Abstract
Jaagsiekte sheep retrovirus (JSRV) is the etiologic agent of a contagious bronchioloalveolar carcinoma of sheep known as sheep pulmonary adenomatosis (SPA; ovine pulmonary carcinoma). JSRV is unique among retroviruses because it transforms the alveolar type II cells and the nonciliated bronchiolar cells (Clara cells) of the lungs; these cells are where JSRV is specifically expressed in both naturally and experimentally SPA-affected sheep. In this study, we investigated the cell specificity of JSRV expression. By transient-transfection assays of 23 different cell lines with a reporter plasmid driven by the JSRV long terminal repeat (LTR), pJS21-luc, we found that the JSRV LTR is preferentially active in cell lines derived from type II pneumocytes and Clara cells (MLE-15 and mtCC1-2 mouse cell lines). Reporter assays using progressive 5′ deletions of pJS21-luc allowed us to establish that the JSRV enhancers are able to activate the JSRV proximal promoter in MLE-15 and mtCC1-2 cells, but they have very low activity in mouse cells of other lineages (e.g., NIH 3T3). The JSRV enhancers are able to activate heterologous promoters in both MLE-15 and 3T3 cells, although optimal activity is achieved in MLE-15 cells only with the homologous JSRV promoter. Thus, JSRV cell-specific LTR activity appears to result from an interaction between the enhancer elements and the JSRV proximal promoter elements. By mutation analysis, we established that an upstream NF-κB-like element appears to be responsible for approximately 50% of the JSRV LTR transcriptional activity in MLE-15 cells. Electrophoretic mobility shift assays showed evidence of a factor(s) that binds to this sequence. Antibody supershift experiments indicated that the factor(s) is not related to NF-κB component p50 or p52. This factor also appeared to be present in cells that do not support a high level of JSRV expression. Finally the JSRV21 LTR contains putative enhancer binding motifs for transcription factors such as hepatocyte nuclear factor 3 (HNF-3) that are involved in lung-specific gene expression. Cotransfection experiments demonstrated that exogenous HNF-3 is able to enhance the expression of pJS21-luc in NIH 3T3 cells, which normally show minimal enhancer activity for the JSRV LTR.
Jaagsiekte sheep retrovirus (JSRV) is the causative agent of a contagious lung cancer of sheep known as sheep pulmonary adenomatosis (SPA; also ovine pulmonary carcinoma) (5, 17, 39, 41, 43, 60). SPA is an animal model of human bronchioloalveolar carcinoma (BAC) (45), a human lung cancer that is not etiologically associated with smoking (29) and that is increasing in prevalence in the United States (6). Lung cancer is the main cause of mortality among cancer patients (32), and the characteristics BAC and SPA have in common suggest that the latter could offer novel insights into pulmonary carcinogenesis. JSRV is the only retrovirus that transforms the differentiated epithelial cells of the lungs: type II pneumocytes (37, 56) and Clara cells (50). To fully understand the pathogenesis of SPA, it is necessary to investigate the nature of the association between JSRV and its target cells for transformation.
In general, the envelope gene (env) and the long terminal repeat (LTR) are the major determinants of retroviral tropism. The env gene encodes the viral glycoprotein that specifically interacts with the cellular receptor(s) necessary for viral entry (57). Retroviruses are able to infect only cells expressing their specific receptor. On the other hand, the LTR contains the viral promoter and enhancer elements that specifically interact with the cellular transcription machinery. After viral entry and integration, the LTR drives viral expression more efficiently in those cells that express transcription factors that interact with the viral enhancer elements (1, 20).
A unique feature of JSRV is its extremely tight restriction of expression in vivo to the induced tumor cells. For most other retroviral systems, there is substantial infection in numerous cell types within the infected host in addition to the resulting tumor cells. However, in sheep with naturally or experimentally acquired SPA, JSRV is abundantly expressed only in tumor cells of the lungs (40), although it is also possible to detect JSRV DNA and RNA by sensitive PCR assays in a variety of lymphoid tissues of SPA-affected sheep (42). Proviral DNA has been found in adherent cells (macrophages), B lymphocytes, and CD4+ and CD8+ T lymphocytes of the mediastinal lymph nodes of SPA-affected animals (27). Therefore, although JSRV is highly expressed only in the epithelial tumor cells of the lungs, it infects many different cell types. In a recent study, we have shown that in vitro JSRV infects several different sheep cell lines of various tissue origins (44). Both of these in vivo and in vitro observations suggest that the cellular receptor for JSRV is common to a variety of cell types; thus, the restriction of viral expression to epithelial tumor cells in the lungs is likely not due to the presence of the JSRV receptor only on these cells. However, it is theoretically possible that there is higher expression of the JSRV receptor on lung epithelial cells. Nevertheless, it seemed possible that the JSRV LTR is preferentially active in type II pneumocytes and Clara cells.
The aim of this study was to investigate whether JSRV-specific expression in the differentiated epithelial cells of the lung is due to lung epithelial cell-specific activation of the viral LTR. We performed reporter assays with several cell lines originating from different cell types with a construct (pJS21-luc) in which the luciferase gene is under the transcriptional control of the JSRV LTR. JSRV LTR function was then dissected by assaying the transcriptional activity of LTR deletion mutants and cotransfections with potentially activating transcription factors. The results support the hypothesis that JSRV expression is strongly influenced by the differentiation state of lung epithelial cells. In addition, potential cellular factors involved in LTR activity were identified and evidence for interaction between enhancer and promoter elements was obtained.
MATERIALS AND METHODS
Cell cultures.
The cell lines used in this study; the tissues, cells, and animal species from which they originated; and their sources, references, or American Type Culture Collection (ATCC) catalog numbers are listed in Table 1. Cells were grown at 37°C with 5% CO2. MLE-15 (provided by J. Whitsett), MLE-12, JS-7, and primary fetal lamb lung (FLL) cells were grown in RPMI 1640 medium (Gibco BRL)–2% fetal bovine serum (FBS)–0.5% insulin-transferrin-sodium selenite (Sigma) modified by the addition of 5 mg of transferrin per ml, 10 mM HEPES, 10−8 M β-estradiol, and 10−8 M hydrocortisone. 293T, OAT, CP-MRI, OA1, mtCC1-2 (provided by F. De Mayo), IC-21, and NIH 3T3 cells were grown in Dulbecco's modified Eagle medium (DMEM; ATCC)–10% FBS. IC-21 and ABI-2 cells were grown in RPMI 1640 medium (Gibco BRL)–10% FBS. FOP, ST3, CP-ATCC, C2C12, and TCMK cells were grown in DMEM–1× nonessential amino acids (Cellgro)–10% FBS. F9 cells were grown in DMEM (ATCC)–7 × 10−6 M mercaptoethanol–1× nonessential amino acids–10% FBS. BV2 and A549 cells were grown in F-12K (Gibco BRL)–10% FBS. H441 and H358 cells were grown in RPMI 1640 medium (Gibco BRL) adjusted to contain 1.5 g of sodium bicarbonate per liter, 4.5 g of glucose per liter, and 10 mM HEPES with 10% FBS.
TABLE 1.
Cell lines used in this study
| Cell line | Origin | Tissue or cell type | ATCC no. or reference |
|---|---|---|---|
| A549 | Human | Lung carcinoma | CCL-185 |
| H441 | Human | BAC | HTB-174 |
| H358 | Human | BAC | CRL-5807 |
| 293T | Human | Embryonic kidney | Lebkowsky et al. (33) |
| OA1 | Sheep | Brain fibroblast | CRL-6538 |
| OAT | Sheep | Sheep testis | CRL-6546 |
| FLL | Sheep | Primary fetal lamb | MRIa |
| JS7 | Sheep | BAC | Jassim (30) |
| CP-MRI | Sheep | Choroid plexus | MRIa |
| CP-ATCC | Sheep | Choroid plexus | CRL-1700 |
| mtCC1-2 | Mouse | Clara cell | Magdaleno et al. (34) |
| BV2 | Mouse | Microglia | A. Tennera |
| F9 | Mouse | Testicular carcinoma | CRL-1720 |
| FOP | Mouse | Mammary carcinoma | J. Hassela |
| IC-21 | Mouse | Peritoneal macrophage | TIB-186 |
| MHS | Mouse | Alveolar macrophage | CRL-2019 |
| C2C12 | Mouse | Myoblast | CRL-1772 |
| ABI-2 | Mouse | Hybridoma | HB-33 |
| MLE-12 | Mouse | Lung epithelium | CRL-2110 |
| TCMK | Mouse | Mouse kidney | CCL-139 |
| NIH 3T3 | Mouse | Mouse embryo | CCL-92 |
| MLE-15 | Mouse | Type II pneumocyte | Wikenheiser et al. (59) |
| ST3 | Mouse | Thymus stroma | Brightman et al. (10) |
Cells were provided directly by an investigator or institution with no reference available. MRI, Moredun Research Institute.
Oligonucleotides.
