Abstract
The envelope protein is a primary pathogenic determinant for T-cell-tropic feline leukemia virus (FeLV) variants, the best studied of which is the immunodeficiency-inducing virus, 61C. We have previously demonstrated that T-cell-tropic, cytopathic, and syncytium-inducing viruses evolve in cats infected with a relatively avirulent, transmissible form of FeLV, 61E. The envelope gene of an 81T variant, which encoded scattered single-amino-acid changes throughout the envelope as well as a 4-amino-acid insertion in the C-terminal half of the surface unit (SU) of envelope, was sufficient to confer the T-cell-tropic, cytopathic phenotype (J. L. Rohn, M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh, J. Virol. 72:2686–2696, 1998). In the present study, we examined the role of the 4-amino-acid insertion in determining viral replication and tropism of FeLV-81T. The 4-amino-acid insertion was found to be functionally equivalent to a 6-amino-acid insertion at an identical location in the 61C variant. However, viruses expressing a chimeric 61E/81T SU, containing the insertion together with the N terminus of 61E SU, were found to be replication defective and were impaired in the processing of the envelope precursor into the functional SU and transmembrane (TM) proteins. In approximately 10% of cultured feline T cells (3201) transfected with the 61E/81T envelope chimeras and maintained over time, replication-competent tissue culture-adapted variants were isolated. Compensatory mutations in the SU of the tissue culture-adapted viruses were identified at positions 7 and 375, and each was shown to restore envelope protein processing when combined with the C-terminal 81T insertion. Unexpectedly, these viruses displayed different phenotypes in feline T cells: the virus with a change from glutamine to proline at position 7 acquired a T-cell-tropic, cytopathic phenotype, whereas the virus with a change from valine to leucine at position 375 had slower replication kinetics and caused no cytopathic effects. Given the differences in the replication properties of these viruses, it is noteworthy that the insertion as well as the two single-amino-acid changes all occur outside of predicted FeLV receptor-binding domains.
Retroviruses are notorious for their high degree of genetic variation. As a consequence of this variation, there is considerable flexibility for the virus population to adapt to different selective pressures. For example, particular virus variants may be better able to spread from host to host whereas others are more suited to persist within a chronically infected host. For retroviruses that cause immunodeficiency, pathogenesis is linked to the emergence of T-cell-tropic, cytopathic (T-tropic) viruses, and it is these host-adapted viruses that appear to cause immunosuppression (18, 22). This pattern of evolution and pathogenesis has been observed for lentiviruses such as human immunodeficiency virus and simian immunodeficiency virus as well as for simple oncoretroviruses such as feline leukemia virus (FeLV). Although FeLV infections are frequently associated with neoplastic diseases, in some cases FeLV infection leads to the emergence of T-tropic variants that are highly immunosuppressive (33).
The transmissible form of FeLV is nonacutely pathogenic, whereas the T-tropic viruses are highly pathogenic and induce immunodeficiency (26, 27). The cytopathic and pathogenic properties of these variants are conferred by the extracellular surface unit (SU) of the envelope glycoprotein (13). The SU is generated through proteolytic cleavage of an envelope precursor protein that also includes a signal peptide and the transmembrane (TM) domain. The processed form of the TM protein anchors the SU to the cell membrane. The SU protein determines cell tropism through specific binding with the cognate cellular receptor. Thus, changes in SU may affect pathogenesis, at least in part by altering the host cell specificity of the virus.
We have previously demonstrated that viruses with altered cell tropism and replication properties evolve within FeLV-infected cats (7, 32). In a cat infected with virus derived from a clone of FeLV that is a prototype transmissible virus, 61E, we identified envelope variants that replicated to higher levels in feline T lymphocytes than did their progenitor virus. These envelope variants (called 81T), when expressed in the context of the parental 61E virus, were highly cytopathic for feline T cells in culture and caused syncytium formation (32). Interference studies suggested that the 81T envelope may have acquired a novel receptor specificity relative to the progenitor viral envelope that could account for both its T-cell tropism and its cytopathic properties (32).
The best-studied T-tropic FeLV variant, 61C, was isolated from cats infected with an uncloned natural isolate called FeLV-FAIDS (26). The transmissible form of FeLV, 61E, was obtained simultaneously from this isolate (26). Analyses of a panel of 61E and 61C chimeras suggested that an insertion in the C-terminal half of SU was a key pathogenic determinant and that other single-amino-acid changes could have an enhancing effect on pathogenesis (13). The 81T variants had acquired an insertion at the same sequence location as 61C with respect to the 61E-progenitor virus, but the insertion in 81T was different in size and sequence from the insertion in 61C. Other scattered single-amino-acid changes were also observed in the 81T envelope SU, some of which were also found in 61C. Because viruses expressing 61C or 81T envelope proteins were both T-cell tropic and cytopathic but only the 81T virus was syncytium inducing, we asked what sequence changes conferred this unique phenotype to the 81T envelope. We found that the insertion itself was not sufficient to create a T-tropic, cytopathic, syncytium-inducing virus. In fact, the insertion impaired envelope protein processing in 61E/81T envelope chimeras. Over time, tissue culture-adapted (tca) variants evolved and were selected in some cultures transfected with the chimeric viruses. This allowed us to identify compensatory single-amino-acid changes in both the N terminus and the very C terminus of SU that restored envelope processing. Interestingly, a virus that had both the amino acid change at the N terminus and the insertion in the C-terminal half of SU was T tropic, cytopathic, and syncytium inducing. In contrast, a virus with the insertion paired with the C-terminal mutation exhibited replication properties more typical of the transmissible 61E virus. These data suggest that multiple domains in the envelope may play a role in determining the tropism and cytopathic properties of FeLV variants.
