segregation of hub regions into neural sink and sources, and a strategic wiring of neuromodulatory nuclei as connector hubs and critical orchestrators of network communication. We also find that the mouse cortical connectome is hierarchically organized along two superimposed cortical gradients reflecting unimodal-transmodal functional processing and a modality-specific sensorimotor axis, recapitulating a phylogenetically conserved feature of higher mammals. These findings advance our understanding of the foundational wiring principles of the mammalian connectome.
INTRODUCTION
Studies examining the structural architecture of the brain have advanced our knowledge of how information is processed and integrated across distributed and specialized neural circuits. Current network theory applied to brain connectomes has greatly contributed to this process, highlighting a series of common organizational principles underlying brain connectivity, many of which appear to be species and scale invariant (1). These include the presence of discrete regional subsystems (termed communities) critically interlinked by a small number of highly connected hub nodes, a configuration optimally suited for effective information processing, and integration of neural signals across sensory and cognitive domains (2). Brain communities and hub regions have been observed at different investigational scales and using multiple connectivity readouts in several species, from the nematode Caenorhabditis elegans to humans (3–5).
Recently, the mesoscale connectome of the mouse brain has been mapped via the use of directional viral tracers, representing one of the best characterized directed mammalian connectome ever described to date (6, 7). The integration of this dataset with gene expression maps and layer-specific viral tracing have advanced our understanding of the wiring principles of the mammalian brain, revealing a network core of highly interconnected and metabolically costly hub nodes (8), and a phylogenetically conserved feedforward-feedback laminar hierarchy in intracortical structure (7). However, most investigations of the mouse connectome to date have been limited by the use of predefined anatomical parcellations in which connectional parameters, from which network attributes are computed, are quantified under the assumption of regional homogeneity (6). This has typically entailed the interrogation of subsets of anatomically aggregated meta-regions [for example, 213 × 213 regions in (8), or 130 × 130 in (5)], an option that greatly increases the computational tractability of the mouse connectome. However, the use of predefined meta-areas is non-ideal, as the sharp inter-areal boundaries that characterize most neuroanatomical parcellations reflect a discretization of otherwise regionally continuous cytoarchitectural or anatomical parameters that may straddle cross-regional network features. Moreover, the use of meta-regions might limit the resolution of topological mapping in the mouse connectome, biasing the ensuing network metrics toward areas that are anatomically larger or more prominent, and potentially obscuring fine-grained or subregional attributes that could be critical to the network organization of the mammalian connectome. The recent release of a voxel-level data-driven model of the mouse connectome (9) offers the possibility of overcoming the limitations or regional-aggregated investigations in this species. This resource entails an improved interpolation model for single tracer injection maps and provides whole-brain coverage, encompassing subcortical districts not covered by state-of-the-art parcellations (6, 9). Moreover, the voxel-wise mouse connectome is characterized by a sampling resolution that is unprecedented for mammalian species, offering the opportunity to probe the topological structure of the mammalian connectome at a regional-scale never investigated so far.
Here, we leverage the voxel-level mouse connectome by Knox et al. (9) to provide a brain-wide, high-resolution description of the network structure and hierarchical organization of the directed mouse connectome, unconstrained by regional partitioning (15,314 × 15,314 matrix, Voronoi voxel volume 0.027 mm3 non-isotropic, mean spatial extension of Voronoi voxels: 242 μm × 323 μm × 336 μm; see Materials and Methods). Our results show that the mouse connectome is characterized by a finer network topography than previously reported, uncovering some previously underappreciated network features of the mammalian connectome. These include a segregation of hub regions into source and sink nodes, pointing at an organizational hierarchy in which higher-order cortical areas serve as primary sources of neural output to the rest of the brain, and basal ganglia are configured as pivotal recipients of incoming projections. Using in silico network attacks, we also uncovered a strategic role of ascending modulatory nuclei as essential orchestrators of network communicability, a connectional property that makes these systems points of vulnerability for network function. We also found a tight interdependence between functional and structural brain organization, entailing the spatial arrangement of mouse cortical areas according to a hierarchy reflecting unimodal-transmodal and modality-specific functional processing, hence broadly reconstituting basic organizational principles of the primate brain. Our findings define a high-resolution structural scaffold linking mesoscale connectome topography to its macroscale functional organization and create opportunities for identifying targets of interventions to modulate brain function in a physiologically accessible species.
RESULTS
Global hubs and rich-club core of the voxel-wise mouse connectome
A defining characteristic of brain connectomes is the presence of spatially localized set of integrative hub regions, characterized by high connectivity density (2). Hub regions serve as focal points of network interaction and exert a tight influence on the structure and dynamics of brain networks (2). To identify regional features exhibiting hub-like properties at the voxel scale, we first mapped voxels exhibiting high connectivity strength using a spatially resampled (15,314 × 15,314) version of the Allen Institute mouse connectome (9), irrespective of the directionality of the connections. We termed the identified regions as global hubs to distinguish them from further hub identification carried out using the directed connectome (described below). This analysis revealed several focal areas exhibiting global hub-like properties (Fig. 1A). Consistent with the high centrality of hub regions, the identified foci were prominently located in associative cortical areas such as the prefrontal, anterior cingulate, posterior parietal, and retrosplenial cortices (Fig. 1A). An additional large cluster of hub voxels was apparent in dorsal portions of the hippocampus. Last, our fine-grained mapping also allowed the recognition of a small set of hub nodes in subregional portions of the basolateral and central amygdala.
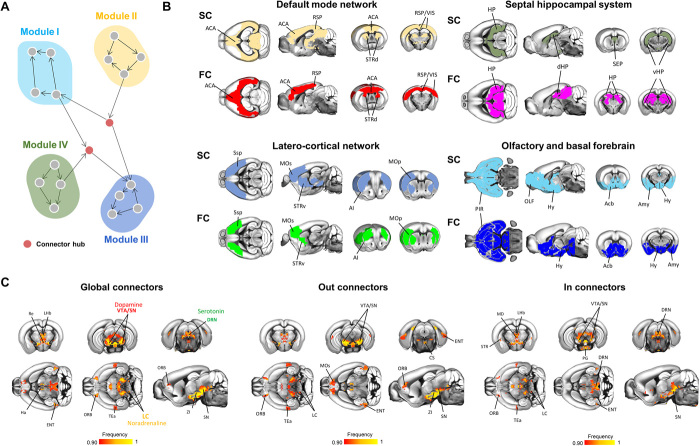
Fig. 1. Global hubs and rich club of the mouse connectome.
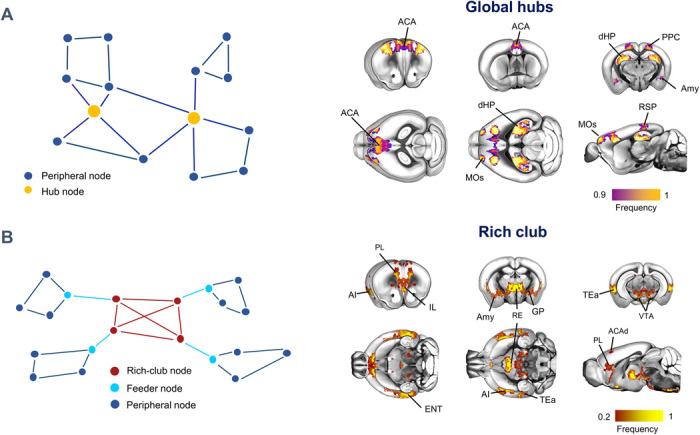
(A) Anatomical distribution of global hubs of the voxel-wise mouse connectome. Global hubs (yellow nodes on the left panel) were defined on the basis of nodal total strength. A frequency map was obtained by computing the fraction of times a node scored among the highest-ranking strength nodes, limiting the visualization to the nodes that were classified as hubs at least 90% of the time. (B) Anatomical distribution of the rich club (red nodes on the left panel) of the voxel-wise mouse connectome. The frequency map indicates fraction of times high-degree nodes were retained as significant with respect to a set of random networks. ACA, anterior cingulate area; ACAd, anterior cingulate area, dorsal part; AI, agranular insular area; Amy, Amygdala; dHP, dorsal hippocampal area; ENT, entorhinal area; GP, globus pallidus; IL, infralimbic area; MOs, secondary motor area; PL, prelimbic area; PPC, posterior parietal cortex; RE, nucleus reuniens; RSP, retrosplenial area; TEa, temporal association areas.
In brain networks, highly connected central hub nodes have a tendency to be tightly interlinked with each other, defining a core network structure, often referred to as rich club, which supports the efficient integration of otherwise segregated neural systems (4, 8). To obtain a description of the mouse brain rich-club unconstrained by preexisting anatomical partitioning, we used the procedure described by (8) on the ipsilateral voxel-wise connectome, benchmarking our mapping against 1000 weighted rewired networks characterized by the same empirical in- and out-degree distribution (10). The obtained map revealed a more extended spatial topography than observed with global hub mapping, encompassing two major antero-posterior integrative axes (Fig. 1B and fig. S3). The first of these included transmodal cortical integrators of sensory input [i.e., insula and temporal association cortex (11)]. The second axis encompassed infralimbic and mid-thalamic components of the fronto-hippocampal gateway (12). Nodal mapping also revealed the participation of midbrain nuclei such as the ventral tegmental area (VTA), pointing at a previously unappreciated involvement of ascending dopaminergic nuclei as integral components of the rich club of the mouse connectome.