For electrophoretic mobility shift assays (EMSAs), the double-stranded oligonucleotide probes used were JS21wt(−267/−247) (TGCGGGGGACGACCCGTGAA) and JS21mt(−267/−247) (TGCGGTTTACGACCCGTGAA [mutated nucleotides are in boldface]). JS21wt(−266/−247) corresponds to positions −266 to −247 of U3 of JSRV21 and includes an NF-κB-like binding site (21) (underlined). JS21mt(−266/−247) has three nucleotide changes with respect to JS21wt(−267/−247) in the NF-κB-like site. Oligonucleotide probes for the consensus sequence of NF-κB were purchased from Geneka and used as positive controls.
The sequences of the oligonucleotides used for the PCR cloning of the plasmids described below are available on request.
Plasmids.
Plasmids pGL3-control, pGL3-promoter, and pGL3-basic where purchased from Promega. pGL3-control expresses the firefly luciferase gene (luc) under the control of the simian virus 40 (SV40) promoter and enhancer regions; pGL3-promoter expresses the luc gene under the control of an SV40 promoter, while pGL3-basic is devoid of eukaryotic promoter and enhancer regions. Plasmid pMLV-luc was obtained by inserting the whole LTR of Moloney murine leukemia virus (M-MuLV) (amplified from plasmid p63.2 [3]) into pGL3-basic by PCR-based cloning techniques. PCRs were performed using Pfu-Turbo polymerase (Stratagene) as recommended by the manufacturer. Plasmid pCMV-luc was derived by inserting the HindIII-BamHI fragment of pGL3-basic containing the luc gene and the poly(A) signal into pCDNA3.1 (Invitrogen).
The LTR of JSRV21 (43) was amplified from pJSRV21 and inserted into the MluI and BglII sites of pGL3-basic. The resulting plasmid was called pJS21-luc. The derivatives of pJS21-luc described below were all cloned into the MluI and BglII sites of pGL3-basic. Progressive 5′ deletions of pJS21-luc were generated by PCR cloning (2) and are shown in Fig. 1B (plasmids c to i). All constructs were checked by nucleotide sequencing and/or restriction digestion.
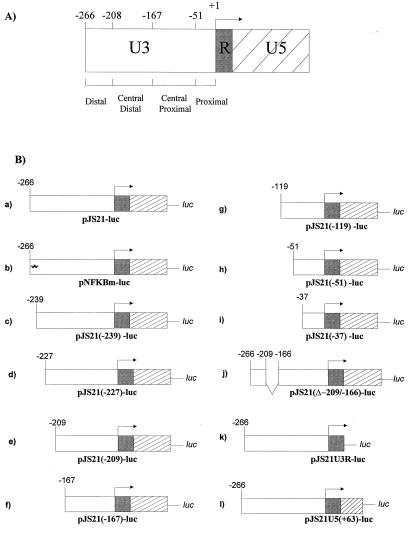
FIG. 1.
JSRV LTR reporter plasmids. (A) Organization of the JSRV LTR. The U3 region was divided into four regions, based on the endpoints of deletions. (B) Reporter plasmids used in this study. pJS21-luc contains the entire JSRV LTR fused upstream of the firefly luciferase gene. A series of nested deletions from the 5′ end of pJS21-luc were generated as shown (plasmids c to i). Additional alterations in the U3 region of pJS21-luc included mutation of the distal NFκB-like site (plasmid b) and deletion of the central distal sequences (plasmid j). Truncations of U5 sequences were also made (plasmids k and l).
The mutant pJS21(Δ−209/−167)-luc (Fig. 1B, plasmid j) contains the whole JSRV LTR, with the exception of a portion of U3 encompassing nucleotides −209 and −166. Plasmid pJS21U3R-luc (Fig. 1B, plasmid k) is composed of the U3 and R regions of the pJS21 LTR. Plasmid pJS21U5(+63)-luc (Fig. 1B, plasmid l) is truncated at position +63 in U5. Plasmid pNFKBm-luc (Fig. 1B, plasmid b) was obtained by amplification of pJS21-luc with primers 3LTR-BglII and NFKBmMluI. Primer NFKBmMluI has incorporated in its sequence the desired mutation of the NF-κB-like binding site present at position −262 of pJS21-luc (GGG → TTT).
Plasmid pMLVp-luc was obtained by inserting the proximal promoter region of M-MuLV (from −150 in U3 to the end of U5) into pGL3-basic (see Fig. 4). Plasmid pMLVp+JSe was derived by inserting the JSRV21 enhancers in the U3 region between −40 and −267 in front of pMLVp-luc. Plasmid pSVp+JSe was derived by inserting the U3 region of JSRV21 between −51 and −267 into pGL3-promoter.
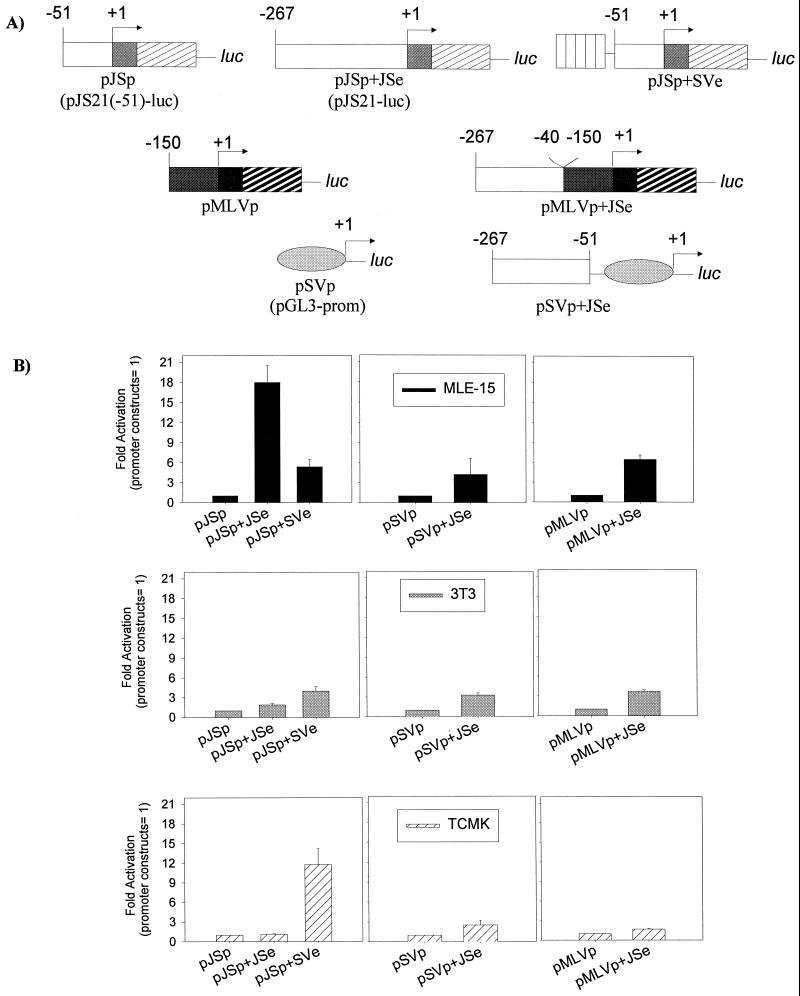
FIG. 4.
Interaction of JSRV LTR elements with other promoters-enhancers. (A) A series of chimeric reporter constructs was generated in which the promoter (p) or enhancer (e) from the JSRV LTR (JS) was combined with corresponding elements from the SV40 (SV) or M-MuLV (MLV) promoter-enhancer. The organizations of these chimeras are shown. (B) Transfections of the chimeric reporter constructs were carried out with MLE-15, NIH 3T3, and TCMK cells. In each of the panels, values for the chimeras relative to that of a plasmid containing only the basal promoter (pJSp, pSVp, or pMLVp) are shown.
Plasmid pJSp+SVe has the SV40 enhancer driving the expression of the JSRV proximal promoter region starting at −51 in U3 and includes the R and U5 regions of the JSRV LTR. pJSp+SVe was derived by inserting the SV40 enhancer region from pGL3-control into pJS21(−51)-luc.
For the transactivation experiments, we used the following expression plasmids. Plasmid pBETNFI-B1f, expressing the NFI-A1.1 isoform driven by the chicken β-actin promoter, and control plasmid pBET, containing only the β-actin promoter, were provided by C. Bachurski and were originally developed by T. Tamura (28). Plasmid pCMV-TTFI, expressing thyroid transcription factor TTF-I, was originally made by R. Di Lauro (8) and provided by G. Suske (Philipps-Universität, Marburg, Germany); pCMV-HNF3α and pCMV-HNF3β, expressing hepatocyte nuclear factor 3α (HNF3α) and HNF-β, respectively, were originally developed by R. H. Costa (14) and provided by G. Suske, as was pEVR2-Sp1, expressing the Sp-1 transcription factor under the control of the cytomegalovirus (CMV) promoter (9).
To adjust the luciferase values (see below) for transfection efficiency and lysate preparations, cotransfections were performed with the following plasmids: pCMV-βgal, expressing the β-galactosidase gene under the control of the CMV promoter; pRL-tk (Promega), expressing renilla luciferase under the control of the herpes simplex virus thymidine kinase promoter; and pRL-null (Promega), a promoterless plasmid containing the renilla luciferase gene.
Transient transfections and luciferase assays.