MATERIALS AND METHODS
Cell culture.
AH927 feline embryonic fibroblasts were maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum. Human 293T cells were maintained in Dulbecco's MEM (DMEM) supplemented with 10% fetal bovine serum. The feline T-cell line 3201 was maintained in 50% Leibovitz's L15–50% RPMI 1640 supplemented with 15% fetal bovine serum. All complete media (e.g., cMEM and cDMEM) contained 100 U of penicillin per ml, 100 μg of streptomycin per ml, 0.25 μg of amphotericin B per ml, and 2 mM l-glutamine.
Construction of chimeric and mutant proviral clones.
The 61E and 61C proviruses were derived from an FeLV-FAIDS-infected cat; the EECC prototype FeLV-FAIDS clone was constructed from the 5′ half of 61E (including the 5′ long terminal repeat [5′LTR] gag and pol) and the 3′ half (envelope 3′LTR) of a replication-defective FeLV-FAIDS clone, 61C (26). The EET109E [previously referred to as EET(TE)-109 in reference 32] virus containing all of an 81T envelope gene (81T clone 109), including the SU and TM domains, in a 61E background was described previously (32). Like the 81T-109 envelope clone, the subclones encoding the 81T-3 and 81T-6 envelope were generated from PCR-amplified envelope gene fragments obtained from the tumor of cat 40681; these clones were described previously (31). To construct the EE(ET)E chimeras, a 0.9-kb gel-purified MamI-RsrII fragment encompassing the 3′ half of the envelope gene of 81T-3 and 81T-6, including coding sequences for the C-terminal half of SU and all of TM, was cloned into a similarly digested 3′ subclone of 61E (called 3′EE [28]). Clones with the correct chimeric structure were identified by restriction enzyme digestion and nucleotide sequence analysis. The full-length provirus was constructed from this chimeric 3′ subclone by introducing a 6.5-kb EcoRI-XhoI fragment carrying the 5′LTR gag and pol of 61E into the 3′ subclone as described previously (27). A similar strategy was employed for constructing the EE(CT)C chimeras, except that the 3′CC (26) subclone rather than the 3′EE subclone was used.
The mutant EE(ET6,V→L)E was also constructed using the two-step cloning strategy used for construction of the EE(ET6)E virus. In this case, the MamI-RsrII fragment was gel isolated from a cell culture-derived envelope clone that encoded the desired V → L mutation. The mutant envelope clone was obtained by amplification of envelope gene sequences from genomic DNA from 3201 cells infected with the tca virus EE(ET6)Etca using methods described previously (31).
An A → C single-base mutation predicted to encode a Q → P change at position 7 in the mature SU was engineered into EE(ET3)E by overlap PCR to generate EE(EQ→PT3)E. The single-base change that was introduced into this mutant corresponds to a change at position 6099 (GenBank, FCVF6A [12]). The overlap PCR method used was similar to a method described previously for engineering mutations in the FeLV-B envelope (6). Briefly, two overlapping fragments were generated with the desired mutation. The 5′ fragment encoded the desired change as a result of a T → G mutation engineered into the 3′ primer used to generate that product. The 3′ fragment encoded the corresponding desired mutation at its 5′ end as a result of an A → C change in the 5′ primer used for that PCR. The overlapping products of these reactions were then combined, along with appropriate primers internal to the extreme 5′ and 3′ sequences, in a second round of PCR. A 1.3-kb fragment from this overlap PCR product was isolated and cloned directly into the EE(ET3)E parental virus using unique XhoI (near the end of the pol gene) and MamI (in the middle of the envelope) restriction sites. The presence of the desired mutation was verified by nucleotide sequence analysis. Multiple clones were analyzed to identify a clone for these studies that did not encode any other predicted amino acid changes as a result of errors during PCR amplification.
For analyses of tca envelope variants, envelope fragments encompassing sequences for the 5′ envelope leader through the U3 region of the LTR were cloned from 3201 cell DNA using primers FeLV-Pol5 and FeLV-U32B and methods described previously (31).
Transfection and infection studies.
To examine whether the EE(ET)E and EE(CT)C chimeras could generate replication-competent virus, the four proviral clones were transfected into the feline T-cell line 3201. The 61E, EECC, and EET109E clones were transfected in parallel, in all cases using electroporation. The production and spread of virus in the culture was monitored by an enzyme-linked immunosorbent assay that detects p27gag in the supernatant (ViraCHECK; Synbiotics). Cell-free virus was harvested when cells were chronically infected, as judged by detection of high levels of p27gag. Cell-free viral supernatants from these cells, diluted to each have the same level of reverse transcriptase (RT) activity, were used to infect naive 3201 T cells. To compare replication kinetics, the cells were infected with an equal dose of each virus and reverse transcriptase activity, cytopathic effects, and syncytium formation were monitored as described previously (32).
For experiments involving the mutant viruses EE(ET6,V→L)E and EE(EQ→PT3)E, virus was generated by introducing the proviral clones into 293T cells by calcium phosphate transfection. The transfection was done in duplicate to permit infection studies with two independent virus supernatants. Cell supernatants were collected and filtered after 48 h. Virus levels were normalized to a control virus of known infectious titer using p27gag enzyme-linked immunosorbent assay, and an amount of virus equivalent to a multiplicity of infection of ∼0.05 was used to infect 3201 cells. Infections were performed in duplicate using supernatants from the transfections, and total viable-cell numbers were averaged. Cells were monitored as described previously (32), except that viable cells were diluted to a density of 5 × 105 cells/ml in a final volume of 4 ml. By day 12 postinfection, cells experiencing cytopathic effects were no longer passaged but viable cell counts continued to be recorded.