The spatial extension of global hubs and rich-club voxels in most cases encompassed only a marginal portion of the corresponding anatomical structure as defined in the Allen Brain Atlas, significantly deviating from corresponding voxel-level distributions at the regional level (fig. S1). In keeping with this notion, a qualitative comparison of voxel-wise mapping with that obtained with a state-of-the-art anatomical parcellation (6) revealed substantial differences in the anatomical distribution of hub-like and rich-club regions (fig. S2). This result corroborates the specificity of our findings, suggesting that previous mapping of hub-like properties in the parcellated connectome might have been resolution-limited.
Hub regions can be directionally segregated into neural sinks and sources
Our initial analyses were aimed at mapping global network features and, as such, were carried out on a nondirected version of the mouse connectome. However, directionality is a fundamental feature of brain connectomes, and most structural brain networks are intrinsically directed because of the monodirectional nature of axonal projections. Thus, directional encoding can critically add key information to the topological organization of brain networks (13), revealing organizational motifs that can be predictive of the information flow and hierarchical organization of the mammalian brain. To probe how the direction of structural connections affects network attributes, we parsed high connectivity strength regions based on their directional profile, resulting in the identification of a set of segregable nodes that we termed source and sink, characterized by high-strength outgoing or incoming connections, respectively (Fig. 2A).
Fig. 2. Source and sink hubs of the mouse connectome are spatially segregable.
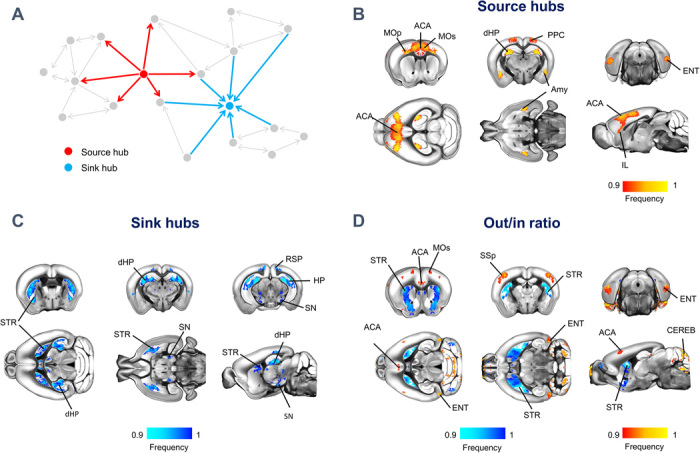
(A) Network schematic illustrating our topological classification of high strength regions into neural sources (red) and sinks (light blue). Source (B) and sink (C) hubs were defined on the basis of the voxel-wise strength of outgoing and incoming connectivity, respectively. Frequency maps were obtained by computing the fraction of times a node scored among the highest-ranking strength nodes, limiting the visualization to the nodes that were classified as hubs at least 90% of the time. (D) Out/in ratio mapping. For each node, we computed the ratio between the strength of the outgoing and incoming connectivity. Frequency maps were obtained by computing the fraction of times a node scored among the highest (red/yellow) or lowest ranking (light blue/blue) nodes as in (C) and (D). ACA, anterior cingulate area; Amy, Amygdala; CEREB, cerebellum; dHP, dorsal hippocampal area; ENT, entorhinal area; HP, hippocampus; MOp, primary motor areas; MOs, secondary motor area; PPC, posterior parietal cortex; RSP, retrosplenial area; SN, substantia nigra; SSp, primary somatosensory area; STR, striatum.
Source node distribution broadly recapitulated the location of global hubs, encompassing higher-order areas such as the anterior cingulate and posterior parietal cortices, amygdala, and dorsal hippocampus, together with posterior entorhinal areas (Fig. 2B). Mapping of sink nodes revealed the involvement of dorsal hippocampal areas along with an additional set of substrates, which comprised the basal ganglia throughout their antero-posterior extent (Fig. 2C). Participation of nuclei within the substantia nigra was also apparent. These results show that high connection strength regions can be segregated on the basis of their directional profile and point at an organizational hierarchy in which higher-order areas, such as the prefrontal cortex, serve as primary sources of neural output to the rest of the brain, while basal ganglia are pivotal recipients of incoming projections. In keeping with what we observed with hub and rich-club regions, the identified sink and source voxel clusters showed prominent subregional distribution with respect to preexisting anatomical subdivision (fig. S1) and a remarkably different spatial organization when computed using an anatomical parcellation (fig. S2), corroborating the specificity of our fine-grained mapping with respect to canonical parcellation-based approaches.
The observation of segregable sink and source high-connection strength areas prompted us to investigate whether such a hierarchy could be expanded to nonhub areas (i.e., to all brain regions, independent of their connection strength), by computing the voxel-wise ratio between outgoing and incoming connection strength, a metric that we term “out/in ratio” (14). This analysis might allow us to differentiate regions characterized by a net connectional imbalance from those exhibiting both high input and output density (e.g., dorsal hippocampus). The resulting out/in ratio map (Fig. 2D) revealed a prominent configuration of basal ganglia as regions characterized by a low ratio of outgoing/incoming connections, corroborating a configuration of these substrates as connectivity sinks. Conversely, foci exhibiting a high out/in connection ratio were identifiable in higher-order cortical areas, such as the anterior cingulate and entorhinal cortices, but also prominently encompassed some nonhub substrates, such as the cerebellum, and primary motor–sensory regions. Together, these results show that the directed connectome is topologically rich and configured according to a global hierarchy that can be used to segregate regions in primary sources or receivers of axonal connections. Furthermore, they provide a fine-grained description of hub-like topography that may guide further targeted manipulations of salient network attributes in this species.
Structural communities of the voxel-wise connectome recapitulate large-scale fMRI networks of the mouse brain
The presence of distinct subnetworks or “communities” of tightly interlinked nodes is a hallmark of all mammalian connectomes mapped to date (15). Structural communities are composed of topologically related neural elements reflecting regularities in wiring diagram, typically delineating groups of brain areas with shared functionality (16). Previous investigations of the community structure of the mouse connectome have been either anatomically biased by the use of meta-regions (5) or limited to the sole cortical mantle (7), preventing a fine-grained description of the community structure of the entire mouse connectome. To identify stable brain-wide communities in the directed connectome with voxel resolution, we used a multiscale modular decomposition approach (fig. S4) (5). This approach revealed five prominent communities, encompassing different combinations of cortical and subcortical regions (Fig. 3). Corroborating the robust structural foundations of resting-state functional magnetic resonance imaging (rsfMRI) network architecture (17, 18), the identified structural communities exhibited a spatial distribution closely recapitulating previously described rsfMRI connectivity communities of the mouse brain (19, 20). The first of these communities comprised transmodal cortico-limbic areas as well as the dorsal striatum and antero-medial thalamus, spatially reconstituting key components of the mouse default-mode network [DMN; (21)]. A second community encompassed latero-cortical motor-sensory areas as well as striatal and thalamic nuclei, which have been previously characterized as component of the mouse latero-cortical network (LCN). This network is considered to be a possible evolutionarily precursor of the human “task-positive” network, as it appears to be tightly anticorrelated to DMN activity (20). A third module encompassed septo-hippocampal areas, while the fourth comprised olfactory areas and basal forebrain regions, once again recapitulating corresponding rsfMRI functional communities (19). Notably, anatomically similar structural connectivity (SC) partitions were also obtained using an agglomerative hierarchical clustering procedure (Dice coefficients of 0.7, 0.8, 0.9, and 0.9 for the DMN, LCN, hippocampal system, and olfactory/basal forebrain moduli, respectively), corroborating the validity of the nodal partitioning reported here. By contrast, community detection in the parcellated connectome revealed three macromodules (fig. S2) whose spatial topography was not directly relatable to corresponding rsfMRI communities, underscoring a closer structural-functional correspondence of the voxel-wise connectome with respect to parcellated connectome.
Fig. 3. Connector hubs encompass key ascending neuromodulatory nuclei.
(A) Network schematic illustrating a graph-based definition of communities and connector hubs. (B) Structural communities anatomically recapitulate functional (rsfMRI) networks of the mouse brain. Structural communities (SC; top row; see Materials and Methods) were matched to corresponding functional communities [FC; bottom row; Liska et al. (19)]. (C) Neuromodulatory nuclei are configured as connector hubs. Global (left), out-connector (middle), and in-connector (right) hubs were computed on the basis of the participation coefficient metric, accounting for outgoing or incoming connections only. ACA, anterior cingulate area; Acb, nucleus accumbens; AI, agranular insular area; Amy, amygdala; CS, superior central nucleus raphe; DRN, dorsal nucleus raphe; ENT, entorhinal area; HP, hippocampus; Ha, habenula; Hy, hypothalamus; LC, locus coeruleus; LHb, lateral habenula; MD, mediodorsal nucleus of the thalamus; MOp, primary motor areas; MOs, secondary motor area; OLF, olfactory areas; ORB, orbital areas; PG, pontine gray; PIR, piriform area; RE, nucleus reuniens; RSP, retrosplenial area; SEP, septal complex; STRd, striatum dorsal region; STRv, striatum ventral region; TEa, temporal association areas; vHP, ventral hippocampal area; VIS, visual areas; VTA, ventral tegmental area; ZI, zona incerta.