Transient transfections were performed on 2 × 105 to 4 × 105 cells plated in six-well plates (Falcon) approximately 24 h prior to transfection. For each well, a total of 2 μg of plasmid DNA (1 μg of reporter plasmid and 1 μg of pCMV-βgal to adjust for transfection efficiency) and 6 μl of Fugene (Boehringer) were used as recommended by the manufacturers. For selected cell lines (MLE-15, mtCC1-2, 3T3, TCMK, ST3, CP-MRI, and CP-ATCC), experiments were performed using the Dual Luciferase Reporter System (Promega) (0.5 μg of reporter plasmid and 0.5 μg or 50 ng of pRL-tk or pRL-null) and the activity of pJS21-luc was compared to those of different neutral promoters (pGL3-control, pMLV-luc, pCMV-luc, and pGL3-basic). For transactivation experiments, we used 200 ng of pJS21-luc, 1 to 200 ng of a transactivating plasmid (or a control plasmid containing the same promoter as the transactivating plasmid), and 100 ng of pRL-null. After 48 h, transfected cells were washed with phosphate-buffered saline, lysed with 400 μl of 1× Reporter Lysis Buffer (Promega) per well, and frozen at −20°C. Luciferase assays were performed on 20 μl of the cleared lysate by rapid addition of Luciferase Assay Reagent (Promega), and light output was integrated over 10 s at room temperature using a Monolight 2010 luminometer (Analytical Luminescence Laboratory). Luciferase activity was normalized for transfection efficiency and cell extract preparation by either assaying 50 μl of each lysate for β-galactosidase activity using Luminescent β-Galactosidase Genetic Reporter System II (Clontech) as recommended by the manufacturer or measuring the renilla luciferase activity driven by pRL-tk and pRL-null in the Dual Luciferase Reporter System (Promega) as recommended by the manufacturer.
The relative activity of pGL3-control adjusted for transfection efficiency was set to 100 for the experiments intended to compare the activities of pJS21-luc across different cell lines. The activity of pJS21-luc in selected cell lines was compared to those of different “neutral” promoters (pGL3-control, pCMV-luc, pMLV-luc, and pGL3-basic) using either pCMV-βgal, pRL-tk, or pRL-null to adjust for transfection efficiency (see Results).
All of the transfections were performed at least six independent times, and results are presented as the mean value for each sample. Values were determined at extract concentrations at which the luciferase assays were in the linear range.
Analysis of putative transcription factor binding sites.
Analysis of putative transcription factor binding elements was done by computer using the MatInspector v2.2 (Genomatix) program (51).
Nuclear extracts and EMSAs.
Nuclear extracts were prepared from TCMK and MLE-15 cells by established procedures with minor modifications (18). The salt concentration of the extraction buffer was 1.2 M KCl; the final concentration was adjusted to 300 mM KCl. NIH 3T3 cell nuclear extract was purchased from Geneka.
EMSAs were performed using the Nushift Kit (Geneka) as recommended by the manufacturer. Five micrograms of nuclear extract was incubated with 0.5 ng of a 32P-end-labeled oligonucleotide probe for 20 min at 4°C with or without a 100-fold excess of an unlabeled competitor. For antibody supershift interference assays, an anti-NFκB p50 (Geneka) or an anti-NF-κB p52 (Santa Cruz) rabbit polyclonal antibody was used. Nuclear extracts (5 μg) and antibodies (2 μl) were incubated for 20 min at 4°C. Bound and free probes were separated by nondenaturing electrophoresis in a 5% polyacrylamide gel.
RESULTS
Type II pneumocytes and Clara cells support preferential expression of the JSRV LTR.
In order to analyze the transcriptional activity of the JSRV LTR, we performed transient-transfection assays with different cell lines using a construct containing the firefly luciferase gene driven by the LTR of JSRV21 (43), an infectious molecular clone of JSRV (pJS21-luc). In each cell line, the relative activity of pJS21-luc was determined with respect to a promoter-enhancer plasmid (pGL3-control) containing a neutral promoter-enhancer (SV40, active in many cell types) driving the same reporter gene. In all cases, cotransfections with a second reporter plasmid expressing a different reporter gene (e.g., pCMV-βgal expressing β-galactosidase) also served to normalize transfection efficiencies between different experiments. We initially concentrated on mouse cell lines because of the availability of cell lines originating from differentiated epithelial cells of the lung that maintain the original phenotype and transcriptional characteristics. In particular, the MLE-15 line is a mouse cell line originating from lung tumors generated in transgenic mice harboring the SV40 large T antigen under the transcriptional control of the promoter-enhancer region from the human surfactant protein C (SP-C) gene (59). mtCC1-2 is a cell line derived from Clara cells from transgenic mice in a similar fashion but with the SV40 T antigen under the transcriptional control of the CC10 promoter-enhancer (34). We compared the activity of the JSRV LTR in lung cell lines versus that in non-lung cell lines, such as those originating from testicular carcinoma, mammary carcinoma, mouse kidney cells, myoblasts, etc. (Table 1). The relative activity of pGL3-control adjusted for transfection efficiency (by cotransfected pCMV-βgal) was set to 100. The results are shown in Fig. 2.
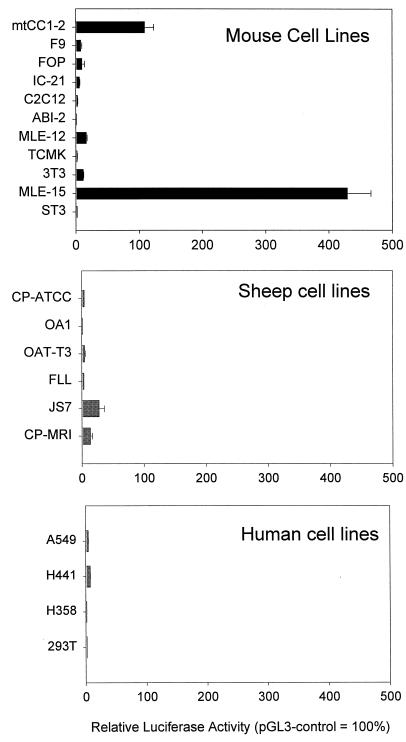
FIG. 2.
JSRV LTR activity in cell lines. The pJS21-luc plasmid was transfected into a series of murine, ovine, and human cell lines. Luciferase activities of pJS21-luc relative to the activity of pGL3, a reporter plasmid driven by the SV40 promoter-enhancer are shown. The activity of pGL3 in each cell line was set at 100%. All transfections included cotransfection with a second reporter plasmid driving a different reporter gene to control for transfection efficiency in different experiments (see Materials and Methods). At least six replicates were tested for each cell line, and the standard error of the mean is indicated by the error bars.
The highest relative luciferase values for pJS21-luc were obtained in MLE-15, mtCC1-2, and MLE-12 cells, with 429, 109, and 16% of the activities of pGL3-control in the respective cell lines. Among the nonpulmonary cell lines, pJS21-luc showed the highest level of activity in NIH 3T3 cells (11%). These results suggested that the JSRV LTR is preferentially expressed in cell lines derived from differentiated epithelial cells of the lungs.
We also tested various cell lines derived from sheep and humans. Generally, low activity was observed, even though several of the cell lines tested were derived from lung epithelial cells, including those that originated from human patients with BAC, A549, H358, and H441 (11, 22), and the JS-7 cell line derived from an SPA tumor from a sheep (30). The fact that these BAC- and SPA-derived cell lines showed relatively low expression of pJS21-luc may be due to the fact that the cell lines generally have lost differentiation properties typical of BAC and SPA tumors and/or lung epithelial cells. The nonepithelial cell line that supported the highest levels of pJS21-luc activity was a sheep choroid plexus cell line (CP-PRI) obtained from the Moredun Research Institute; note that another sheep choroid plexus cell line obtained from the ATCC (CP-ATTC) supported the expression of pJS21-luc very inefficiently.
While hydroxycortisone was added to the medium used for MLE-15 cells, it was unlikely that this was solely responsible for the high activity of the JSRV LTR since other cell lines that showed relatively low activity (e.g., MLE-12, JS-7, and FLL cells) were also cultured in the same medium. Likewise, mtCC1-2 cells that showed high LTR activity were cultured in standard medium without added hydroxycortisone.
One challenge in comparing transcriptional activities of pJS21-luc across different cell lines is that a neutral promoter-enhancer (equally active in all cell types) must be used as a reference point. This is important in order to allow adjustment for differences in transfection efficiency among different cell lines and between different experiments. However, no mammalian promoter-enhancer has exactly the same activity in every cell line. To confirm the results of Fig. 2, for which pGL3-control (SV40 promoter-enhancer) was used as the reference neutral promoter-enhancer, we repeated the luciferase assays with selected murine and ovine cell lines using the M-MuLV LTR, the CMV immediate-early promoter, or pGL3-basic (SV40 promoter but no enhancer) as neutral promoters-enhancers (Table 2). Transfection efficiency was adjusted with either pCMV-βgal (experiment series 1) or pRL-tk (experiment series 2), and the relative activity of the neutral promoter was set to 100. In another set of experiments (3 and 4), the relative activity of pJS21-luc was compared among the selected cell lines by simply dividing the pJS21-luc (0.5 μg/well) firefly luciferase values by the cotransfected pRL-tk or pRL-null (50 ng/well) renilla luciferase value without additional normalization against a neutral promoter-enhancer. The cell lines studied included MLE-15 and mtCC1-2 cells because they support high expression of pJS21-luc; 3T3 cells were the murine non-lung epithelial cells that supported the highest levels of pJS21-luc expression, while TCMK and ST3 gave the lowest levels of expression. The ovine CP-MRI and CP-ATCC lines were also tested to compare murine and ovine cell lines.
