Abstract
RNA polymerase complexes were purified from Cryptosporidium parvum, a parasitic protozoan known to infect many species of mammals including humans. Western blot analysis revealed the association of the complexes with two different proteins, encoded by large and small segments of viral double-stranded RNAs. Each complex was found to contain only double-stranded RNA, both double- and single-stranded RNA, or only single-stranded RNA. Maximum RNA-dependent RNA polymerase activity was observed within the complexes containing both double- and single-stranded RNAs. These complexes possessed both transcriptase and replicase polymerase activities. Virus-like particles with a diameter of 31 nm were copurified with RNA polymerase complexes, and buoyant density and polymerase studies suggest that C. parvum harbors a putative double-stranded RNA virus which separately encapsidates the large and small RNA segments. The mechanism of replication and other characteristics of this virus are similar to those of the viruses of the family Partitiviridae, previously identified only in fungi and plants.
Cryptosporidium parvum is an intestinal intracellular parasitic protozoan of the phylum Apicomplexa which infects a wide variety of mammalian species. Infection results in diarrhea that is self-limiting in most cases but may be life threatening in immunocompromised individuals. C. parvum is one of the opportunistic infections in AIDS patients, and thus far, there is no effective treatment for cryptosporidiosis (8). Because of the lack of adequate treatment, other types of therapeutic targets are being sought, and extrachromosomal genetic elements are considered potential targets. For example, the extranuclear plastid genomes of Plasmodium and Toxoplasma spp., both parasites distantly related to C. parvum, are essential for parasite survival, and inhibition of plastid replication blocks the multiplication of parasites (9).
All isolates of C. parvum tested thus far are known to harbor both large (L) and small (S) extrachromosomal viral double-stranded RNA (dsRNA) segments (17, 19). The cDNAs of both dsRNAs have been cloned and sequenced, and only a single long open reading frame (ORF) has been identified in each dsRNA. The deduced protein sequence of the L-dsRNA (1,786 nucleotides [nt]) has similarity with viral RNA-dependent RNA polymerases (RDRP). The sequence of the putative protein encoded by the S-dsRNA (1,374 nt) has no significant similarity with polypeptide sequences in databases (17). The presence of RDRP activity in crude lysates of C. parvum oocysts, as well as replicative intermediates (RI) and full-size plus strands, which may represent mRNAs of the dsRNAs, suggests that these molecules are transcribed in the parasite (18). Additional indirect evidence of protein coding capacity of the dsRNAs exists as a result of a comparative analysis of the sequences of dsRNAs from a number of isolates of C. parvum with two distinct genotypes. The majority of nucleotide substitutions occur in the third positions of the codons in putative ORFs and fail to change the protein sequences (19). However, to date, there is no direct evidence for the presence of proteins encoded by the dsRNAs, nor have virus particles been identified in the previous studies (17–19).
Extrachromosomal dsRNAs are very common in protozoa (24, 28, 29), fungi (2, 7, 11, 30), and plants (21). Although many of these genomes have no “visible” effect on the host, some dsRNAs do appear to influence host biology (20). For example, yeasts from several genera contain dsRNA viruses encoding toxins that kill other strains not synthesizing the protein (26, 30). Infection of the chestnut blight fungus, Cryphonectria parasitica, with dsRNA associated with membranous vesicles causes reduction of virulence (hypovirulence) of the fungus (20, 23). However, the effect of extrachromosomal dsRNAs on the biology of C. parvum and the routes of transmission are as yet unknown. The purpose of the present study was to further examine dsRNAs, in respect to protein coding capacity, complex formation, and replication mechanism, with the aim of helping determine the relationship of the segments with other known dsRNA genetic elements and to help understand the role of these dsRNAs within the parasite.
MATERIALS AND METHODS
Parasite and oocyst purification.
The KSU-1 isolate of C. parvum was used in this study. Oocysts were purified from feces of infected 5-day-old calves by CsCl gradient centrifugation as described previously (27).
RNA polymerase assay.