The close topographical overlap between voxel-wise structural communities and corresponding rsfMRI functional networks prompted us to probe the relationship between SC and functional connectivity (FC) at the level of individual co-registered voxels. To this purpose, we carried out a correlation analysis between SC and FC for the DMN, LCN, and hippocampal networks—three well-characterized distributed mouse rsfMRI networks (22) for which we identified unambiguous structural correlates. In keeping with recent investigations in primates (23), we found that voxel-wise correlation between FC and SC was nonlinear, reflecting connection length–dependent contributions (fig. S5). Specifically, functional-structural correlation was moderate to high (Spearman’s rho = 0.35, 0.45, and 0.34 for DMN, LCN, and the hippocampal network, respectively) for relatively short connections (<1 mm; e.g., the scale of mouse cortical width), but lower for longer-range links (>2 mm, Spearman’s rho = 0.26, 0.38, and 0.17 for DMN, LCN, and hippocampal network, respectively). Consistent with the neural mass nature of rsfMRI fluctuations, the correlation between FC and SC was robustly linear when both quantities were resampled at a lower spatial resolution using an anatomical parcellation (Pearson’s r = 0.59, 0.55, and 0.54, P < 0.0001, for the DMN, LCN, and hippocampal network, respectively). Together, these findings underscore the robust structural foundations of functional network activity as inferred from rsfMRI and suggest that spontaneous fMRI signal fluctuations underscoring macroscale rsfMRI coupling reflect the pooled activity of large ensembles of neurons, exceeding the finer spatial scale of the voxel-wise mouse connectome (24–26).
Ascending modulatory nuclei are configured as between-network connector hubs
The observation of tightly overlapping structural and functional communities prompted us to investigate the topological structure and anatomical location of network nodes configured as connector hubs. These are nodal components critically configured as key orchestrators of intermodular communication, enabling the dynamic interaction of lower- and higher-order networks to control complex behavioral and cognitive functions (27).
We first located connector hubs irrespective of the directionality of the connections and termed the identified connector nodes global connectors (Fig. 3C). We found global connector hubs to be mainly localized in midbrain, hypothalamic, and medio-dorsal thalamic regions, with only a marginal cortical involvement limited to orbitofrontal and temporal association areas. Midbrain connector hubs focally encompassed three major sets of ascending neuromodulatory nuclei, namely, the VTA and substantia nigra (dopamine), dorsal raphe nuclei (serotonin), and a set of voxels encircling the locus coeruleus (norepinephrine). Accounting for the directionality of the connections revealed evidence of a negligible topological segregation for most of the identified connector nodes (Fig. 3C). As observed with other hub subtypes, most connector hub nodes exhibited a significant subregional distribution with respect to a predefined high-resolution anatomical parcellation (fig. S1B). Moreover, anatomical mapping of connector hubs using a parcellated connectome failed to reveal the involvement of key neurotransmitter nuclei (e.g., VTA, raphe areas, and locus coeruleus), hence concealing a key topological attribute of the mouse connectome. Collectively, these results reveal that ascending modulatory systems are strategically wired as connector regions. The robust topographical correspondence between functional and structural modules supports a role for the identified connector hubs as strategic orchestrators of brain-wide network activity (28), a notion consistent with emerging evidence pointing at a pivotal contribution of catecholaminergic neurotransmission in modulating functional network activity and dynamics (29, 30).
Connector hubs are critical mediators of network communicability
Graph theory postulates a critical contribution of hub regions to network integrity and stability, a notion supported by computational modeling of the human brain connectome (31). These properties support a theoretical framework in which hub regions serve as putative points of vulnerability for network disruption in the mammalian connectome (2). To test whether these assumptions hold for the voxel-wise mouse connectome, we performed a series of targeted in silico nodal attacks and assessed how these virtual lesions affect the ensuing network properties (Fig. 4). The effect of hub (or random node) removal was assessed using two well-characterized global network attributes: (i) the size of the giant component, i.e., the largest subgraph in the network, a proxy for the network’s integrity (31), and (ii) global network efficiency, a measure of the ability of a network to efficiently route information (32). This latter attribute is a measure of integration closely related to characteristic path length, based on the intuition that short path lengths in a network will facilitate rapid and efficient communication. A globally efficient network is therefore a network in which information can be efficiently routed in a cost-effective way. Targeted removal of sources and out-connector hub nodes did not produce appreciably larger network fragmentation than observed with random nodal attacks (Fig. 4). Similarly, the removal of central nodes had an overall marginal impact in decreasing network efficiency, producing a fragmentation that was, on average, only ~1.5% greater than random node removal (P < 0.01; Fig. 4). These results are in line with previous observations suggesting that, independent of its sampling resolution (fig. S6A), the topological structure of the mouse connectome does not recapitulate a canonical scale-free organization (6, 33). They also support the notion that, irrespective of their classification and directionality, hub nodes of the voxel-wise connectome are not critical for the integrity and efficiency of brain network, making the mouse connectome highly resilient to targeted perturbations.
Fig. 4. Connector hubs are critical effectors of network communicability.
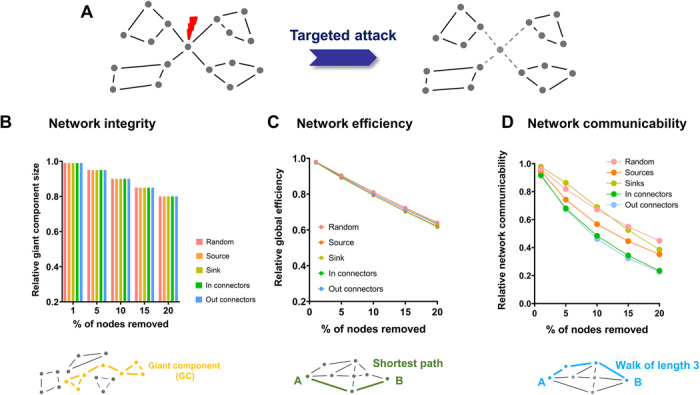
(A) Schematic illustration of targeted node removal and its effect on network integrity. (B to D) Effect of targeted hub removal on different network properties.
We therefore next probed whether hub regions could be key to a different communication mechanism other than the routing of information through shortest paths (i.e., as measured by network efficiency). To this purpose, we measured the effect of virtual hub lesioning using total network communicability (34, 35). This graph attribute measures the network’s capacity for parallel information transfer, i.e., by taking into account the ability of information to disperse equally across all paths and walks in the network, i.e., not necessarily and exclusively through the shortest paths. This property has therefore been equated to a measure of “bandwidth” or “broadcasting capacity” (34) for information transfer within a network. Notably, we found that removal of connector hubs markedly reduced network communicability with respect to random node deletion (P < 0.001; Fig. 4). A replication of virtual lesioning using a state-of-the-art parcellation (6) produced similar results (fig. S6B), suggesting that the resiliency of the mouse connectome in terms of network integrity and efficiency, as well as its vulnerability in terms of network communicability, are resolution-invariant properties. Together, these findings suggest that connector hubs, and the neurotransmitter nuclei therein contained, besides acting as pivotal orchestrators or intermodular communication, are also configured as key effectors of network communicability, enabling effective routing of information across regions via multiple parallel pathways.
The voxel-wise mouse connectome is organized along two superimposed cortical gradients
Recent functional and structural investigations in humans and primates have shown that the spatial arrangement of cortical connectivity reflects two superimposed gradients along which cortical locations are ordered according to their similarity in connections to the rest of the cortex (18). A first dominant cortical gradient is anchored in sensorimotor regions and radiates toward higher-order transmodal areas; a second gradient exhibits instead an axis of differentiation between sensorimotor modalities (18). The organization of the unimodal-transmodal gradient is thought to define a hierarchy of increasing functional integration that guides the propagation of sensory inputs along multiple cortical relays into transmodal regions (36).
The fine-grained sampling of the voxel-wise connectome is ideally suited to probe the structural foundations of these organizational axes at an unprecedented spatial resolution. To this aim, we first probed whether a similar organization is phylogenetically conserved in rodents. We therefore applied diffusion map embedding (37) to the directed cortical connectome (Fig. 5). To account for the directional encoding of the connectome, the procedure was applied to a matrix mapping the connectional profile of each node, i.e., incorporating the information provided by both incoming and outgoing connections. Notably, we found that the structural connectome exhibits two spatial gradients of connectivity broadly recapitulating organizational principles observed in primates. Specifically, a dominant gradient (gradient A) involved a sensory-fugal transition between unimodal motor–sensory regions of the mouse LCN and transmodal components of the mouse DMN (Fig. 5A). A second gradient (gradient B) extended across unimodal visual and auditory cortices up to primary motor–sensory regions, hence providing a regional differentiation between sensorimotor modalities. The spatial topography of cortical gradients appeared to be seemingly preserved when computed using a state-of-the-art parcellation (fig. S2G). However, resolution-dependent discrepancies in the topology of gradient B were apparent when a parcellation scheme was used, encompassing an atypical involvement of associative areas that departs from the modality-specific structure of analogous cortical gradients in higher mammals (fig. S2).
Fig. 5. Gradients of SC and FC in the mouse cortex exhibit comparable topology.
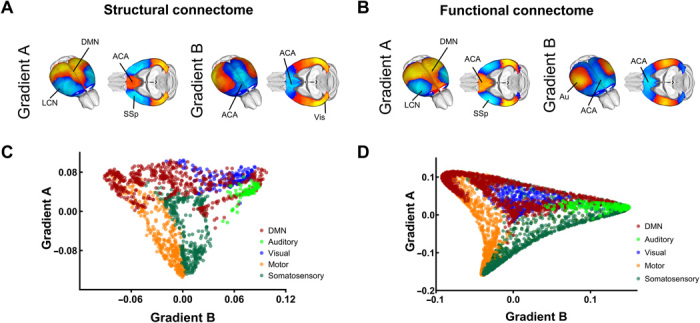
Structural (A) and functional (B) gradients of cortical organization in the mouse connectome. Gradient A encompasses a unimodal-polymodal spectrum of cortical regions extending from motor-sensory LCN (light blue/blue) to the DMN (yellow/red). Gradient B extends antero-posteriorly across primary sensorimotor (yellow) and transmodal associative regions (blue). (C and D) Regional scatter plots of gradient organization for SC (C) and FC (D). ACA, anterior cingulate area; Au, auditory area; DMN, default mode network; LCN, latero-cortical network; SSp, primary somatosensory area; Vis, Visual areas.