TABLE 2.
Comparison of relative luciferase activities of pJS21-luc in selected cell lines by using different reference promoters and/or cotransfected plasmids
| Cell line | Avg relative luciferase activity ± SEM (95% CI)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Set 1
|
Set 2
|
Set 3 (pJS21luc/pRL-tk ratio)f | Set 4 (pJS21luc/pRL-null ratio)f | |||||||
| SVb | CMVc | MLVd | Basice | SV | CMV | MLV | Basic | |||
| MLE-15 | 223 ± 19.5 | 9.7 ± 1.2 | 22.8 ± 0.1 | 11.2 ± 1.4 | 187 ± 5.4 | 52.1 ± 0.8 | 50 ± 1 | 11.6 ± 0.22 | 77.2 ± 1.3 | 56.1 ± 1.9 |
| mtCC1-2 | 51.3 ± 7.5 | 15.2 ± 3.2 | 22.7 ± 5 | 12.4 ± 1.6 | 90 ± 2.4 | 93.6 ± 2.4 | 76.3 ± 3 | 5.6 ± 0.4 | 62.9 ± 0.8 | 29.9 ± 0.6 |
| NIH 3T3 | 5.9 ± 0.4 | 2.4 ± 0.2 | 8.8 ± 0.4 | 4.5 ± 0.4 | 5.9 ± 0.1 | 46 ± 1.2 | 24 ± 0.5 | 2.5 ± 0.1 | 3.3 ± 0.1 | 5.4 ± 0.1 |
| TCMK | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST3 | 2.9 ± 0.3 | 3.1 (0.3) | 14.3 ± 1.2 | 0.8 ± 0.1 | 6.5 ± 0.2 | 15 ± 1.1 | 34.7 ± 1.4 | 1 ± 0.1 | 11.6 ± 1.5 | 6.8 ± 0.1 |
| CP-MRI | 7.1 (1.6) | 6.6 ± 1 | 4 (0.3) | 13.2 ± 1 | 29.2 ± 1 | 46.2 ± 3 | 34 ± 1.1 | 9.4 ± 0.1 | 26.8 ± 1.5 | 76.5 ± 2.5 |
| CP-ATCC | 2.2 ± 0.1 | 1.8 ± 0.2 | 0.9 ± 0.1 | 6.1 ± 1 | 6.6 ± 0.1 | 10.4 (0.3) | 9.9 ± 0.3 | 3.5 ± 0.2 | 24 ± 0.1 | 13 ± 1 |
Results were expressed by taking as a value of 1 (in italics) the activity of pJS21-luc relative to each neutral promoter in TCMK cells. CI, confidence interval. Experiments in sets 1 and 2 also included cotransfections of each luciferase reporter (e.g., pJS21-luc) with a second plasmid (pCMV-βgal, set 1; pRL-tk, set 2) to allow normalization for DNA transfection efficiency among different experiments. The values shown are averages of at least six independent experiments. In boldface are the two highest values in each series of experiments.
SV, pGL3-control (SV40 promoter-enhancer).
CMV, pCMV-luc.
MLV, pMLV-luc.
Basic, pGL3-basic (promoterless luciferase plasmid).
All cotransfections contained 500 ng of pJS21-luc/well and 50 ng of pRL-tk or pRL-null/well.
In Table 2, sets 1 and 2, the relative activities of pJS21-luc in the different cell lines relative to the four neutral promoters-enhancers are shown. Depending on the reference promoter-enhancer, there was variation in the relative strength of the JSRV LTR in the different cell lines. For instance, the activity of pJS21-luc in MLE-15 cells was approximately 200-fold greater than in TCMK cells using the SV40 promoter-enhancer as the neutral promoter but there was only a 20- to 50-fold difference in the same cell lines with the M-MuLV LTR as the neutral promoter. Nevertheless, of all of the murine cell lines, MLE-15 and mtCC1-2 consistently showed the highest activities for pJS21-luc regardless of the reference neutral promoter-enhancer. In sets 3 and 4, where pJS21-luc was cotransfected with a renilla luciferase expression plasmid driven by either the herpes simplex virus thymidine kinase promoter or pRL-null and the values were divided without further correction, the results were also consistent with the results of sets 1 and 2 in that MLE-15 and mtCC1-2 cells showed the highest levels of pJS21-luc activity. Overall, the results supported the implications of Fig. 2 that the two murine lung epithelium-derived cell lines supported the highest JSRV LTR transcriptional activity.
Table 2 also shows expanded studies of the relative activities of pJS21-luc in the sheep choroid plexus cell lines CP-MRI and CP-ATCC. These two lines generally showed higher pJS21-luc activities than the non-lung epithelial murine cell lines and in some cases also with respect to the murine lung cell lines, which might reflect higher activity of the JSRV LTR in ovine than in murine cell lines. It was also noteworthy that, overall, CP-MRI cells supported higher pJS21-luc activity than did CP-ATCC cells, consistent with the initial results of Fig. 2. We also carried out similar experiments in which the concentrations of the cotransfected transfection control plasmids were reduced in order to rule out the possibility that promoter-enhancer elements on the cotransfected plasmids were titrating cellular transcription factors. Results essentially the same as those shown in Table 2 were obtained (data not shown).
The JSRV enhancers are particularly active in MLE-15 cells.
To map transcriptional control elements in the JSRV LTR, a series of overlapping 5′ truncations of pJS21-luc were prepared as shown in Fig. 1. These truncations delineated four regions from the U3 region of the LTR: (i) a distal region (−208 to −266), (ii) a central distal region (−167 to −208), (iii) a central proximal region (−51 to −167), and (iv) a promoter-proximal region (0 to −51). The deletions were then tested for transcriptional activity by transfection into various murine and ovine cell lines as shown in Fig. 3. The activities of the deletions are shown as fold activation relative to pJS21(−37)-luc, a plasmid containing the JSRV LTR truncated at position −37, in each cell line. This plasmid would contain the putative basal promoter elements of the JSRV LTR, including the TATA box (position −23), as well as the R and U5 regions. The results obtained with the murine cell lines (solid bars) showed that the central and distal elements were able to enhance the transcriptional activity of the basal JSRV promoter in the murine MLE-15 and mtCC1-2 cell lines, with the strongest evidence for enhancer activity in MLE-15 cells. On the other hand, the other murine cell lines showed little evidence for enhancer activity for the JSRV LTR, with, at most, a twofold difference between full-length pJS21-luc and basal pJS21(−37)-luc. These results were very consistent with the results of Fig. 2 and Table 2 in that the two lung epithelial cell lines that showed the highest levels of JSRV LTR transcriptional activity also showed evidence of functional enhancer elements.
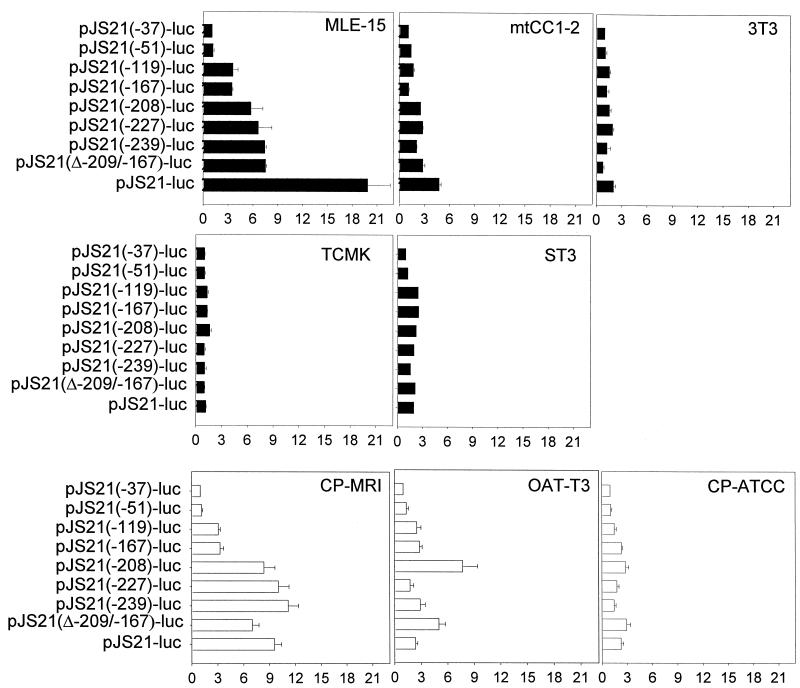
FIG. 3.
Activity of JSRV LTR deletions. The U3 deletions of pJS21-luc were transfected into different murine cell lines (top two rows, closed bars) and ovine cell lines (bottom row, open bars). For each cell line, the activities of each plasmid are shown relative to that of pJS21(-37)-luc, a plasmid containing only the JSRV basal promoter elements (given a value of 1).