The FeLV viral chimeras discussed above were also analyzed using single-cycle viral infection assays, similar to those described previously (9). Briefly, virus particles were generated by cotransfecting the FeLV proviral clones with a murine retroviral vector encoding β-galactosidase, pRT43.2Tnlsβ-gal-1 (36), into 293T cells. Cell-free viral supernatants were collected after 48 h and filtered through 0.2-μm-pore-size filters. Serial dilutions of viral supernatants were added to cMEM in total volumes of 1 ml each. Virus was applied to AH927 feline fibroblasts, which were plated in 24-well plates at 2 × 104 cells per well in cMEM the day before infection. At 48 h later, infected cells were detected by adding 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), a substrate for β-galactosidase. Blue nuclei were counted, and, based on the various serial dilutions analyzed, the β-galactosidase FFU per milliliter of viral supernatant were calculated.
RT assays.
For analysis of RT activity, cell-free viral supernatants were harvested at 2- to 3-day intervals during the 4-week infection study and stored at −70°C. Aliquots (10 μl) of supernatant, and serial dilutions thereof, were assayed essentially as described previously (15). Briefly, we measured the ability of lysed virion preparations to catalyze the incorporation of radiolabeled nucleotide [α-32P]dTTP (NEN), using the polyribonucleotide template poly(A)p(dT)10 sodium salt (Sigma) to which an oligodeoxyribonucleotide primer was annealed. The intensity of radioactivity was quantitated by PhosphorImager analysis, and the assigned PhosphorImager units (PIU) were adjusted by subtracting the background radiation on the filter. PIUs for cell-free supernatants from the duplicate 3201 infections were averaged. The same procedure was used to assess the RT activity of the FeLV/β-galactosidase viral vectors that were generated in 293T cells and used for subsequent single-cycle infection studies.
Metabolic labeling and RIPA of cellular lysates.
Radioimmunoprecipitation analysis (RIPA) was performed with minor modification of a method described previously (8, 30). Cells were pulse-labeled for 2 h in methionine- and cysteine-deficient DMEM supplemented with 100 μCi of [35S]methionine-[35S]cysteine per ml (Trans-Label; ICN). After being labeled, the cells were chased for 3 h in cDMEM. The cells were then washed twice in cold phosphate-buffered saline and lysed in cold lysis buffer (25 mM Tris [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride [37]). Lysates were clarified by centrifugation at 15,000 × g at 4°C for 15 min followed by preclearing with protein A-Sepharose beads (Sigma) overnight with rocking at 4°C. Incorporated radioactivity was determined by precipitation with trichloroacetic acid. An equal number of trichloroacetic acid-precipitable counts were added to each RIPA reaction mixture.
Anti-SU monoclonal antibody C11D8 (16) and anti-TM monoclonal antibody PF6J-2A were obtained from Custom Monoclonals, Sacramento, Calif. Antibody was premixed with protein A-Sepharose beads for 1 h at 4°C. The beads were then washed three times in wash buffer (50 mM Tris [pH 7.2], 150 mM NaCl, 0.1% SDS, 0.1% Triton X-100). Cell lysates were added to antibody-protein A beads and incubated with rocking for 3 h at 4°C. The RIPA reaction mixtures were then washed five times with 1 ml of cold wash buffer on ice and resuspended in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. The eluted proteins were analyzed by SDS-PAGE (19) and fluorography.
RESULTS
Determinants of T-cell tropism, syncytium formation, and cytopathic effects.
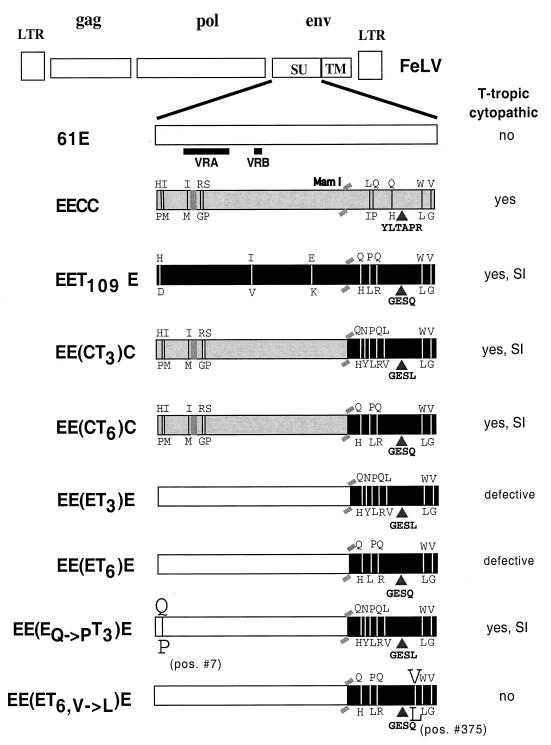
The 81T envelope gene variants were cloned from a thymic tumor of a 61E-infected cat (40681). Although more than 90% of the envelope clones derived from the tumor encoded a predicted 4-amino-acid insertion, none of these clones were identical in sequence (31, 32). For example, we detected viruses that differed at one amino acid position within the 4-amino-acid insertion and viruses that differed at the very N terminus of the mature envelope SU. We chose envelope clones encoding the two different insertions that were otherwise representative of the consensus sequence of the 81T-tumor derived clones (81T-3 [GESL] and 81T-6 [GESQ]) (Fig. 1) (31) to generate the chimeric proviral constructs for this study. For reference, a schematic of the 81T envelope variant (EET109E) that was recently shown to confer a cytopathic, syncytium-inducing phenotype when expressed in a 61E proviral context is shown (32). The FAIDS variants EECC and the progenitor virus for the 81T variants, 61E, are also included in Fig. 1.