Gradients of SC and FC in the mouse cortex exhibit comparable topology
The close topographical overlap between structural and functional communities observed in our modular analyses prompted us to probe the presence of a similarly tight relationship between anatomical and rsfMRI gradient organization in the mouse brain (Fig. 5B). To this aim, we mapped cortical gradients with voxel resolution in the mouse functional connectome. This analysis revealed that the functional connectome is organized into a unimodal-transmodal gradient (DMN-LCN, gradient A) and a modality-specific gradient (gradient B, Fig. 5B) closely recapitulating key topographical features of the structural voxel-wise connectome (spatial correlation, Spearman’s rho = 0.83, P < 0.01 for gradient A and rho = 0.60, P < 0.05 for gradient B, corrected for spatial autocorrelation). An anatomical classification of the regional constituents of the identified gradients revealed that the topographical organization of trans-modal and unimodal areas was broadly comparable across modalities (Fig. 5, C and D), although a small rearrangement in the spatial organization of modality-specific areas in functional gradient B was apparent, peaking in auditory-somatosensory regions as opposed to auditory-visual areas (fig. S7B). Notwithstanding these modality-specific differences, unimodal versus polymodal cortical arrangement in the functional and axonal connectomes appeared to be largely similar, pointing at a common hierarchical organization for the functional and structural mouse connectome. Together, these results reveal a robust structural foundation for cortical gradient organization in the mammalian cortex and show that the mouse brain connectome recapitulates phylogenetically conserved architectural principles observed in higher mammalian species.
Gradients of SC reflect cortico-cortical laminar hierarchy and constrain fMRI network dynamics
Human studies have linked the organization of cortical gradients to hierarchical structure inferred from patterns of laminar cortical connectivity (38). The recent description of a feedforward-feedback laminar hierarchy in cortical regions of the mouse brain (7) allowed us to probe whether a similar organizational principle could explain the architectural organization of some of the gradients identified in the connectome. By computing the correlation between laminar hierarchy from (7) and structural gradient topography in a set of corresponding cortical regions, we found that the regional organization of the modality-specific gradient (gradient B) was robustly correlated with intracortical laminar hierarchy (Fig. 6A; Spearman’s rho = 0.49, P < 0.01 corrected for spatial autocorrelation), hence linking mouse cortical gradient (B) organization to patterns of laminar connectivity. Intralaminar cortical hierarchy was instead not predictive of unimodal-polymodal gradient (gradient A) topography (Fig. 6A; Spearman’s rho = 0.17, P = 0.33, corrected for spatial autocorrelation).
Fig. 6. Gradients of SC reflect cortico-cortical laminar hierarchy and constrain fMRI network dynamics.
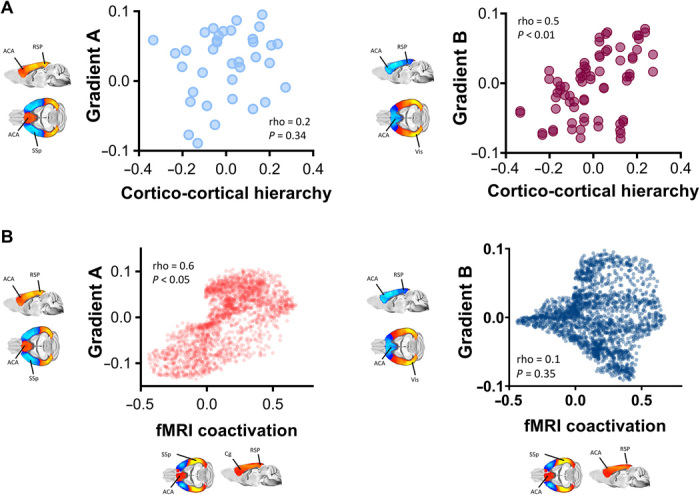
(A) Modality-specific gradient B (right), but not polymodal-unimodal gradient A (left), reflects hierarchical intra-laminar organization of the mouse cortex. (B) Unimodal-polymodal DMN-LCN gradient A, but not modality-specific gradient B, closely recapitulates the spatial topography of dominant cortical CAPs governing fMRI dynamics in the mouse. ACA, anterior cingulate area; RSP, retrosplenial area; SSp, primary somatosensory area; Vis, Visual areas.
We finally noted that the anatomical organization of the unimodal-polymodal gradient A was anatomically consistent with the topography of dominant patterns of blood oxygen level–dependent (BOLD) fMRI coactivation [coactivation patterns (CAPs)] recently described in the mouse (24). CAPs serve as recurring “building blocks” of spontaneous fMRI network dynamics and are characterized by a distinct anatomical topography involving infraslow oscillatory transitions differentially affecting unimodal latero-cortical areas and midline polymodal regions (24, 39). The observation of a possible anatomical overlap between cortical gradient organization and dominant CAP topography may therefore explain the so far unaccounted specific anatomical organization of these large-scale dynamic fluctuations (24). In keeping with this notion, we found a strong spatial correspondence (Fig. 6B; Spearman’s rho = 0.60 and P < 0.05, corrected for spatial autocorrelation) between gradient A and dominant CAP topography. Conversely, the modal-specific gradient B did not show a significant relationship with the spatiotemporal structure of this dominant CAP (Fig. 6B, right panel; Spearman’s rho = 0.09 and P = 0.35, corrected for spatial autocorrelation). These results suggest that the hierarchical organization of the cortical connectome may critically shape and constrain spontaneous patterns of fMRI network dynamics.
DISCUSSION
Here, we provide a fine-grained description of salient architectural motifs of the mouse connectome, without the imposed limits of discrete regional parcellations. Departing from regional-constrained studies, we find that hub regions and core network components of the voxel-wise mouse connectome exhibit a rich topography encompassing key cortical and subcortical relay regions. We also typify regional substrates based on their directional topology into sink or source regions and report a previously unappreciated role of modulatory nuclei as critical effectors of intermodular and network communicability. Last, we demonstrate a close spatial correspondence between the mesoscale topography of the mouse connectome and its functional macroscale organization and show that, like in primates and humans, the mouse cortical connectome is organized along two major topographical axes that can be linked to hierarchical patterns of laminar connectivity and shape the topography of fMRI dynamic states, respectively.
Our regionally unconstrained mapping of hub-like regions complements and expands previous investigation of the mouse connectome, providing a spatially precise identification of network features and hierarchical motifs that may guide future manipulations of nodal properties in this species (5). These include a fine-grained localization of hub-like properties in subregional components of large integrative areas, such as the dorso-lateral hippocampus or the central and basolateral amygdala, which were previously been considered as regionally homogeneous (5). Similarly, our rich-club mapping revealed a more detailed spatial topography than previously reported (8), revealing two major organizational axes of high relevance for sensory integration and higher cognitive functions, and which recapitulate organizational features observed also in nonmammalian species (40). Notably, perturbational studies support the biological relevance of our findings, as chemogenetic inactivation of the nucleus reuniens of the thalamus—a pivotal component of the mouse rich club—has been recently shown to impair hippocampal-dependent cognitive function in mice (41). These results suggest that subcortical relay stations are core components of nodal rich clubs across evolution, serving as critical integrators between top-down and bottom-up functional processing. Our analyses also show that hub-like network attributes in the voxel-wise mouse connectome are neuroanatomically segregable. This finding suggests that the network structure of the mammalian connectome is the result of converging evolutionary pressure, resulting in a regional organization in which spatially distinct hub-like regions delineate a hierarchy between higher-order highly interconnected associative regions and bottom-up input from neuromodulatory areas configured as critical effectors of interregional communication.
Our results also revealed previously unappreciated organizational features of the mouse connectome that advance our understanding of the fundamental wiring principles of the mammalian brain in three main directions. First, the use of a high-resolution and directed connectome enabled us to segregate hub regions into source and sink areas. The ensuing classification revealed the emergence of a global hierarchy in which higher-order cortical areas and hippocampal regions serve as primary sources of neural input to the rest of the brain, and basal ganglia (plus focal mesencephalic nuclei) are wired as major receivers of distributed neural input. This hierarchical configuration follows a phylogenetic gradient in the arrangement of SC and is optimally designed for the execution of rapid motor responses in response to salient external stimuli (42). Such a hierarchical configuration could also be expanded to nonhub regions via a brain-wide computation of the ratio of outgoing and incoming connectivity strength, defining a related organizational axis with motor-related nuclei, such as the cerebellum and basal ganglia, being located at its extremes. Notably, most of the network and topological attributes we describe in the present work appear to be resolution specific (fig. S2), suggesting that previous topological mapping of the mouse connectome may have been biased by the coarser resolution of existing regional parcellations.