The results in Fig. 3 allowed the localization of enhancer activity within the JSRV LTR for MLE-15 and mtCC1-2 cells. In particular, in MLE-15 cells, approximately 40% of the enhancer activity was associated with elements between positions −51 and −240 while the remaining 60% was associated with the distal elements between −240 and −267. We generated an internal deletion of the JSRV LTR [pJS21(Δ−209/−166)] lacking the central distal elements and found that it had approximately 40% activity in MLE-15 cells. This is consistent with the importance of the central distal elements of the JSRV LTR for enhancer activity in these cells.
When the sheep cell lines were tested with the truncation series (Fig. 3, open bars), the CP-MRI cells showed substantial evidence of enhancer activity while there was little activity in CP-ATCC cells and intermediate levels in OAT-T3 cells. This was consistent with the higher JSRV LTR activity in CP-MRI cells (Fig. 2 and Table 2). It would have been desirable to study JS7 cells, since they showed the greatest ability to support JSRV LTR transcription (Fig. 2) in sheep. However, these cells are quite difficult to transfect, so extensive transfections with the entire set of truncation plasmids were not performed. Unfortunately, as mentioned above, ovine cell lines that retained the differentiation properties of lung epithelial cells have not been reported.
We also tested the contribution of elements downstream from the transcriptional start site (i.e., R and U5) for the activity of the JSRV LTR. The U5 region appears to contain elements necessary for optimal expression, since deletion of the U5 region from pJS21-luc (construct pJS21U3R-luc) resulted in 20 to 50% activity relative to pJS21-luc in all of the murine cell lines tested (MLE-15, mtCC1-2, NIH 3T3, and TCMK; data not shown). Addition of the first 63 nucleotides of U5 to this construct [pJS21(+63)-luc] fully or partially restored activity to the same levels as pJS21-luc in NIH 3T3 and TCMK cells, but these nucleotides did not increase expression to pJS21-luc levels in MLE-15 or mtCC1-2 cells (not shown). Deletion of both the R and U5 sequences from pJS21-luc reduced transcriptional activity to the background level given by pGL3-basic. Further studies are required to elucidate the roles of the R and U5 sequences in JSRV LTR-driven transcription.
Interaction between promoter and enhancer elements for optimal expression from the JSRV LTR.
As shown in Fig. 3, both distal and central elements in the U3 region of the JSRV LTR contribute to optimal expression. It seemed that the most likely explanation was that the JSRV LTR contains enhancer elements that are particularly active in lung epithelium-derived cells. We therefore asked whether JSRV enhancer elements (positions −51 to −260) could confer cell specificity on heterologous promoters. As shown in Fig. 4A, we generated a series of luciferase reporter constructs in which the JSRV enhancers were inserted in front of the basal SV40 or M-MuLV promoter. These constructs were tested for activity in MLE-15, NIH 3T3, and TCMK cells (Fig. 4B, middle and right panels). The activity of pJSp (or pSV-p or pMLV-p), after normalization for transfection efficiency with pCMV-βgal, was taken as a value of 1 and compared to the activity of constructs containing heterologous enhancers. As expected, the JSRV enhancers were able to enhance expression from the SV40 and M-MuLV promoters in MLE-15 cells. Also as expected, the JSRV enhancers were unable to enhance expression from the same promoters and TCMK cells, where there was little evidence of enhancer activity (Fig. 3). The results obtained with NIH 3T3 cells were somewhat unexpected, in that the JSRV enhancers were able to enhance the expression of the SV40 and M-MuLV promoters (about fourfold); this enhancement was greater than the difference between the basal JSRV promoter and the full-length JSRV LTR in these cells (pJSp versus pJSp+JSe, left panel) and equivalent to the level of enhancement of the SV40 enhancers on the SV40 basal promoter (not shown). Thus, in NIH 3T3 cells, the JSRV enhancers apparently are active but are more efficient at activating transcription from the heterologous SV40 and M-MuLV promoters than from the basal JSRV promoter (less than twofold; left panel). Similar results were obtained when a slightly larger portion of the JSRV LTR (−32 to −266) was placed in front of the basal SV40 promoter (not shown).
The fact that the JSRV enhancers combined with the JSRV basal promoter showed less enhancement in NIH 3T3 cells than when they were placed in front of the heterologous promoters raised the possibility that the JSRV promoter is not active in NIH 3T3 (and/or TCMK) cells. To investigate this, we also prepared a chimeric luciferase reporter construct in which the SV40 enhancers were placed in front of the basal JSRV promoter (pJSp+SVe; Fig. 4A) and tested its activity relative to that of the basal JSRV promoter and the full-length JSRV LTR (Fig. 4B, left panels). The results indicated that the SV40 enhancers are able to activate expression of the basal JSRV promoter in all three cell lines. Thus, the low expression of the native JSRV LTR in NIH 3T3 (or TCMK) cells cannot be attributed to lack of basal promoter activity. These results suggest that high-level expression of the JSRV LTR in MLE-15 cells requires not only active enhancer and basal promoter elements but appropriate interaction between these elements. Indeed, in MLE-15 cells, the SV40 enhancers are less efficient at activating the basal JSRV promoter than are the JSRV enhancers, while in NIH 3T3 and TCMK cells, the converse is true.
The JSRV LTR responds to cellular transcription factors involved in the expression of lung SPs.
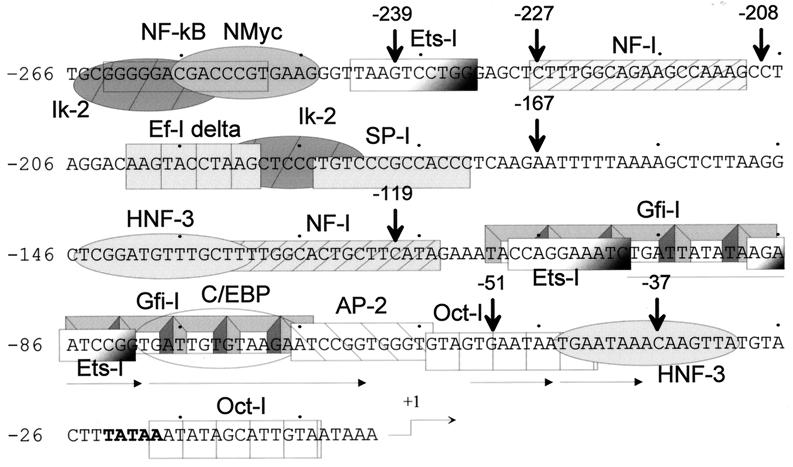
In light of the demonstration that the JSRV LTR is preferentially active in lung epithelium-derived cell lines, we surveyed the U3 region of the JSRV LTR for potential binding sites for transcription factors known to be important for expression of genes in these cells. Figure 5 shows potential factor binding sites in this region. In particular, there are two putative HNF-3 binding sites; members of the HNF-3/forkhead family of nuclear transcription factors have been shown to be important in the regulation of surfactant gene expression (13, 25, 36, 58). It has also been reported that other transcription factors such as NF-1, SP-1, and members of the octamer family cooperate with HNF-3/forkhead proteins in lung-specific expression, and these binding elements are also present in the JSRV LTR (4, 9, 54).
FIG. 5.
Potential factor binding sites in the JSRV LTR. The U3 sequences in the JSRV LTR were analyzed for potential factor binding sites by the program MatInspector v2.2 (Genomatix) (51). The sites with the best matches to consensus sequences are shown.
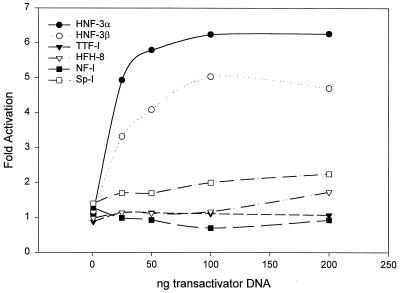
To test if putative binding elements in the JSRV LTR are important for expression, we cotransfected pJS21-luc into NIH 3T3 cells along with expression plasmids for a series of transcription factors: TTF-1, HNF-3α, HNF-3β, SP-1, HFH-8, and NF-1. The activation of JS21-luc by the various transcription factors was calculated by comparing the relative activity of pJS21-luc cotransfected with either a plasmid expressing the tested transcription factor driven by the CMV promoter or a plasmid with the CMV promoter alone. Transfection efficiency was normalized using pRL-null. As shown in Fig. 6, when the different amounts of the transcription factor expression plasmids were cotransfected with a standard amount of pJS21-luc, HNF-3α and HNF-3β stimulated luciferase expression in a dose-dependent fashion. In contrast, HFH-8, another member of the HNF-3/forkhead family, did not activate expression of the JSRV LTR. It is interesting that HNF-3α and HNF-3β are expressed in type II pneumocytes and Clara cells (14, 61) while HFH-8 expression is restricted to the epithelium and fibroblasts of the alveolar sac (47). It was also interesting that the JSRV LTR did not respond to cotransfection with the TTF-1 expression plasmid.
FIG. 6.