FIG. 1.
Schematic of the structures of SU encoded by various FeLV viral chimeras. The open boxes denote regions derived from 61E, the shaded boxes denote regions derived from 61C, and the black boxes denote regions derived from 81T. Amino acid differences are shown by a line, and the amino acid found at that position in 61E is shown above the box; the corresponding amino acid found in the indicated variant is shown below the box. The position of the line approximates where the amino acid difference occurs in the linear sequence of SU. The sequence of the 61C or 81T insertion is shown below the triangle that indicates where the insertion occurs relative to 61E. The small shaded rectangle placed at the N terminus of EECC, EE(CT3)C, and EE(CT6)C indicates the presence of a 6-amino-acid deletion relative to 61E; in addition, these variants contain TM and 3′ LTR sequences derived from 61C. Relative positions of VRA and VRB are identified by bold black lines below 61E SU. Variants that are syncytium inducing are identified by SI.
To determine whether the 4-amino-acid 81T insertion, which occurs at position 352 in the 61E envelope SU, could functionally substitute for the 6-amino-acid 61C insertion, we designed chimeric proviral clones EE(CT3)C and EE(CT6)C. These constructs encode the C-terminal quarter of the 81T envelope SU, which includes the insertion, in the context of an EECC provirus; EECC carries the envelope and 3′ LTR of 61C and the 5′ LTR, gag, and pol of 61E (Fig. 1). Viruses generated from both constructs were cytopathic in 3201 T cells (data not shown) [see also Fig. 3 for EE(CT3)C]. This result suggests that either of the 4-amino-acid insertions in 81T SU (GESL and GESQ) can substitute as a cytopathic determinant for the 6-amino-acid 61C insertion. While there are two other single-amino-acid differences in the 81T-derived C-terminal portions of EE(CT3)C and EE(CT6)C (N → Y and Q → R [Fig. 1]), these do not affect the cytopathic phenotype of the viruses, further supporting a role for the insertion in determining viral phenotype. These chimeras also induced syncytium formation (data not shown), which is characteristic of 81T but not 61C SU. This suggests that the C-terminal portion of 81T SU includes determinants for syncytium formation. However, this does not exclude the possibility that N-terminal sequences in 61C SU serve as cytopathic determinants.
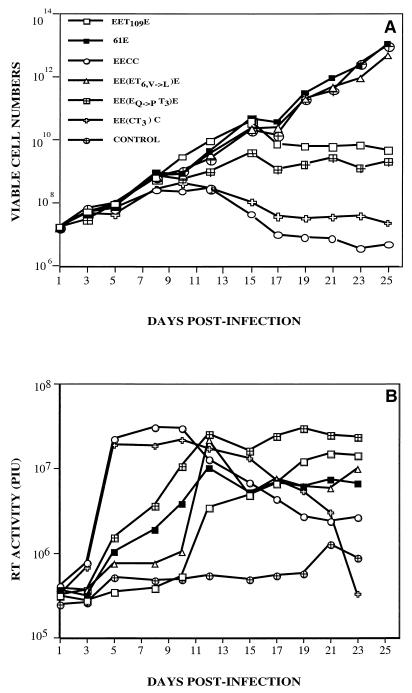
FIG. 3.
Replication kinetics and RT activity following infection of feline 3201 T cells with the viral chimeras EE(EQ→PT3)E and EE(ET6,V→L)E. (A) Viable cell numbers calculated after exclusion of the vital dye trypan blue; values were averaged (log scale). (B) Total RT activity in the culture supernatants per 10 μl, expressed in terms of arbitrarily adjusted PIU; units were averaged (log scale).
Chimeras EE(ET3)E and EE(ET6)E (Fig. 1) were constructed to determine whether the C-terminal portion of 81T SU is sufficient to confer the cytopathic, T-tropic phenotype in the context of a 61E virus. These chimeras were apparently replication defective, based on our inability to detect the production of viral p27gag antigen in the supernatant of 3201 T cells that had been transfected with these clones and subsequently cultured for several weeks to allow virus spread. However, in one of nine transfection experiments for each construct, infectious virus was recovered after several weeks of cell passage. We examined the genotype of the replicating virus in these cultures by amplifying and cloning multiple envelope genes from the DNA of transfected cells and analyzing the nucleotide sequence of these clones. We confirmed that the predominant proviral genotype in these cells was derived from the original transfected chimeras, since all sequences analyzed encoded the predicted 81T insertion (data not shown).