A second notable feature is our observation of a strategic configuration of ascending modulatory systems as connector hubs and essential effectors of network communicability. Previous investigations of the regionally segregated mouse connectome have produced a largely cortico-centric description of connector hubs, involving cingulate, orbitofrontal, and posterior association cortices, together with the basal ganglia and regionally undifferentiated midbrain regions (5). Our results shift the focus from the cortex to subcortical relay stations and document that ascending neurotransmitter systems are central to the mouse connectome and are configured as intermodular connector hubs. The observed spatial correspondences between the structural and functional topography of the mouse connectome argue for a critical role for these neuromodulatory nuclei in shaping large-scale neural activity. This notion is consistent with the observation that catecholaminergic and serotonergic activities critically control functional network topography and dynamics (29, 43). Together with the observation that connector hub removal critically diminishes network communicability, these results suggest that the ascending modulatory systems are strategically wired as central orchestrators of large-scale intermodular communication, enabling the parallel routing of large chunks of information across the network. This network property might be key in ensuring the effective and finely tuned control of complex behavioral and physiological states exerted by these systems. At the same time, these properties might render these nodes key points of vulnerability for functional network disruption in brain disorder, a notion consistent with emerging evidence linking neuromodulatory dysfunction to neurodegenerative pathologies (44). Targeted removal of global hubs did only negligibly affect measurements of network integrity and efficiency when compared to random node deletion. This finding suggests that the mammalian connectome is structurally highly resilient and argues against a role for this class of hub regions as critical mediators of network integrity in the mouse connectome. This result is partly supported by analogous investigations of the human connectome. For example, Crossley et al. (45) reported a linear decrease in global efficiency in a targeted attack for human structural networks, analogously to what we observed for the mouse connectome. Similarly, Alstott et al. (46) found functional network fragmentation to occur only when about 75% of the high strength nodes were removed. It should, however, be noted that other reports seem to be at odds with these results, suggesting a significant vulnerability of the human connectome against targeted attacks in human networks [see (31) for a recent overview on the topic]. Whether these discrepancies reflect modality- and resolution-related discrepancies, or a lower proportion of long-range integrative fibers in rodents owing to evolutionary scaling of white/gray matter ratio (47), remains to be established.
Last, our voxel-wise description of two principal axes of cortical organization in the mouse connectome, and their topological linking with cortico-laminar organization and patterns of spontaneous fMRI dynamics, establishes a direct link between the mesoscale topography of the mouse connectome and its functional macroscale organization. These results suggest that the spatial arrangement of cortical areas along unimodal-polymodal and modality-specific gradients represents a general evolutionarily conserved principle governing the hierarchical organization of the mammalian cortex across evolution, and one that might intimately constrain the emergence and topology of spontaneous brain activity. This notion is consistent with a recent description of a cortical hierarchy in the parcellated mouse brain, as assessed by using an imaging marker of intracortical myelin content (48). Our findings expand these previous observations by providing cross-modal and voxel-wise evidence of two superimposing functional and structural cortical gradients broadly recapitulating organizational principles observed in the human and primate brain. These include a hierarchical organization reflecting a well-characterized feedforward-feedback laminar hierarchy (7) and a spectrum between unimodal regions and transmodal areas. It should be noted, however, that in the mouse the latter are known to exhibit a much lower degree of regional specialization than in primates, an observation that explains a categorization of latero-posterior visual and auditory territories as polymodal components of the posterior parietal cortex (49, 50). Our results also revealed that a dominant cortical gradient spatially shapes the emergence of prevailing patterns of cortical coactivation governing spontaneous fMRI dynamics, further relating the topography of the connectome with the structure and temporal evolution of spontaneous cortical activity (24). The notion of a tight constraining effect of the structural connectome on functional network topography was further corroborated by evidence of largely overlapping functional and structural communities. This finding expands previous investigations of the mouse functional connectome (51, 52), by highlighting a robust structural basis for distributed fMRI networks of the mouse brain such as the DMN and LCN. Such a close spatial overlap, however, does not appear to comprise hub topography, as previous voxel-wise mapping of functional hubs in the mouse only partly recapitulated the rich connectional features reported here (19). This incongruity might reflect the fact that the spontaneous fMRI signal is a neural mass phenomenon, reflecting local and remote contributions that are negligibly constrained by more fine-grained topological features of the structural connectome.
Multiple lines of future research that expand and complement our work can be envisaged. Rich-club mapping was carried out here and in previous studies (8) on the ipsilateral connectome to enable computational tractability. Further testing of rich-club topography using a whole-brain connectome may be warranted to corroborate the validity of these findings. Similarly, future network investigations using the full-resolution, nonsymmetrized connectome from (9) may complement our work by revealing attributes especially sensitive to the microscale properties of the mouse connectome. Last, future differentiations of the excitatory or inhibitory connectional output for each of the mapped network features via cell type–specific tracing could greatly expand the scope of our findings, especially for cerebellar and striatal areas characterized by high density of inhibitory populations.
In summary, here, we provide a precise characterization of the network structure of the mouse connectome, with voxel resolution. Our results reveal a high-resolution structural scaffold linking mesoscale connectome topography to its macroscale functional organization and create opportunities for identifying targets of interventions to modulate brain function and its network structure in a physiologically accessible species.
MATERIALS AND METHODS
Construction of the structural connectome
Our work leverages a high-resolution models of the mouse brain connectome (100 μm3) previously released by Knox and colleagues (9). The Knox connectome is based on 428 viral microinjection experiments in C57BL/6J male mice obtained from the Allen Mouse Brain Connectivity Atlas (http://connectivity.brain-map.org/). The connectome data were derived from imaging enhanced green fluorescent protein (eGFP)–labeled axonal projections that were then registered to the Allen Mouse Brain Atlas and aggregated according to a voxel-wise interpolation model (9). All the additional computational steps detailed below were implemented to make this resource computationally tractable and suited to the topological analyses we described in our manuscript. Before constructing the SC matrix, we ensured symmetry along the right-left axis for all the major macrostructures of the mouse brain. This step was required because the computation of most graph-based metrics requires the use of a square connectivity matrix as input. To this purpose, we flipped each macrostructure (isocortex, hippocampal formation, subcortical plate, pallidum, striatum, pons, medulla, midbrain, thalamus, hypothalamus, cerebellum, and olfactory bulb) along the sagittal midline (once for the right hemisphere and once for the left hemisphere) and we took the intersection with the respective nonflipped macrostructure This procedure resulted in the removal of a set of nonsymmetric voxel (total fraction, 8.6%), the vast majority of which reside in fringe white/gray matter or cerebrospinal fluid/gray matter interfaces. The removal of these nonsymmetric voxels did not substantially affect the network structure of the resampled connectome, as assessed with a spatial correlation analysis between the symmetrized and nonsymmetrized right ipsilateral (i.e., squared) connectome (spatial correlation, Spearman’s rho = 0.98, 0.93, and 0.97 for global, incoming, and outgoing connectivity strength, respectively). We then filtered out fiber tracts and ventricular spaces, and estimated SC using a resampled version of the recently published voxel scale model of the mouse structural connectome (9), to make the original matrix computationally tractable. Resampling of the Knox et al. connectome was carried out by aggregating neighboring voxels according to a Voronoi diagram based on Euclidean distance between neighboring voxels (fig. S8). Voronoi-based resampling allowed us to spatially weight voxels with respect to neighboring areas, preserving the intrinsic architectural foundation of the connectome (9). Moreover, this procedure allowed us to minimize spatial blurring and boundary effects between ontogenically distinct neuroanatomical divisions of the mouse brain, or white/gray matter, and parenchymal/ventricular interfaces. Last, by averaging the connectivity profile of neighboring voxels based on their relative spatial arrangement, this strategy has also the advantage of mitigating limitations related to the enforced smoothness of source space used by the original kernel interpolation used by (9).
The used Voronoi-based aggregation strategy entails the identification, for each voxel of the mouse connectome, of its 27 closest neighbors as per Euclidean distance, and the subsequent averaging of their connectivity profiles into a single value. We also made sure not to include the same 27 voxels in the computation of a new down-sampled voxel, to avoid spatial redundancy and oversampling. As a trade-off between spatial resolution and computational tractability, we decided to reduce spatial resolution by a factor of 3, therefore aggregating the connectivity profile of 27 voxels into a single one. To keep the estimation of SC consistent with the procedure of (9) and minimize mixing or cross-regional anatomical features, a Voronoi diagram was computed for each of the 12 same major brain divisions of the Allen Institute atlas separately and for one hemisphere only, flipping the resulting diagram across the sagittal midline to ensure Voronoi grid symmetry across the two hemispheres. Given that connectome resampling was carried out on the original 100-μm resolution connectome from (9), the resulting averaged Voronoi voxels are characterized by a total volume of 0.027 mm3 but are not necessarily isotropic or regular. The average spatial extension of the obtained Voronoi voxels in each plane corresponds to 242 μm × 323 μm × 336 μm in the x (sagittal), y (horizontal), and z (coronal) planes, respectively.
To probe whether the used Voronoi resampling procedure (and the resulting anisotropic Voronoi voxels) would affect the connectional and spatial properties of the mouse connectome, we computed, for each Voronoi voxel, the average spatial correlation between the connectivity profile of each of its 27 original constituting 100-μm3 voxels across 12 macrostructures. This computation yielded an average Spearman rho correlation of 0.98, 0.96, 0.99, 0.99, 0.99, 0.99, 0.99, 0.998, 0.97, 0.98, 0.98, and 0.99 for the cerebellum, subcortical plate, hippocampal formation, isocortex, medulla, midbrain, olfactory areas, pallidum, pons, striatum, and thalamus, respectively, corroborating the specificity of the used resampling strategy and suggesting that the resampled connectome offers a reliable fine-grained representation of the most salient connectional features of the original Knox et al. connectome.
A whole-brain connectome was then built under the assumption of brain symmetry (5). Forty-four dangling nodes (i.e., nodes with no outgoing connectivity) were next removed from the resulting matrix, resulting in a final weighted and directed 15,314 × 15,314 matrix composed of 0.027-mm3 aggregate Voronoi voxels. The obtained Voronoi diagram allowed us to map the results back into the original 100-μm three-dimensional coordinate system of the Allen Institute mouse brain connectome [CCFv3 (53)].