Transactivation of the JSRV LTR by known transcription factors. pJS21-luc was cotransfected into NIH 3T3 cells (that do not efficiently support JSRV enhancer activity) along with expression plasmids for various human transcription factors. Different amounts of the transcription factor expression plasmids were cotransfected with a set amount (1 μg) of pJS21-luc DNA. The amounts of luciferase activity for the different cotransfections are shown as fold activation over that of pJS21-luc transfected by itself into the same cell line.
An NFκB-like binding site is important for expression of the JSRV LTR.
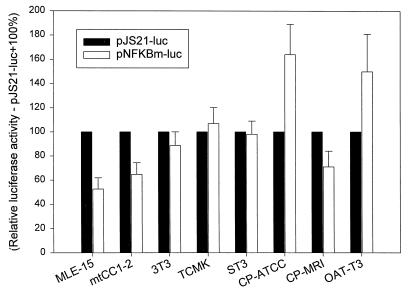
As shown in Fig. 3, approximately one-half of the enhancer activity of the JSRV LTR in MLE-15 and mtCC1-2 cells could be attributed to elements in the distal region (−239 to −266). This region contains an NFκB-like binding side with one mismatch (in boldface: 5′-GGGACGACC-3′) from the canonical NFκB consensus binding sequence (5′-GGGPuNNPyPyCC-3′). To test if this binding side was important for the enhancer activity in the distal region of the JSRV LTR, we generated a version of pJS21-luc in which the NFκB-like site was mutated (pNFκBm-luc; Fig. 1). The activity of pNFKBm-luc was compared to that of pJS21-luc in various cell lines, as shown in Fig. 7; the relative activity of pJS21-luc was set as 100.
FIG. 7.
Effect of mutation of the distal NFκB-like site in different cell lines. pJS21-luc and pNFKBm-luc (containing mutations in the distal NFκB-like site) were transfected into various murine and ovine cell lines. For each of the cell lines, the activity of pJS21-luc was set as 100% (solid bars). The activity of pNFKBm-luc relative to that of pJS21-luc (open bars) is shown for each cell line.
Mutation of the NFκB-like site from the JSRV LTR reduced transcriptional activity in MLE-15 and mtCC1-2 cells, while it did not affect the level of expression in the other murine cell lines. Thus, these results supported the idea that the NFκB-like element is important for the high-level expression of the JSRV LTR in lung epithelium-derived cells while it is not important for low-level expression in non-lung epithelial cells. Figure 7 also shows results of studies with three ovine cell lines, and it was interesting that the only cells in which mutation of the NFκB-like site showed a negative effect were CP-MRI cells, which also show the highest expression of the JSRV LTR.
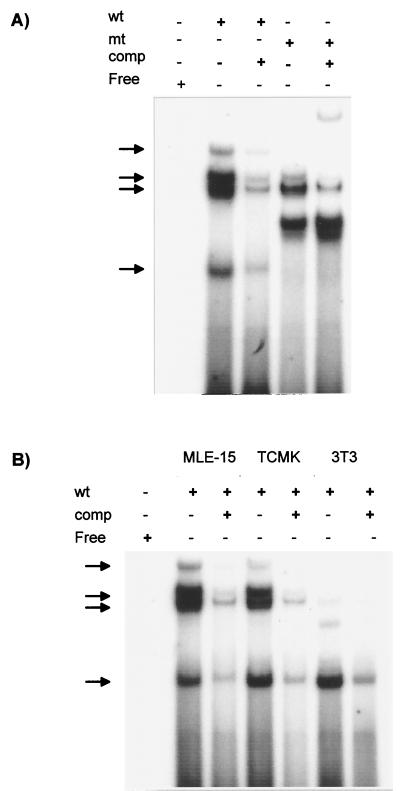
In light of the importance of the NFκB-like site for expression of the JSRV LTR in MLE-15 and mtCC1-2 cells, we tested for the presence of nuclear factors that could bind to this sequence by EMSAs (Fig. 8A). Four major complexes with different mobilities were detected in extracts from MLE-15 cells when they were incubated with a labeled oligonucleotide containing the wild-type NFκB-like sequence, and these complexes could all be competed with excess wild-type oligonucleotide. When the same nuclear extracts were incubated with a mutant oligonucleotide corresponding to the mutation in pNFKBm-luc, two of the wild-type complexes were absent (the slowest and the most rapidly migrating ones) while two complexes were still detected. A novel complex with intermediate mobility was also observed with the mutant oligonucleotides, and this was not competed by excess unlabeled wild-type oligonucleotide. The complexes bound by the wild-type but not the mutant oligonucleotides seemed most likely to represent factors important in expression of the JSRV LTR. We tested for the presence of the NFκB site binding proteins in NIH 3T3 and TCMK cells, which do not support high-level expression of the JSRV LTR (Fig. 8B). Somewhat surprisingly, these cells also generated complexes that comigrated with the complexes unique to the wild-type oligonucleotide from MLE-15 cells (although NIH 3T3 cells showed low levels of the slowly migrating complex). Thus, the factor(s) that binds to the JSRV NFκB-like sequences may be ubiquitously expressed.
FIG. 8.
Detection of cellular factors binding to the JSRV NFκB-like site. (A) MLE-15 nuclear extracts were incubated with radioactively labeled oligonucleotides corresponding to the wild-type (wt) or mutant (mt) NFκB-like site as described in Materials and Methods. EMSAs were carried out as shown. A 100-fold excess of a labeled wild-type oligonucleotide was added to some reaction mixtures as a competitor (comp). The positions of protein-oligonucleotide complexes are indicated by the arrows. (B) Similar EMSAs were carried out with TCMK and NIH 3T3 nuclear extracts. The same complexes obtained from the MLE-15 extracts were observed for these two cell lines, even though they do not show evidence of in vivo activity of the distal NFκB-like site. Darker exposures of the gel shown indicated that the two more slowly migrating bands were present in the NIH 3T3 reaction mixtures.
We used antibodies to the p50 and p52 members of the NFκB protein complex in attempts to supershift or inhibit complex formation in nuclear extracts from MLE-15 cells. However, neither antibody showed inhibition or a supershift of any of the complexes. Thus, the protein(s) that binds to the NFκB-like site in the JSRV LTR may be a previously unidentified NFκB-like protein(s) or an unrelated factor(s). It should be noted that the NFκB-like site also overlaps an Ik-2-like binding site for Ikaros-related proteins. However, the gene for Ikaros is expressed only in hematopoietic cells (38) and there have been no reports of its expression in differentiated lung cells.
DISCUSSION
In this study, we have shown that the JSRV LTR is preferentially active in type II pneumocytes and Clara cell lines. This conclusion has been drawn from several results. First, in transient-transfection assays, the JSRV LTR showed preferential activation in mouse cell lines derived from type II pneumocytes (MLE-15) and Clara cells (mtCC1-2). Second, analysis of pJS21-luc deletion mutants showed that the JSRV21 enhancers strongly activate the JSRV21 proximal promoter in MLE-15 cells. Third, the JSRV21 U3 region contains putative enhancer binding motifs for transcription factors such as HNF-3 that are involved in lung-specific expression of SPs and of Clara cell protein CC10 (25, 58). Finally, cotransfection experiments demonstrated that exogenous HNF-3 is able to enhance expression of pJS21-luc in NIH 3T3 cells, which normally show minimal enhancer activity for the JSRV LTR.
These data point to the LTR as an important determinant of the tropism of JSRV for type II pneumocytes and Clara cells. The restriction of JSRV expression in other cell types (both in vivo and in vitro) may be due to the lack of lung-specific transcription factors necessary for JSRV LTR activation (or the presence of transcription repressors) (27, 42, 44). This transcriptional specificity may also explain the observed difficulty in obtaining an efficient tissue culture system for the propagation of JSRV. The most suitable cells for JSRV replication in vitro would be type II pneumocytes and Clara cells derived from sheep. However, both of these cell types are extremely difficult to isolate and grow in vitro and they lose their differentiated state within a few hours in culture. For example, type II pneumocytes lose their characteristic lamellar body inclusions, apical microvilli, the production of phospholipids, and SP synthesis (37); thus, the transcriptional machinery of these cells is likely altered upon in vitro culture. The availability of mouse cell lines such as MLE-15 and mtCC1-2 that retain their differentiated phenotype (34, 35, 59) made this study possible. Thus, it was necessary to focus this study on mouse cell lines. MLE-15, a type II pneumocyte-derived cell line, synthesizes abundant SP-B and low levels of SP-A and SP-C (59). mtCC1-2, a Clara cell-derived line, synthesizes abundant SP-B and low levels of CC10. Although the conclusions of this study are largely based on murine cell lines (instead of sheep cell lines), they nevertheless provide a starting point from which we can begin to understand JSRV cell tropism. It is interesting that both MLE-15 and mtCC1-2 express high levels of SP-B and lower levels of other SPs. Future studies on JSRV expression might focus on transcription factors implicated in the activation of the SP-B promoter.