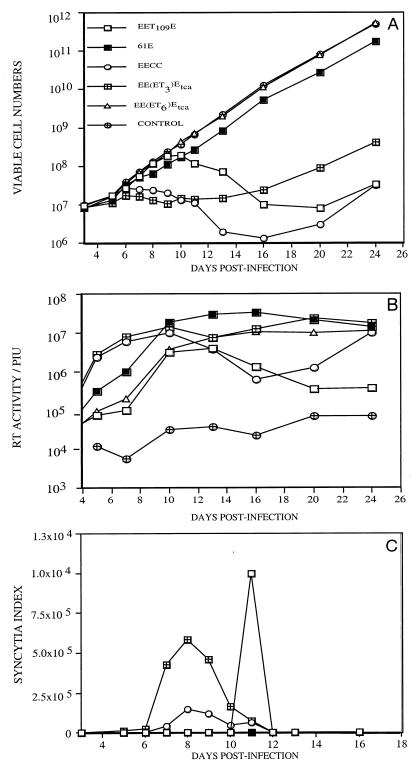
To examine the replication properties of these tca variants, EE(ET3)Etca and EE(ET6)Etca, the virus recovered from the transfected cells was used to infect naive 3201 T cells. The tca virus EE(ET6)Etca replicated with kinetics like that of 61E; replication of this virus did not affect cell viability or lead to syncytium formation (Fig. 2). Intriguingly, the other tca variant, EE(ET3)Etca, was rapidly replicating, cytopathic, and T tropic; this virus was also syncytium inducing (Fig. 2). Thus, tissue culture adaptation had led to the emergence of 81T-derived viruses with different cytopathic properties.
FIG. 2.
Viral replication kinetics and cytopathic effects, RT activity, and syncytium formation after infection of 3201 feline T cells with tca viruses, EE(ET3)tca and EE(ET6)Etca. (A) Viable cell numbers, as assessed by the exclusion of the vital dye trypan blue (log scale). (B) Total RT activity in the culture supernatants, expressed in terms of arbitrary adjusted PIU per microliter (log scale). (C) Syncytium index, which is the quotient of the number of large syncytia visible in one defined field of the microscope divided by the cell density at that time point (linear scale).
Defining the determinants for adaptation in tissue culture.
To examine the molecular basis for the biological differences in the tca viruses, we examined the nucleotide sequence of multiple envelope gene clones derived from each culture, including the region encoding both the SU and the TM domains of the envelope. Interestingly, nucleotide sequence analysis of the envelope gene clones showed that the adapted variants had evolved several amino acid changes, including one that was invariably detected in all clones examined. EE(ET3)Etca evolved a predicted Q → P change at position 7 of the mature envelope SU, near the N terminus in four of four clones examined. This mutation is adjacent to a site where other cytopathic clones such as 61C encode a proline change relative to the noncytopathic clone, 61E (Fig. 1). EE(ET6)Etca evolved a predicted V → L change at position 375 in the mature SU, near the C terminus, and this mutation was found in 11 of 11 envelope clones examined (data not shown). We hypothesized that these changes evolved because they structurally compensate for changes in conformation caused by the insertion.
To test this hypothesis, two chimeras were constructed that incorporated the specific amino acid changes of the culture-adapted viruses into the original parental chimeras from which they evolved. Thus, these constructs encode a chimeric envelope with the C terminus derived from 81T SU and the N terminus derived from 61E, with either an additional N-terminal change [EE(EQ→PT3)E] or a C-terminal change [EE(ET6,V→L)E] (Fig. 1). Unlike the original chimeras from which they evolved in culture, the EE(EQ→PT3)E and EE(ET6,V→L)E mutants replicated in 3201 T cells (Fig. 3). Moreover, each virus exhibited the characteristics of its uncloned tca counterpart, EE(ET3)Etca and EE(ET6)Etca, respectively. The EE(EQ→PT3)E chimera encoding a Q → P change at amino acid 7 was cytopathic and syncytium inducing like EE(ET3)Etca, whereas EE(ET6,V→L)E, which encodes a V → L change at position 375, was noncytopathic like EE(ET6)Etca. Infection of 3201 T cells with EE(ET6,V→L)E did not induce syncytium formation, similar to what has previously been observed with 61E.
To assess the efficiency of virus replication, the RT activity of the cell-free viral supernatants over the time course of infection was determined (Fig. 3). As previously shown, EET109E appeared to replicate and/or spread in infected T cells more slowly than did EECC but approximately at levels similar to those of 61E (32). EE(ET6,V→L)E, while exhibiting similar replication kinetics to 61E, had RT activity that increased more slowly but to a higher peak than that of 61E. In contrast, EE(EQ→PT3)E replicated more rapidly and to higher levels than either EET109E, 61E, or clone EE(ET6,V→L)E.
Analyses of infectivity using a single-cycle replication assay.
We also examined the infectivity of the viruses in this study for feline fibroblast cells using a single-cycle infection assay. For this purpose, FeLV particles containing murine retroviral genomes encoding β-galactosidase were used and infection was monitored by measuring the expression of the β-galactosidase protein. Thus, this assay requires that envelope be able to bind its cognate receptor and initiate entry. To provide a quantitative measure of virus that is independent of infectivity, we examined the RT activity of the viruses (Table 1). Both the EE(EQ→PT3)E and EE(ET6,V→L)E viruses were infectious for AH927 cells in this assay. In this regard, they both resemble 61E. When normalized to RT, the relative infectivity of EE(ET6,V→L)E was approximately sevenfold lower than that of EE(EQ→PT3)E, suggesting that the ratio of infectious to noninfectious virus may be different for these tca variants. Neither of these two mutant clones were able to infect AH927 cells at levels comparable to the prototype FeLV, 61E, but both were able to infect at levels similar to EET109E. Thus, the changes in the tca viruses appear to affect both T-cell tropism and infection in non-T-cell lines. Interestingly, both of the tca variants were more infectious for AH927 cells than either EECC or the 61C/81T envelope chimeras were, suggesting that the N-terminal 61C-derived sequences may negatively affect replication in feline fibroblast cells.