Both the original voxel-wise connectome from (9) and our resampled version were almost 100% dense, raising the issue of how to account for and remove weak or irrelevant connections. To address this problem, we used a recently developed method based on graph percolation (54). Briefly, this procedure consists of iteratively removing the weakest connections until the giant component of the graph starts breaking apart. The threshold that maximizes sparsity without breaking the giant component (i.e., the largest integral graph within the matrix) is the one that should be selected for the analysis. Bordier et al. (54) showed that the threshold obtained through percolation analysis maximizes information on the network community structure. This sparsification procedure resulted in a network density of 22%. A comparison of multiple topological attributes and metrics (i.e., global, sink, source hubs, in/out ratio, modules, and gradients) at different thresholds (percolation threshold, 30 and 40%, respectively) revealed that all the probed parameters are highly robust to thresholding (Dice coefficient > 0.97 for all modules and hubs at all thresholds; Spearman rank correlation of >0.99 for out/in ratio and >0.97 for all gradients, at all thresholds).
Regional quantifications of network properties and correlations between structural and functional attributes were carried out using three main sets of predefined anatomical parcellations of the mouse connectome. To quantify the subregional localization of network attributes (fig. S1), we used one of the finest parcellation available of the mouse connectome [i.e., the lowest hierarchical level in the Allen Mouse Brain Atlas, excluding layer encoding; (53)]. This parcellation was volumetrically matched to the sampling dimension of our voxels by discarding small nuclei whose spatial extension was—for either hemisphere—lower than the resolution of our voxel-wise connectome (45 of 323 regions; table S1). Regional quantifications of subregional localizations were then limited to the remaining set of 278 areas. Correlation between FC and SC was carried out on a set of meta-regions to reduce spatial resolution and maximize the contrast with corresponding correlations at the voxel level. The list of the 89 regions used for these comparisons is reported in table S2. Meta-regions were selected to cover the anatomical distribution of the functional modules described in (19). Regional quantification of structural gradient features and cortical hierarchy were carried out using the original cortical parcellation described in (7), corresponding to the isocortical subset in table S1. Last, to probe the robustness and resolution dependence of the topological properties we described in the present work, we recomputed all the network attributes and metrics using the state-of-the-art parcellated mouse connectome described in (6) (426 × 426 connectivity matrix).
Hub and rich-club mapping
Normalized out-strength (source), in-strength (sink), and in + out strength (global) hub regions of the voxel-wise connectome were computed at the percolation threshold. To map the anatomical extension of hub-like areas and obtain a “hubness” index for individual voxels, for each metric, we first iteratively identified and labeled the highest-ranking voxels at increasing percentile threshold (50th to 99th). The obtained information was then combined into a single frequency (%) map by plotting the number of times a voxel was labeled as hub across varying percentile thresholds. We limited the visualization to the nodes that were classified as hubs at least 90% of the time, with the aim to capture top strength nodes and produce heatmaps where hotter colors indicate hub-like voxels corresponding to highest percentile ranking. This approach ultimately led to the final representation of nodes exceeding the 94% strength percentile for all hub categories. No correlation between global connectivity strength (total, incoming, outgoing) and injection density or distance inferred from Knox et al. was observed (P > 0.6, r < 0.17, all six correlation pairs), arguing against a confounding contribution of regional injection inhomogeneity in our subregional hub mapping.
The network core or “rich club” of the mouse connectome was mapped using with the weighted variant described in (8), limiting the analysis to the weighted ipsilateral connectome to ensure the computational tractability of the corresponding null models (8). Specifically, we first obtained a percolation threshold specific for the ipsilateral connectome and we then computed the normalized rich-club coefficient, defined as the ratio between the empirical rich-club coefficient and the rich-club coefficient obtained from an ensemble of 1000 rewired networks where each network maintained the empirical in and out degree, together with the total wiring length of each node [as assessed by Euclidean distance; (10, 55)]. Because of the high computational demands of the rewiring procedure, we left a margin of 5% error on nodal wiring length constraint. Instead of testing all possible degree configurations, which usually range from 1 to k, with k being the highest degree found in the network, we restricted the mapping between 6720 and 8143, corresponding to the 90th and 99th percentile, respectively, of the total degree distribution. This choice was motivated to both reduce the influence of low degree nodes, unlikely to represent hubs of the network, and to reduce the computational demands associated with rich-club mapping with our high-resolution matrix. Statistical significance (P < 0.05) was assessed by obtaining a P value directly from this null distribution. Across all normalized rich-club coefficients, we next computed for each node the fraction of times it was included in the rich club to produce a frequency map, similarly to the procedure described for the definition of source, sinks, and global hubs. Given the more restricted percentile range explored for the rich club with respect to global hubs, the rich club (Fig. 1B) was mapped over a wider frequency range (0.2 to 1) than the other metrics. We observed that all rich-club coefficients tested in the abovementioned range yielded statistically significant results (P < 0.001).
Multiscale modular decomposition and participation coefficient
We analyzed the network structure of the weighted and directed mouse structural connectome using the Louvain algorithm as implemented in the Brain Connectivity Toolbox (32). Similarly to the procedure outlined in (5), we systematically varied (from 0.3 to 3.0 in 0.1 step, 100 repetitions at each step) the resolution parameters controlling the size of the modules, performing consensus clustering (56) and thus obtaining a representative community subdivision for each of the tested resolution setting. As in (5), we next sought to identify a range of gamma yielding topographically stable partitions (fig. S4). To this purpose, we computed adjusted mutual information to assess the spatial similarity between the modular partitions obtained at different gamma values, producing a γ × γ matrix.
We next computed, for increasing mutual information thresholds (ranging from 0.9 to 1.0 to 0.005 step), a modular partition of the corresponding γ × γ matrix via a consensus clustering (56). The identified modules in these γ × γ matrices define a discrete γ interval within which modular partitions of the connectome are topographically comparable. We finally obtained a single agreement matrix by computing the binarized fraction of times each pair of nodes (i.e., gamma values) would be classified as part of the same module, for each modular partition of the thresholded γ × γ matrices. The resulting final γ × γ matrix (fig. S4) is assumed to provide an optimized representation of the discrete gamma intervals yielding stable modular partitions across spatial resolution hierarchies. Given the focus of this work on brain-wide network organization and its relationship with previous community partitioning of large-scale FC, for all subsequent analyses, we focused on the first stable hierarchy level (0.6 < Ƴ < 1.1). Using normalized mutual information index [as in (5)] instead of the adjusted mutual information yielded similar results, with the highest hierarchy level being identical across the two measures. Within the chosen 0.6 < Ƴ < 1.1 interval, we selected γ = 1 as representative resolution parameter, and at the selected spatial scale, we run 500 independent iterations of the Louvain algorithm, followed again by consensus clustering. A computation of the Dice coefficient for all the structural community across all the 0.6 < Ƴ < 1.1 interval (0.1 step) produced mean values of 0.98, 0.98, 0.98, 0.93, and 0.94 for the cerebellar-pontine, basal-olfactory, hippocampal, default-mode, and latero-cortical modules, respectively, supporting the validity of our gamma selection, and corroborating the notion of a stable partition topography within the selected 0.6 < γ < 1.1 range. We finally probed the statistical significance of the final partition against 1000 randomly rewired networks characterized by the same empirical in- and out-degree distribution, and by maintaining the total wiring length of each node (8, 55). Specifically, we used the total connectivity strength within each module as significant variable, reasoning that the internal cohesion of a given partition should be higher than expected by chance. We found that the total connectivity strength of each module always exceeded the total connectivity strength of the 1000 rewired networks, suggesting that the degree sequences and the total wiring length of each node cannot adequately account for the spatial organization of the communities of the mouse structural connectome.
Module topography in the structural connectome was further corroborated using an agglomerative hierarchical clustering procedure of a matrix obtained by computing between-nodes similarity (as by Spearman rank correlation) based on the connectivity profile of each node. A comparison of the obtained clusters using the Dice coefficient revealed an overall high concordance between the results obtained with these two procedures. We found a Dice coefficient of 0.7 for the DMN, 0.82 for the LCN, 0.91 for the hippocampal module, and 0.92 for the olfactory-basal forebrain community. Last, we found that the pontine-cerebellar module was almost equally represented by two clusters, one encompassing the cerebellum and the other covering pons and medulla (Dice coefficient of 0.66 and 0.56, respectively).
Our module detection procedure led to the identification of N = 7 modules, including two symmetric monohemispheric DMN and two olfactory-basal forebrain components, which we have joined into a single module for consistency with functional mapping and before computing their significance. The functional (rsfMRI) modules described in Fig. 2 were obtained from (19). The procedure for functional module detection has been extensively described in the original work (19). To better match SC and FC modules, the basal forebrain and ventral midbrain modules identified in (19) were merged together to constitute a single ventral brain community. Last, to map the anatomical extension of global (in + out), in-connector, and out-connector hubs, we carried out a voxel-wise computation of participation coefficient [i.e., a network measure of connection diversity; (57)] and iteratively identified and labeled the highest-ranking voxels at increasing percentile threshold (50th to 99th). The obtained information was then combined into a single frequency (%) map by plotting the number of times a voxel was labeled as hub across varying percentile thresholds. We limited the visualization to the nodes that were classified as hubs at least 90% of the time.
Virtual lesion mapping
The role of hubs for the network global functioning was probed by means of targeted virtual attacks. For each of the metrics of interest (in and out strength, and global participation coefficient), we removed a given fraction of the highest-ranking nodes (from 5 to 40%, in 5% step by zeroing all the incoming and outgoing connections), comparing the size of the giant component, global efficiency (measured as the average inverse shortest path length), and total network communicability, here limited to map path length ≤ 3 corresponding to a polysynaptic connectome (34). Metrics were computed before and after attack, and changes with respect to these indices were expressed as a percentage of the intact network’s value. For each fraction of removed nodes, we compared targeted hub deletion to 1000 random attacks, assessing statistical significance (P < 0.05) by obtaining a P value directly from the null distribution. To limit inferences of virtual lesions to a nodal range that is biologically meaningful, we restricted the illustration of in silico lesions to the 5 to 20% range (Fig. 4).