Elements of the JSRV LTR located both upstream and downstream of the TATA box were important for the optimal function and cell specificity of the JSRV LTR. We have shown that the JSRV enhancers (central and distal elements) are able to activate heterologous promoters (from SV40 and M-MuLV), but optimal activation in MLE-15 was achieved only with the homologous JSRV promoter. We also observed that deletion of all of U5 or a portion of it reduces pJS21-luc expression, suggesting that sequences downstream of the transcriptional start site also influence JSRV transcription. However, R and U5 are not by themselves capable of conferring tissue specificity, since a JSRV reporter construct with the promoter-proximal elements R and U5 [pJS21(−51)-luc] was also activated by SV40 enhancers in TCMK cells, where the native JSRV LTR is poorly active. Further studies are necessary to establish the role of R and U5 in JSRV LTR expression. The R region has been shown to be important for transcription for human and primate lentiviruses through an interaction between the tat protein and the transactivation response region (52) and also in simpler retroviruses such as murine leukemia virus, mouse mammary tumor virus, bovine leukemia viruses, and chicken reticuloendotheliosis virus (15, 31, 49, 53).
It is noteworthy that the JSRV LTR contains two putative binding site for HNF-3, an important factor in lung-specific SP expression (8, 9, 14, 36, 58). By cotransfection experiments with NIH 3T3 cells, which normally show low enhancer activity for the JSRV LTR, we found that the JSRV LTR can be activated by HNF-3α and HNF-3β, consistent with a role for these factors in JSRV expression in lung epithelial cells. However, more studies are necessary to firmly establish the role of HNF-3 in JSRV LTR activity in vivo.
In vitro and in vivo footprinting studies to identify those sites in JSRV U3 that are actually occupied by transcription factors in differentiated epithelial lung cells will be interesting. Future studies to identify the protein(s) that interacts with the NF-κB-like binding site present in the U3 distal element will be valuable, since this site appears to be responsible for half of the enhancing activity of the JSRV LTR in MLE-15 cells. Moreover, since EMSA analysis has shown that this factor is present both in cell lines that are permissive and in those that are nonpermissive for JSRV expression, some form of posttranslational modification (e.g., phosphorylation) or interaction with another lung-specific transcription factor(s) might be important for activity.
Interestingly, the JSRV21 LTR was not particularly active in other type II pneumocyte- or Clara cell-derived lines such as human H441, H358, and A549 cells or the JS-7 cell line (derived from the tumor cells of a sheep with SPA [30]). H441 expresses SP-A but not CC10 or HNF-3β (7), although this cell line demonstrates a Clara cell phenotype and maintains the ability to express reporter genes driven by the CC10 promoter. H358 also expresses only SP-A, and A549 expresses neither SP-A nor SP-B. No data are available for the surfactant production of JS-7 cells, but they have lost the morphological phenotype of type II pneumocytes after passage in vitro (30). As mentioned above, the best correlation was between cell lines that express SP-B and the JSRV LTR.
It is interesting that in lung sections of sheep with SPA, JSRV expression can be detected by immunohistochemistry in tumor cells, but the great majority of adjacent nontransformed type II pneumocytes and Clara cells do not show detectable amounts of JSRV antigens. This might reflect the fact that type II pneumocytes and Clara cells are cells with a very low mitotic index; therefore, they might not be able to support JSRV DNA integration, since simple retroviruses generally require passage of the infected cells through mitosis to allow breakdown of the nuclear membrane and entry of the viral DNA into the nucleus (12). On the other hand, JSRV-transformed cells might express some transcription factors more abundantly than untransformed type II pneumocytes or Clara cells and/or express additional factors necessary for optimal JSRV expression. Another possibility is that the target cells for JSRV transformations are stem cells of the respiratory epithelium (19)—precursors of both type II pneumocytes and Clara cells. These stem cells would be rarer than terminally differentiated type II pneumocytes and Clara cells, and they might have the transcriptional machinery that is optimal for JSRV LTR expression. Alternatively, the stem cells might have a higher mitotic index than differentiated type II pneumocytes or Clara cells. Infection of stem cells might also explain the decrease in susceptibility of adult sheep to experimental infection by JSRV in comparison to young animals and, conversely, the very short incubation time for disease (4 to 8 weeks) observed in newborn lambs inoculated with lung fluid. Presumably, newborn animals have higher concentrations of stem cells.
In addition to providing insights into JSRV biology, these experiments also have practical implications (23, 24, 48). The JSRV LTR might be useful in retroviral vectors designed for gene therapy of lung diseases; efficient transduction and expression in lung cells have proven difficult.
We conclude with a note on the terminology used. So far, we have referred to the disease induced by JSRV as SPA. Because of the similar acronyms of the various lung SPs (SP-A, -B, etc.), we will now refer to the disease as ovine pulmonary carcinoma, a term that has been used by other investigators (16, 26, 46, 55).
ACKNOWLEDGMENTS
We are grateful to C. Lee and L. Chun for help with tissue culture. We thank J. Whitsett (Cincinnati, Ohio), Franco and Janet De Mayo (Houston, Tex.), G. Suske (Marburg, Germany), T. Tamura (Chiba City, Japan), and J. Molkentin (Cincinnati, Ohio) for providing some of the cell lines and plasmids described here. We thank C. Bachursky (Cincinnati, Ohio) and A. Malkinson (Denver, Colo.) for useful suggestions.
M.P. was a recipient of a Wellcome Prize Traveling Research Fellowship from the Wellcome Trust and a recipient of an American Cancer Society Ray and Estelle Spehar Fellowship. This work was supported by NIH grant RO1CA82564. Support from the UCI Cancer Research Institute and the Chao Family Comprehensive Cancer Center is acknowledged.
REFERENCES
- 1.Athas G B, Starkey C R, Levy L S. Retroviral determinants of leukemogenesis. Crit Rev Oncog. 1994;5:169–199. doi: 10.1615/critrevoncog.v5.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 2.Ausbel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 3.Bacheler L T, Fan H. Multiple integration sites for Moloney murine leukemia virus in productively infected mouse fibroblasts. J Virol. 1979;30:657–667. doi: 10.1128/jvi.30.3.657-667.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachurski C J, Kelly S E, Glasser S W, Currier T A. Nuclear factor I family members regulate the transcription of surfactant protein-C. J Biol Chem. 1997;272:32759–32766. doi: 10.1074/jbc.272.52.32759. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Zhu R-Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barsky S H, Cameron R, Osann K E, Tomita D, Holmes E C. Rising incidence of bronchioloalveolar lung carcinoma and its unique clinicopathologic features. Cancer. 1994;73:1163–1170. doi: 10.1002/1097-0142(19940215)73:4<1163::aid-cncr2820730407>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 7.Bingle C D, Hackett B P, Moxley M, Longmore W, Gitlin J D. Role of hepatocyte nuclear factor-3 alpha and hepatocyte nuclear factor-3 beta in Clara cell secretory protein gene expression in the bronchiolar epithelium. Biochem J. 1995;308:197–202. doi: 10.1042/bj3080197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohinski R J, Di Lauro R, Whitsett J A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun H, Suske G. Combinatorial action of HNF3 and Sp family transcription factors in the activation of the rabbit uteroglobin/CC10 promoter. J Biol Chem. 1998;273:9821–9828. doi: 10.1074/jbc.273.16.9821. [DOI] [PubMed] [Google Scholar]
- 10.Brightman B K, Chandy K G, Spencer R H, Fan H. A T lymphoid cell line responds to a thymic stromal cell line by expression of Thy-1 and CD4. J Immunol. 1989;143:2775–2782. [PubMed] [Google Scholar]
- 11.Brower M, Carney D N, Oie H K, Gazdar A F, Minna J D. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- 12.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 13.Bruno M D, Bohinski R J, Huelsman K M, Whitsett J A, Korfhagen T R. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. . (Erratum, 270:16482, 1995.) [DOI] [PubMed] [Google Scholar]
- 14.Clevidence D E, Overdier D G, Peterson R S, Porcella A, Ye H, Paulson K E, Costa R H. Members of the HNF-3/forkhead family of transcription factors exhibit distinct cellular expression patterns in lung and regulate the surfactant protein B promoter. Dev Biol. 1994;166:195–209. doi: 10.1006/dbio.1994.1307. [DOI] [PubMed] [Google Scholar]
- 15.Cupelli L, Okenquist S A, Trubetskoy A, Lenz J. The secondary structure of the R region of a murine leukemia virus is important for stimulation of long terminal repeat-driven gene expression. J Virol. 1998;72:7807–7814. doi: 10.1128/jvi.72.10.7807-7814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMartini J C, Rosadio R H, Lairmore M D. The etiology and pathogenesis of ovine pulmonary carcinoma (sheep pulmonary adenomatosis) Vet Microbiol. 1988;17:219–236. doi: 10.1016/0378-1135(88)90067-3. [DOI] [PubMed] [Google Scholar]