TABLE 1.
Single-cycle infection of AH927 feline fibroblast cells
| Infection | β-Galactosidase level (FFU/ml) | RT activitiy (PIU/μl) | Relative infectivitya (PIU/FFU) |
|---|---|---|---|
| EET109E | 1.70 × 104 | 4.1 × 105 | 0.11 |
| 61E | 9.50 × 105 | 2.5 × 106 | 1 |
| EECC | 48 | 1.7 × 106 | 7.5 × 10−5 |
| EE(ET6, V→L)E | 6.30 × 103 | 1.9 × 106 | 0.009 |
| EE(EQ→PT3)E | 6.70 × 104 | 2.7 × 106 | 0.07 |
| EE(ET3)E | 4 | 1.4 × 106 | 7.6 × 10−6 |
| EE(ET6)E | 2 | 1.4 × 106 | 3.8 × 10−6 |
| EE(CT3)C | 4 | 2.5 × 106 | 4.3 × 10−6 |
| EE(CT6)C | 8 | 1.6 × 106 | 1.3 × 10−5 |
| Mock | 0 | 0b | 0 |
The ratio of PIU to FFU for 61E was assigned a value of 1, and all other ratios were assigned values relative to this normalized value.
Background level of 104 subtracted from other sample values.
We occasionally detected a few cells that expressed the β-galactosidase viral genomic marker upon exposure to the apparently replication-defective chimeras, EE(ET3)E and EE(ET6)E (Table 1). This observation would suggest that low levels of reverse transcription may occur as a result of a very low level of infectious virus expressed from these proviral genomes. The process of reverse transcription would allow the generation of mutations that could have led to the production of the tca variants. Certainly, any replication-competent mutants, when they occurred, would be highly selected. However, because the levels of infectivity that we observed with the EE(ET3)E and EE(ET6)E chimeras were so low, it is difficult to extrapolate from these data to argue that such rare infectious variants were the progenitor viruses for the tca variants.
Envelope processing is affected by the insertion and restored by the N-terminal and C-terminal changes.
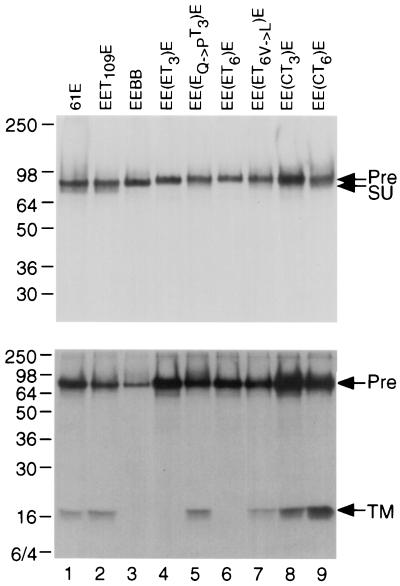
The observation that EE(ET3)E and EE(ET6)E viral chimeras were replication defective suggested that the C-terminal insertions in 81T SU may affect envelope structure or stability in the context of a 61E SU. To examine this possibility, we performed RIPA of cells transfected with these proviral clones. As shown in Fig. 4, the 61E and EET109E gp85 envelope precursor proteins were expressed at similar levels and were processed to SU (gp70) and TM (p15E) (lanes 1 and 2). While the EE(ET3)E and EE(ET6)E chimeras also expressed high levels of gp85 in transfected cells, these envelope precursors were not cleaved to detectable levels (lanes 4 and 6). Indeed, the envelope-processing profile of these chimeras was almost identical to that of the 61B envelope, which we have previously shown to be defective for envelope processing (8) (lane 3). These results indicate that the replication defect in the EE(ET3)E and EE(ET6)E chimeras is due to a defect in envelope processing.
FIG. 4.
Immunoprecipitation of FeLV envelope proteins. 293T cells were transfected with the proviral clones indicated above each lane. At 48 h later, cells were pulse-labeled with [35S]methionine plus [35S]cysteine for 2 h and chased for 3 h in standard media. Molecular mass markers (in kilodaltons) are labeled to the left of the blot, and the unprocessed envelope precursor (Pre), SU, and TM are indicated to the right. (Top) Cellular lysates immunoprecipitated with the envelope antibody C11D8 directed to an epitope in the SU and analyzed by SDS-PAGE (8.5% polyacrylamide). (Bottom) Identical lysates immunoprecipitated with the envelope antibody PF6J-2A directed to an epitope in the TM and analyzed by SDS-PAGE (12% polyacrylamide).
Because the tca variants EE(ET3)Etca and EE(ET6)Etca were replication competent, we predicted that they evolved changes which restored proper processing of the chimeric envelope protein. RIPA of cells transfected with EE(EQ→PT3)E and EE(ET6,V→L)E shows that the single-amino-acid changes evolved in the tca viruses restore proper processing of the envelope protein. This can be most clearly detected with the antibody against the envelope TM protein, although a faint smear characteristic of heavily glycosylated gp70 can also be detected with the anti-SU antibody (Fig. 4, lanes 5 and 7). Our data suggest that other amino acid changes in the 81T and 61C envelopes compensate for structural changes induced by the insertions in these viruses. In support of this hypothesis, we found that EE(CT3)E and EE(CT6)E, which contain the 81T-3 and 81T-6 insertions, respectively, in the context of a 61C envelope, exhibit proper envelope processing (lanes 8 and 9), and these viruses are replication competent.