Functional and structural gradients
Gradient computations were explicitly aimed at probing the presence of evolutionarily relevant mouse cortical topographies capturing the polymodal-unimodal and modality-specific organization of cortical connectivity previously reported for the human and primate brain (18, 58). To this aim, we applied diffusion map embedding on SC and FC as previously described (18, 37). Briefly, this nonlinear dimensionality reduction technique seeks to project high dimensional connectivity data into a lower dimensional Euclidean space, identifying spatial gradients in connectivity patterns. The cortical SC (FC) matrix is first mapped into an affinity matrix that represent the similarity of connectivity profiles across nodes. The eigenvectors describing the diffusion operator formed on the normalized graph Laplacian of the affinity matrix identify gradients in connectivity patterns over space.
To compute SC gradients, we first extracted from the nonthresholded whole-brain connectome the nodes belonging to the isocortex, and we next computed a new threshold via percolation analysis resulting in a density of 7%. The structural affinity matrix was then built on the basis of the connectional profile of each node, i.e., by incorporating the information provided by both incoming and outgoing connections. The functional affinity matrix was built using the same steps described by (18). In reporting the results, we explicitly looked for gradients capturing polymodal-unimodal sensory-fugal differentiation as well as a modality-specific organization of cortical connectivity as described in recent human and primate work (18, 58). To this purpose, we first ranked SC and FC gradients based on explained variance (fig. S7). We next visually inspected the top three ranking SC and FC gradients and found that the SC and FC gradient #1 were characterized by a clear unimodal-polymodal differentiation (gradient A in Fig. 5, A and B), whereas the second-ranked functional and the third-ranked structural gradients delineated a comparable modality-specific spatial configuration of cortical connectivity (gradient B). In keeping with this observation, these pairs of FC and SC gradients exhibited highly concordant topographies (fig. S7; Spearman’s rho = 0.83 and 0.78, respectively).
We additionally computed the correlation between SC gradients spatial maps and a dominant rsfMRI CAPs published by (24), in an attempt to establish a link between the organization of the structural connectome and FC dynamics. In their work, Gutierrez-Barragan et al. (24) described three pairs of recurring oscillatory states that account for more than 60% of rsfMRI variance. Notably, two of these oscillating patterns are characterized by a conserved cortical topography entailing the opposing engagement of latero-cortical and DMN regions reminiscent of the mapped cortical gradients, the main difference between them being a differential involvement of subcortical structures (i.e., hippocampus). To correlate the topography of these dominant CAPs with that of the structural gradients, we therefore generated a mean cortical CAP out of these two fluctuating states, using the mean value across the hemispheres. We did not consider the third pair of states [CAPs 3 and 4 in (24)] owing to its more widespread cortical topography and strong coherence with fMRI global signal, implicating the involvement of a possible global external input to the emergence of this meta-state. Last, we also computed the correlation between SC gradients and cortical hierarchy scores computed on the basis of feedforward-feedback laminar connectivity patterns of the mouse brain as described and computed in (7), using the same set of cortical brain regions described by the authors. For all the spatial correlational analyses involving gradients, we accounted for the spatial autocorrelation using Moran spectral randomization as implemented in the BrainSpace toolbox, using Euclidean distance between nodes as input for computing the Moran eigenvector maps (37).
rsfMRI data
The rsfMRI dataset used in this work consists of N = 15 scans in adult male C57BL/6J mice that are publicly available (22, 24). All in vivo experiments were conducted in accordance with the Italian law (DL 2006/2014, EU 63/2010, Ministero della Sanità, Roma) and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animal research protocols were reviewed and consented by the animal care committee of the Italian Institute of Technology and Italian Ministry of Health. Animal preparation, image data acquisition, and image data preprocessing for rsfMRI data have been recently described in greater detail elsewhere (19, 24, 59). Briefly, rsfMRI data were acquired with a 7.0-T scanner (Bruker BioSpin, Ettlingen) equipped with BGA-9 gradient set, using a 72-mm birdcage transmit coil, and a four-channel solenoid coil for signal reception. Single-shot BOLD echo planar imaging time series were acquired using an echo planar imaging sequence with the following parameters: repetition time/echo time, 1000/15 ms; flip angle, 30°; matrix, 100 ×100; field of view, 2 × 2 cm2; 18 coronal slices; slice thickness, 0.50 mm; 500 (n = 21) or 1500 (n = 19) volumes; and a total rsfMRI acquisition time of 30 min.
Acknowledgments
Funding: This work has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (#DISCONN; no. 802371 to A.G.). A.G. also acknowledges funding by the Simons Foundation (SFARI 400101, A.G.), the Brain and Behavior Foundation (2017 NARSAD, Independent Investigator Grant 25861), the NIH (1R21MH116473-01A1), and the Telethon Foundation (GGP19177). M.P. is supported by European Union’s Horizon 2020 research and innovation program (Marie Sklodowska-Curie Global Fellowship - CANSAS, GA845065). This work was also supported, in part, by NIH grant R01AG047589 to J.A.H. Author contributions: A.G. conceived the project and supervised research. L.C. designed and carried out the computational analyses, with input from M.P. and B.B. J.D.W. and J.A.H. provided input on the use and interpretation of the voxel-scale anatomical connectivity data. A.G. and L.C. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Correspondence and material request should be addressed to alessandro.gozzi@iit.it. All graph theoretical analyses were performed using the Brain Connectivity Toolbox (32). All the other analyses were carried out using custom Matlab/Python scripts, FSL, and ANTS, unless otherwise stated. The Voronoi resampled structural connectome and all functional and structural maps are available for download at http://dx.doi.org/10.17632/dxtzpvv83k.2. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/51/eabb7187/DC1
REFERENCES AND NOTES
- 1.van den Heuvel M. P., Sporns O., A cross-disorder connectome landscape of brain dysconnectivity. Nat. Rev. Neurosci. 20, 435–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Heuvel M. P., Sporns O., Network hubs in the human brain. Trends Cogn. Sci. 17, 683–696 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Wang X., Lin Q., Xia M., He Y., Differentially categorized structural brain hubs are involved in different microstructural, functional, and cognitive characteristics and contribute to individual identification. Hum. Brain Mapp. 39, 1647–1663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Towlson E. K., Vértes P. E., Ahnert S. E., Schafer W. R., Bullmore E. T., The rich club of the C. elegans neuronal connectome. J. Neurosci. 33, 6380–6387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinov M., Ypma R. J. F., Watson C., Bullmore E. T., Wiring cost and topological participation of the mouse brain connectome. Proc. Natl. Acad. Sci. U.S.A. 112, 10032–10037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh S. W., Harris J. A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A. M., Mortrud M. T., Ouellette B., Nguyen T. N., Sorensen S. A., Slaughterbeck C. R., Wakeman W., Li Y., Feng D., Ho A., Nicholas E., Hirokawa K. E., Bohn P., Joines K. M., Peng H., Hawrylycz M. J., Phillips J. W., Hohmann J. G., Wohnoutka P., Gerfen C. R., Koch C., Bernard A., Dang C., Jones A. R., Zeng H., A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris J. A., Mihalas S., Hirokawa K. E., Whitesell J. D., Choi H., Bernard A., Bohn P., Caldejon S., Casal L., Cho A., Feiner A., Feng D., Gaudreault N., Gerfen C. R., Graddis N., Groblewski P. A., Henry A. M., Ho A., Howard R., Knox J. E., Kuan L., Kuang X., Lecoq J., Lesnar P., Li Y., Luviano J., Conoughey S. M., Mortrud M. T., Naeemi M., Ng L., Oh S. W., Ouellette B., Shen E., Sorensen S. A., Wakeman W., Wang Q., Wang Y., Williford A., Phillips J. W., Jones A. R., Koch C., Zeng H., Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulcher B. D., Fornito A., A transcriptional signature of hub connectivity in the mouse connectome. Proc. Natl. Acad. Sci. U.S.A. 113, 1435–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knox J. E., Harris K. D., Graddis N., Whitesell J. D., Zeng H., Harris J. A., Shea-Brown E., Mihalas S., High-resolution data-driven model of the mouse connectome. Netw. Neurosci. 3, 217–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslov S., Sneppen K., Specificity and stability in topology of protein networks. Science 296, 910–913 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Zingg B., Hintiryan H., Gou L., Song M. Y., Bay M., Bienkowski M. S., Foster N. N., Yamashita S., Bowman I., Toga A. W., Dong H.-W., Neural networks of the mouse neocortex. Cell 156, 1096–1111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vertes R. P., Differential projections of the infralimbic and prelimbic cortex in the rat. Synapse 51, 32–58 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Kale P., Zalesky A., Gollo L. L., Estimating the impact of structural directionality: How reliable are undirected connectomes? Netw. Neurosci. 2, 259–284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Heuvel M. P., Scholtens L. H., de Reus M. A., Topological organization of connectivity strength in the rat connectome. Brain Struct. Funct. 221, 1719–1736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubinov M., Constraints and spandrels of interareal connectomes. Nat. Commun. 7, 13812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betzel R. F., Bassett D. S., Multi-scale brain networks. Neuroimage 160, 73–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckner R. L., DiNicola L. M., The brain’s default network: Updated anatomy, physiology and evolving insights. Nat. Rev. Neurosci. 