- 17.DeMartini J C, York D F. Retrovirus-associated neoplasms of the respiratory system of sheep and goats. Ovine pulmonary carcinoma and enzootic nasal tumor. Vet Clin N Am Food Anim Pract. 1997;13:55–70. doi: 10.1016/s0749-0720(15)30364-9. [DOI] [PubMed] [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emura M. Stem cells of the respiratory epithelium and their in vitro cultivation. In vitro Cell. Dev Biol Anim. 1997;33:3–14. doi: 10.1007/s11626-997-0015-4. [DOI] [PubMed] [Google Scholar]
- 20.Fan H. Influences of the long terminal repeats on retrovirus pathogenicity. Semin Virol. 1990;1:165–174. [Google Scholar]
- 21.Fujita T, Nolan G P, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappaB. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 22.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W O. In vitro cultivation of human tumors: establishment of cell lines from a series of solid tumors. J Natl Cancer Inst. 1972;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 23.Goldman M J, Lee P S, Yang J S, Wilson J M. Lentiviral vectors for gene therapy of cystic fibrosis. Hum Gene Ther. 1997;8:2261–2268. doi: 10.1089/hum.1997.8.18-2261. [DOI] [PubMed] [Google Scholar]
- 24.Grubb B R, Pickles R J, Ye H, Yankaskas J R, Vick R N, Engelhardt J F, Wilson J M, Johnson L G, Boucher R C. Inefficient gene transfer by adenovirus vector to cystic fibrosis airway epithelia of mice and humans. Nature. 1994;371:802–806. doi: 10.1038/371802a0. [DOI] [PubMed] [Google Scholar]
- 25.Hay J G, Crystal R G. Lung-specific gene expression. In: Crystal R G, West J B, Weibel E R, Barnes P J, editors. The lung: scientific foundations. 2nd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 277–304. [Google Scholar]
- 26.Hod I, Zimber A, Klopfer U, Helder A W, Novel T A, Perk K. Pulmonary carcinoma (Jaagsiekte) of sheep: pathologic findings and comparison in multiple-case and case-free herds. J Natl Cancer Inst. 1974;53:103–110. doi: 10.1093/jnci/53.1.103. [DOI] [PubMed] [Google Scholar]
- 27.Holland M J, Palmarini M, Garcia-Goti M, Gonzalez L, McKendrick I, de las Heras M, Sharp J M. Jaagsiekte retrovirus is widely distributed in both T and B lymphocytes and in mononuclear phagocytes of sheep with naturally and experimentally acquired pulmonary adenomatosis. J Virol. 1999;73:4004–4008. doi: 10.1128/jvi.73.5.4004-4008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue T, Tamura T, Furuichi T, Mikoshiba K. Isolation of complementary DNAs encoding a cerebellum-enriched nuclear factor I family that activates transcription from the mouse myelin basic protein promoter. J Biol Chem. 1990;265:19065–19070. [PubMed] [Google Scholar]
- 29.Ives J C, Buffler P A, Greenberg S D. Environmental associations and histopathologic patterns of carcinoma of the lung: the challenge and dilemma in epidemiologic studies. Am Rev Respir Dis. 1983;128:195–209. doi: 10.1164/arrd.1983.128.1.195. [DOI] [PubMed] [Google Scholar]
- 30.Jassim F A. Ph.D. thesis. Edinburgh, Scotland: University of Edinburgh; 1988. [Google Scholar]
- 31.Kiss-Toth E, Unk I. A downstream regulatory element activates the bovine leukemia virus promoter. Biochem Biophys Res Commun. 1994;202:1553–1561. doi: 10.1006/bbrc.1994.2108. [DOI] [PubMed] [Google Scholar]
- 32.Landis S H, Murray T, Bolden S, Wingo P A. Cancer statistics, 1999. CA Cancer J Clin. 1999;49:8–31. doi: 10.3322/canjclin.49.1.8. [DOI] [PubMed] [Google Scholar]
- 33.Lebkowsky J S, Clancy S, Calos M P. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature. 1985;317:169–171. doi: 10.1038/317169a0. [DOI] [PubMed] [Google Scholar]
- 34.Magdaleno S M, Wang G, Jackson K J, Ray M K, Welty S, Costa R H, DeMayo F J. Interferon-gamma regulation of Clara cell gene expression: in vivo and in vitro. Am J Physiol. 1997;272:L1142–L1151. doi: 10.1152/ajplung.1997.272.6.L1142. [DOI] [PubMed] [Google Scholar]
- 35.Malkinson A M, Dwyer-Nield L D, Rice P L, Dinsdale D. Mouse lung epithelial cell lines—tools for the study of differentiation and the neoplastic phenotype. Toxicology. 1997;123:53–100. doi: 10.1016/s0300-483x(97)00108-x. [DOI] [PubMed] [Google Scholar]
- 36.Margana R K, Boggaram V. Functional analysis of surfactant protein B (SP-B) promoter. Sp1, Sp3, TTF-1, and HNF-3alpha transcription factors are necessary for lung cell-specific activation of SP-B gene transcription. J Biol Chem. 1997;272:3083–3090. doi: 10.1074/jbc.272.5.3083. [DOI] [PubMed] [Google Scholar]
- 37.Mason R J, Shannon J M. Alveolar type II pneumocytes. In: Crystal R G, West J B, Weibel E R, Barnes P J, editors. The Lung: scientific foundations. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 543–555. [Google Scholar]
- 38.Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmarini M, Cousens C, Dalziel R G, Bai J, Stedman K, DeMartini J C, Sharp J M. The exogenous form of Jaagsiekte retrovirus is specifically associated with a contagious lung cancer of sheep. J Virol. 1996;70:1618–1623. doi: 10.1128/jvi.70.3.1618-1623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmarini M, Dewar P, De las Heras M, Inglis N F, Dalziel R G, Sharp J M. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for Jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 41.Palmarini M, Fan H, Sharp J M. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 42.Palmarini M, Holland M J, Cousens C, Dalziel R G, Sharp J M. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J Gen Virol. 1996;77:2991–2998. doi: 10.1099/0022-1317-77-12-2991. [DOI] [PubMed] [Google Scholar]
- 43.Palmarini M, Sharp J M, De las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmarini M, Sharp J M, Lee C, Fan C. In vitro infection of ovine cell lines by Jaagsiekte sheep retrovirus. J Virol. 1999;73:10070–10078. doi: 10.1128/jvi.73.12.10070-10078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perk K, Hod I. Sheep lung carcinoma: an endemic analogue of a sporadic human neoplasm. J Natl Cancer Inst. 1982;69:747–749. [PubMed] [Google Scholar]
- 46.Perk K, Michalides R, Spiegelman S, Schlom J. Biochemical and morphologic evidence for the presence of an RNA tumor virus in pulmonary carcinoma of sheep (Jaagsiekte) J Natl Cancer Inst. 1974;53:131–135. doi: 10.1093/jnci/53.1.131. [DOI] [PubMed] [Google Scholar]
- 47.Peterson R S, Lim L, Ye H, Zhou H, Overdier D G, Costa R H. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech Dev. 1997;69:53–69. doi: 10.1016/s0925-4773(97)00153-6. [DOI] [PubMed] [Google Scholar]
- 48.Pickles R J, McCarty D, Matsui H, Hart P J, Randell S H, Boucher R C. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol. 1998;72:6014–6023. doi: 10.1128/jvi.72.7.6014-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierce J, Fee B E, Toohey M G, Peterson D O. A mouse mammary tumor virus promoter element near the transcription initiation site. J Virol. 1993;67:415–424. doi: 10.1128/jvi.67.1.415-424.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plopper C G, Dallas M H, Buckpitt A R. Clara cells. In: Crystal R G, West J B, Weibel E R, Barnes P J, editors. The lung: scientific foundations. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 517–533. [Google Scholar]
- 51.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabson A B, Graves B J. Synthesis and processing of viral RNA. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 205–261. [PubMed] [Google Scholar]
- 53.Ridgway A A, Kung H J, Fujita D J. Transient expression analysis of the reticuloendotheliosis virus long terminal repeat element. Nucleic Acids Res. 1989;17:3199–3215. doi: 10.1093/nar/17.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawaya P L, Stripp B R, Whitsett J A, Luse D S. The lung-specific CC10 gene is regulated by transcription factors from the AP-1, octamer, and hepatocyte nuclear factor 3 families. Mol Cell Biol. 1993;13:3860–3871. doi: 10.1128/mcb.13.7.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stunzi H, Head K W, Nielsen S W. International histological classification of tumours of domestic animals: tumours of the lung. Bull W H O. 1974;50:9–19. [PMC free article] [PubMed] [Google Scholar]
- 56.Voelker D R, Mason R J. Alveolar type II epithelial cells. In: Massaro D, editor. Lung cell biology. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 358–487. [Google Scholar]
- 57.Weiss R A. Retrovirus receptors. Cell. 1995;82:531–533. doi: 10.1016/0092-8674(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 58.Whitsett J A, Glasser S W. Regulation of surfactant protein gene transcription. Biochim Biophys Acta. 1998;1408:303–311. doi: 10.1016/s0925-4439(98)00076-3. [DOI] [PubMed] [Google Scholar]
- 59.Wikenheiser K A, Vorbroker D K, Rice W R, Clark J C, Bachurski C J, Oie H K, Whitsett J A. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90:11029–11033. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the Jaaksiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou L, Lim L, Costa R H, Whitsett J A. Thyroid transcription factor-1, hepatocyte nuclear factor-3beta, surfactant protein B, C, and Clara cell secretory protein in developing mouse lung. J Histochem Cytochem. 1996;44:1183–1193. doi: 10.1177/44.10.8813084. [DOI] [PubMed] [Google Scholar]