DISCUSSION
A small, 6-amino-acid insertion in the C-terminal half of the FeLV-FAIDS variant envelope protein was previously shown to be a critical determinant of its highly pathogenic, immunosuppressive phenotype (13, 26). Although this insertion appeared to be the result of an imperfect duplication of adjacent sequences, the precise progenitor of the FeLV-FAIDS variants is unknown (26). In contrast, the progenitor for the 81T variants is known because these envelope sequences were obtained from a cat infected with a virus derived from a molecularly cloned, transmissible form of FeLV, 61E (31). These 81T variants had acquired an insertion of unknown origin in the same position (position 352 in SU [31]) as in the 61C virus. Here we show that viruses encoding the C-terminal portion of SU of 81T, including the 4-amino-acid insertion, and the N-terminal portion of 61C SU are T-cell tropic and cytopathic. Of note, tumor-derived 81T variants encoding two different 4-amino-acid insertions (GESL and GESQ) were equally cytopathic when combined with the C terminus of 61C SU, suggesting that the single-amino-acid difference in the insertion does not affect T-cell tropism and cytopathicity. Moreover, our data suggest that the 4-amino-acid insertion(s) in 81T and the 6-amino-acid insertion in 61C are functionally interchangeable despite their difference in size and sequence. A chimera encoding a segment of the 81T envelope containing the insertion in a background of the 61C envelope [e.g., EE(CT3)C] was also syncytium inducing, which is a unique property of the 81T virus. The observation that 4- or 6-amino-acid insertions with very different sequences can function as determinants of T-cell tropism suggests that the inserted sequences may not be altering replication and cell tropism by a direct interaction with the cell surface receptor. Rather, these data are more consistent with a model in which the insertion has a conformational effect that affects receptor-binding determinants elsewhere on the envelope protein.
The domains of the FeLV envelope that are important for receptor specificity have been defined through analyses of viral envelope chimeras. To date, these studies have focused on subgroup B FeLVs because the receptor for this FeLV was the first to be identified (24, 35). These studies suggest that sequences in the N terminus of FeLV SU are important for receptor recognition (6, 34). This region of the envelope encompasses the variable region A and B (VRA and VRB) domains, which have also been described as receptor-binding domains for the related murine leukemia viruses (MuLVs) (2–5, 10, 11, 17, 21, 23, 25, 29). It is noteworthy that the insertion at position 352 that plays a key role in defining the replication properties of the T-tropic FeLV variants is C terminal to the described receptor recognition domains for FeLV-B and MuLV. These findings suggest that segments of envelope outside of VRA and VRB may also play an important role in determining cell tropism and, by extension, may play a role in determining receptor specificity.
The insertion and scattered nearby mutations in the SU of the 81T envelope impairs envelope protein processing within a background of the parental 61E envelope protein. The emergence of replication-competent tca viruses in cells expressing proviruses encoding chimeric 61E/81T SU proteins allowed us to identify two independent compensatory mutations that restored envelope protein processing, one at the very N terminus of SU and another near the C terminus of SU. Viruses encoding these changes were infectious in feline fibroblast cells, as judged by single-cycle infection assays. Interestingly, 61E/81T chimeric viruses encoding these amino acid changes displayed very distinct phenotypes in feline T cells. A virus with the C-terminal SU change coupled with the 4-amino-acid insertion replicated with similar kinetics to that of the avirulent 61E virus. Thus, the replication properties of this mutant virus suggest that the insertion alone cannot confer the T-tropic, cytopathic phenotype of 81T. We cannot determine from these studies whether differences in replication and T-cell tropism may directly impact the cytopathic properties of the virus. Interestingly, the N-terminal change, when coupled with the insertion, rendered the virus highly cytopathic and syncytium inducing in feline T cells. In addition, this virus was as infectious in feline fibroblast cells as a virus encoding the complete 81T SU (e.g., EET109E). We have previously provided evidence that the 81T variants are dually tropic; they exhibited interference properties that suggest that they use a receptor specific for the T-tropic 61C virus as well the more ubiquitously expressed FeLV-61E receptor, perhaps at reduced efficiency (32). The studies described here suggest that the N-terminal change, which makes the virus both T-cell tropic and able to infect fibroblast cells, may be a key determinant for the proposed dual-receptor specificity of the virus.
The N-terminal proline change that evolved in the tca virus clusters with similar mutations in the original 81T viruses isolated from the cat thymic tumor. Thus, mutations in this region of SU evolved independently in viruses selected for replication in an infected cat and in viruses selected for replication in feline T cells in culture. Moreover, 61C and other FeLV-FAIDS-derived variants also have amino acid differences relative to 61E in this very N-terminal portion of envelope SU. Proline is a particularly common amino acid, occurring at position 6 in the mature SU of several FeLV-FAIDS variants (27) and in some of the 81T variants (31, 32). FeLV-61E encodes a histidine at position 6. However, some of the 81T variants, including the 81T-109 virus previously shown to be T tropic, encode an aspartic acid at this position, suggesting that amino acid changes other than proline may also alter replication and processing in the context of certain envelope sequence. Interestingly, a histidine residue at position 8 of the MuLV SU affects the efficiency of receptor binding and/or postbinding events (1, 20, 38). Together, these data suggest that the very N terminus of the FeLV and MuLV envelopes may play a critical role in defining the replication properties of the virus.