20, 593–608 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Margulies D. S., Ghosh S. S., Goulas A., Falkiewicz M., Huntenburg J. M., Langs G., Bezgin G., Eickhoff S. B., Castellanos F. X., Petrides M., Jefferies E., Smallwood J., Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc. Natl. Acad. Sci. U.S.A. 113, 12574–12579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liska A., Galbusera A., Schwarz A. J., Gozzi A., Functional connectivity hubs of the mouse brain. Neuroimage 115, 281–291 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Sforazzini F., Schwarz A. J., Galbusera A., Bifone A., Gozzi A., Distributed BOLD and CBV-weighted resting-state networks in the mouse brain. Neuroimage 87, 403–415 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Gozzi A., Schwarz A. J., Large-scale functional connectivity networks in the rodent brain. Neuroimage 127, 496–509 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Grandjean J., Canella C., Anckaerts C., Ayrancı G., Bougacha S., Bienert T., Buehlmann D., Coletta L., Gallino D., Gass N., Garin C. M., Nadkarni N. A., Hübner N. S., Karatas M., Komaki Y., Kreitz S., Mandino F., Mechling A. E., Sato C., Sauer K., Shah D., Strobelt S., Takata N., Wank I., Wu T., Yahata N., Yeow L. Y., Yee Y., Aoki I., Chakravarty M. M., Chang W.-T., Dhenain M., Elverfeldt D., Harsan L.-A., Hess A., Jiang T., Keliris G. A., Lerch J. P., Meyer-Lindenberg A., Okano H., Rudin M., Sartorius A., Van der Linden A., Verhoye M., Weber-Fahr W., Wenderoth N., Zerbi V., Gozzi A., Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis. Neuroimage 205, 116278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hori Y., Schaeffer D. J., Gilbert K. M., Hayrynen L. K., Cléry J. C., Gati J. S., Menon R. S., Everling S., Comparison of resting-state functional connectivity in marmosets with tracer-based cellular connectivity. Neuroimage 204, 116241 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez-Barragan D., Basson M. A., Panzeri S., Gozzi A., Infraslow state fluctuations govern spontaneous fMRI network dynamics. Curr. Biol. 29, 2295–2306.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deco G., Jirsa V. K., Robinson P. A., Breakspear M., Friston K., The dynamic brain: From spiking neurons to neural masses and cortical fields. PLOS Comput. Biol. 4, e1000092 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breakspear M., Dynamic models of large-scale brain activity. Nat. Neurosci. 20, 340–352 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Gratton C., Sun H., Petersen S. E., Control networks and hubs. Psychophysiology 55, e13032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon E. M., Lynch C. J., Gratton C., Laumann T. O., Gilmore A. W., Greene D. J., Ortega M., Nguyen A. L., Schlaggar B. L., Petersen S. E., Dosenbach N. U. F., Nelson S. M., Three distinct sets of connector hubs integrate human brain function. Cell Rep. 24, 1687–1695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Brink R. L., Nieuwenhuis S., Donner T. H., Amplification and suppression of distinct brainwide activity patterns by catecholamines. J. Neurosci. 38, 7476–7491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van den Brink R. L., Pfeffer T., Warren C. M., Murphy P. R., Tona K.-D., van der Wee N. J. A., Giltay E., van Noorden M. S., Rombouts S. A. R. B., Donner T. H., Nieuwenhuis S., Catecholaminergic neuromodulation shapes intrinsic MRI functional connectivity in the human brain. J. Neurosci. 36, 7865–7876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aerts H., Fias W., Caeyenberghs K., Marinazzo D., Brain networks under attack: Robustness properties and the impact of lesions. Brain 139, 3063–3083 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Rubinov M., Sporns O., Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Henriksen S., Pang R., Wronkiewicz M., A simple generative model of the mouse mesoscale connectome. eLife 5, e12366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benzi M., Klymko C., Total communicability as a centrality measure. J. Complex Netw. 1, 124–149 (2013). [Google Scholar]
- 35.Avena-Koenigsberger A., Misic B., Sporns O., Communication dynamics in complex brain networks. Nat. Rev. Neurosci. 19, 17–33 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Mesulam M. M., From sensation to cognition. Brain 121, 1013–1052 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Vos de Wael R., Benkarim O., Paquola C., Lariviere S., Royer J., Tavakol S., Xu T., Hong S.-J., Valk S. L., Misic B., Milham M. P., Margulies D. S., Smallwood J., Bernhardt B. C., BrainSpace: A toolbox for the analysis of macroscale gradients in neuroimaging and connectomics datasets. Commun. Biol. 3, 103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burt J. B., Demirtaş M., Eckner W. J., Navejar N. M., Ji J. L., Martin W. J., Bernacchia A., Anticevic A., Murray J. D., Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat. Neurosci. 21, 1251–1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yousefi B., Shin J., Schumacher E. H., Keilholz S. D., Quasi-periodic patterns of intrinsic brain activity in individuals and their relationship to global signal. Neuroimage 167, 297–308 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunst M., Laurell E., Mokayes N., Kramer A., Kubo F., Fernandes A. M., Förster D., Dal Maschio M., Baier H., A cellular-resolution atlas of the larval zebrafish brain. Neuron 103, 21–38.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Vetere G., Kenney J. W., Tran L. M., Xia F., Steadman P. E., Parkinson J., Josselyn S. A., Frankland P. W., Chemogenetic interrogation of a brain-wide fear memory network in mice. Neuron 94, 363–374.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 42.G. Baldassarre, D. Caligiore, F. Mannella, in Computational and Robotic Models of the Hierarchical Organization of Behavior, G. Baldassarre, M. Mirolli, Eds. (Springer Berlin Heidelberg, 2013), pp. 237–270. [Google Scholar]
- 43.Giorgi A., Migliarini S., Galbusera A., Maddaloni G., Mereu M., Margiani G., Gritti M., Landi S., Trovato F., Bertozzi S. M., Armirotti A., Ratto G. M., De Luca M. A., Tonini R., Gozzi A., Pasqualetti M., Brain-wide mapping of endogenous serotonergic transmission via chemogenetic fMRI. Cell Rep. 21, 910–918 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Weinshenker D., Long road to ruin: Noradrenergic dysfunction in neurodegenerative disease. Trends Neurosci. 41, 211–223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crossley N. A., Mechelli A., Scott J., Carletti F., Fox P. T., Mcguire P., Bullmore E. T., The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137, 2382–2395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alstott J., Breakspear M., Hagmann P., Cammoun L., Sporns O., Modeling the impact of lesions in the human brain. PLOS Comput. Biol. 5, e1000408 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mota B., Dos Santos S. E., Ventura-Antunes L., Jardim-Messeder D., Neves K., Kazu R. S., Noctor S., Lambert K., Bertelsen M. F., Manger P. R., Sherwood C. C., Kaas J. H., Herculano-Houzel S., White matter volume and white/gray matter ratio in mammalian species as a consequence of the universal scaling of cortical folding. Proc. Natl. Acad. Sci. U.S.A. 116, 15253–15261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fulcher B. D., Murray J. D., Zerbi V., Wang X.-J., Multimodal gradients across mouse cortex. Proc. Natl. Acad. Sci. U.S.A. 116, 4689–4695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iurilli G., Ghezzi D., Olcese U., Lassi G., Nazzaro C., Tonini R., Tucci V., Benfenati F., Medini P., Sound-driven synaptic inhibition in primary visual cortex. Neuron 73, 814–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meijer G. T., Mertens P. E. C., Pennartz C. M. A., Olcese U., Lansink C. S., The circuit architecture of cortical multisensory processing: Distinct functions jointly operating within a common anatomical network. Prog. Neurobiol. 174, 1–15 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Grandjean J., Zerbi V., Balsters J. H., Wenderoth N., Rudin M., Structural basis of large-scale functional connectivity in the mouse. J. Neurosci. 37, 8092–8101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stafford J. M., Jarrett B. R., Miranda-Dominguez O., Mills B. D., Cain N., Mihalas S., Lahvis G. P., Lattal K. M., Mitchell S. H., David S. V., Fryer J. D., Nigg J. T., Fair D. A., Large-scale topology and the default mode network in the mouse connectome. Proc. Natl. Acad. Sci. U.S.A. 111, 18745–18750 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q., Ding S.-L., Li Y., Royall J., Feng D., Lesnar P., Graddis N., Naeemi M., Facer B., Ho A., Dolbeare T., Blanchard B., Dee N., Wakeman W., Hirokawa K. E., Szafer A., Sunkin S. M., Oh S. W., Bernard A., Phillips J. W., Hawrylycz M., Koch C., Zeng H., Harris J. A., Ng L., The allen mouse brain common coordinate framework: A 3D reference atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bordier C., Nicolini C., Bifone A., Graph analysis and modularity of brain functional connectivity networks: Searching for the optimal threshold. Front. Neurosci. 11, 441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samu D., Seth A. K., Nowotny T., Influence of wiring cost on the large-scale architecture of human cortical connectivity. PLOS Comput. Biol. 10, e1003557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lancichinetti A., Fortunato S., Consensus clustering in complex networks. Sci. Rep. 2, 336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guimerà R., Nunes Amaral L. A., Functional cartography of complex metabolic networks. Nature 433, 895–900 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oligschläger S., Xu T., Baczkowski B. M., Falkiewicz M., Falchier A., Linn G., Margulies D. S., Gradients of connectivity distance in the cerebral cortex of the macaque monkey. Brain Struct. Funct. 224, 925–935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pagani M., Bertero A., Liska A., Galbusera A., Sabbioni M., Barsotti N., Colenbier N., Marinazzo D., Scattoni M. L., Pasqualetti M., Gozzi A., Deletion of autism risk gene shank3 disrupts prefrontal connectivity. J. Neurosci. 39, 5299–5310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/51/eabb7187/DC1