Our studies suggest that sequence determinants of FeLV T-cell tropism, which include the N-terminal change and a C-terminal insertion, lie outside previously defined VRA and VRB receptor domains. Together, these data indicate that conformational changes outside of envelope domains that may not specifically contact the receptor could significantly influence receptor specificity. There have been several studies of envelope receptor interactions lacking N-terminal or C-terminal sequences outside of VRA and VRB (5, 10, 11, 17). Our studies suggest that such analyses, while important for identifying residues directly involved in binding, may not provide a broader view of the envelope-receptor interactions in the context of an infecting virus. Indeed, the crystal structure that has been solved for a fragment of MuLV SU does not include the C-terminal half of SU (14). Thus, it will be of interest to determine how an insertion in that portion of SU could influence envelope structure and subsequent viral tropism. The FeLV variants described here, and particularly the tissue culture-derived viruses exhibiting distinct replication properties, will allow us to further examine the role of sequences outside of VRA and VRB in cell tropism and receptor interactions.
ACKNOWLEDGMENTS
We thank Maribeth Eiden for providing pRT43.2Tnlsβ-gal-1 and Maria Anderson for technical assistance and helpful discussions.
This work was supported by NIH grant CA 51080. F.C.H. is supported in part by NIH training grant 5 T32 RR07019. A.S.L. is supported in part by the Poncin Scholarship Fund.
Footnotes
This paper is dedicated to the memory of our friend and colleague, Samuel Rudolph Gwynn.
REFERENCES
- 1.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini J L, Danos O, Heard J M. Definition of a 14-amino-acid peptide essential for the interaction between the murine leukemia virus amphotropic envelope glycoprotein and its receptor. J Virol. 1998;72:428–435. doi: 10.1128/jvi.72.1.428-435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Danos O, Heard J M. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J Virol. 1995;69:713–719. doi: 10.1128/jvi.69.2.713-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini J L, Rodrigues P, Muller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boomer S, Gasper P, Whalen L R, Overbaugh J. Isolation of a novel subgroup B feline leukemia virus from a cat infected with FeLV-A. Virology. 1994;204:805–810. doi: 10.1006/viro.1994.1597. [DOI] [PubMed] [Google Scholar]
- 8.Burns C C, Poss M L, Thomas E, Overbaugh J. Mutations within a putative cysteine loop of the transmembrane protein of an attenuated immunodeficiency-inducing feline leukemia virus variant inhibit envelope protein processing. J Virol. 1995;69:2126–2132. doi: 10.1128/jvi.69.4.2126-2132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chackerian B, Haigwood N L, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213:386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- 10.Davey R A, Hamson C A, Healey J J, Cunningham J M. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J Virol. 1997;71:8096–8102. doi: 10.1128/jvi.71.11.8096-8102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey R A, Zuo Y, Cunningham J M. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donahue P R, Hoover E A, Beltz G A, Riedel N, Hirsch V M, Overbaugh J, Mullins J I. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J Virol. 1988;62:722–731. doi: 10.1128/jvi.62.3.722-731.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M C, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 15.Goff S, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant C K, Ernisse B J, Jarrett O, Jones F R. Feline leukemia virus envelope gp70 of subgroups B and C defined by monoclonal antibodies with cytotoxic and neutralizing functions. J Immunol. 1983;131:3042–3048. [PubMed] [Google Scholar]
- 17.Heard J M, Danos O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J Virol. 1991;65:4026–4032. doi: 10.1128/jvi.65.8.4026-4032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimata J T, Kuller L, Anderson D B, Dailey P, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–541. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lavillette D, Ruggieri A, Russell S J, Cosset F L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miedema F, Meyaard L, Koot M, Klein M R, Roos M L, Groenink M, Fouchier R A M, Van't-Wout A B, Tersmette M, Sciellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 23.Morgan R A, Nussbaum O, Muenchau D D, Shu L, Couture L, Anderson W F. Analysis of the functional and host range-determining regions of the murine ectropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:4712–4721. doi: 10.1128/jvi.67.8.4712-4721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Saas P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 25.Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 27.Overbaugh J, Hoover E A, Mullins J I, Burns D P W, Rudensey L, Quackenbush S L, Stallard V, Donahue P R. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology. 1992;188:558–569. doi: 10.1016/0042-6822(92)90510-v. [DOI] [PubMed] [Google Scholar]
- 28.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukemia virus in vitro. Nature. 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 29.Peredo C, O'Reilly L, Gray K, Roth M J. Characterization of chimeras between the ecotropic Moloney murine leukemia virus and the amphotropic 4070A envelope proteins. J Virol. 1996;70:3142–3152. doi: 10.1128/jvi.70.5.3142-3152.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poss M L, Mullins J I, Hoover E A. Posttranslational modifications distinguish the envelope glycoprotein of the immunodeficiency disease-inducing feline leukemia virus retrovirus. J Virol. 1989;63:189–195. doi: 10.1128/jvi.63.1.189-195.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohn J L, Linenberger M L, Hoover E A, Overbaugh J. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol. 1994;68:2458–2467. doi: 10.1128/jvi.68.4.2458-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohn J L, Moser M S, Gwynn S R, Baldwin D N, Overbaugh J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol. 1998;72:2686–2696. doi: 10.1128/jvi.72.4.2686-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohn J L, Overbaugh J. Pathogenic feline retroviruses: feline leukemia virus and feline immunodeficiency virus. In: Chen I S Y, Ahmed R, editors. Persistent viral infections. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 379–408. [Google Scholar]
- 34.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ting Y T, Wilson C A, Farrell K B, Chaudry G J, Eiden M V. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72:9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavorotinskaya T, Albritton L M. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J Virol. 1999;73:5034–5042. doi: 10.1128/jvi.73.6.5034-5042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]