The assay was carried out in 50-μl reaction volumes containing 50 mM Tris-HCl, pH 7.4; 100 mM NaCl; 10 mM MgCl2; 10 mM dithiothreitol; 1 mM (each) GTP, CTP, and ATP; 10 μCi of [α-32P]UTP (3,000 Ci/mmol); 0.5 mg of actinomycin D per ml; and aliquots of crude lysate of C. parvum oocysts or fractions from sucrose or CsCl gradients. After incubation at 37°C for 1 h, reactions were terminated by addition of 50 μl of stop solution (0.2% sodium dodecyl sulfate [SDS], 50 mM EDTA, 0.5 mg of proteinase K, 3 μg of total yeast RNA) and incubated at 37°C for 10 min. In pulse-chase experiments, and before termination of the reaction with stop solution, cold UTP was added to a final concentration of 1 mM and the mixture was incubated for an additional 30 min at 37°C. Nucleic acids from one-half of these reaction mixtures were twice precipitated with 2 M NH4OAc and 3 volumes of ethanol. Pellets were washed with 70% ethanol, dried in a vacuum, dissolved in 20 μl of TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA), and then resolved on 0.8% agarose gels. After electrophoresis, gels containing 32P-labeled products of the RNA polymerase assays were incubated for 20 min in 10% trichloroacetic acid and then dried for 3 h between filter paper prior to exposure to X-ray film at −70°C with an intensifying screen. Quantitation of incorporation of 32P into nucleic acids was estimated by counting the radioactivity of the second aliquot of the reaction mixture after trichloroacetic acid precipitation as described previously (18).
Northern blot hybridization.
Agarose gel electrophoresis on 1.2% nondenaturing gels and Northern blot hybridization at high stringency with 32P-labeled RNAs were performed as described previously (10).
Expression of ORFs of dsRNA in Escherichia coli and preparation of antiserum to recombinant proteins.
All standard DNA manipulations were performed as previously described (25). The numbering is based on the published sequences of the L- and S-dsRNAs (17). DNA fragments, containing entire coding sequences including start and terminator codons (nt 134 to 1708 and 248 to 1207 for L- and S-dsRNAs, respectively), were generated on total nucleic acids purified from oocysts by reverse transcription-PCR using avian myeloblastosis virus reverse transcriptase (Promega) and High Fidelity Taq DNA polymerase (Boehringer Mannheim). The set of primers for L-dsRNA was as follows: sense, LVN-1, 5′-ctggatccATGAAGTTTGTCAATATCTATG, and antisense, LVC-1, 5′-gggtcgacTTATCCATAAATTTTGTGACTC; the set for S-dsRNA was as follows: sense, SVN-1, 5′-ctggatccATGATTACAAGTTTTGAATCAA, and antisense, SVC-1, 5′-aagtcgacCTAATGGGAGCGATCTGCGCTA. Lowercase letters represent nucleotides used for cloning purposes. The sites for restriction endonuclease BamHI are underlined, and the sites for SalI are double underlined. Start codons in sense primers and stop codons in antisense primers are italicized. The amplified products were digested with the restriction endonucleases BamHI and SalI and ligated into the bacterial expression vector pET-28(a+) (Novagen) previously digested with the same enzymes. This bacterial expression vector was chosen because it has previously been shown to allow for high expression of C. parvum genes (16). The recombinant plasmids were introduced into the E. coli strain BL21(DE3) (Novagen). The fusion proteins, containing N-terminal peptides (34 amino acids) with six histidine residues (His tag) encoded by vector DNA and polypeptides encoded by the ORF of the dsRNA, were synthesized in E. coli after induction with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Recombinant proteins were found in inclusion bodies and purified on His Bind columns under denaturing conditions according to the manufacturer's recommendations (Novagen). Purified recombinant proteins were dialyzed overnight against phosphate-buffered saline (PBS) and used to produce polyclonal antibodies in rats as described previously (15). Antibodies against some E. coli proteins were removed by adsorption of antiserum with concentrated lysates of E. coli strain BL21(DE3) containing only the cloning vector as described previously (25).
Western blot analysis.
Proteins were resolved by electrophoresis on SDS–10% polyacrylamide gels and then electrotransferred to NitroPure membrane (Micron Separations Inc.). Blots were blocked for 2 h in PBS containing 5% nonfat dry milk and 0.05% Tween 20 (blocking solution). Blots were then incubated for 2 h with the rat antiserum (diluted 1:100 in blocking solution) to recombinant proteins. The blots were then washed six times for 5 min each in PBS and incubated for 1 h with a horseradish peroxidase-coupled goat anti-rat secondary antibody (Sigma). Blots were developed with 4-chloro-1-naphthol as substrate.
Isolation of VLPs.
Ten billion purified CsCl oocysts were excysted by incubation at 37°C for 90 min in 5 ml of PBS. Mixtures of sporozoites and nonexcysted oocysts were harvested by low-speed centrifugation (2,000 × g, 3 min), resuspended in 15 ml of buffer A (50 mM Tris-HCl, pH 7.4; 100 mM NaCl; 10 mM MgCl2; 10 mM dithiothreitol), and frozen at −70°C. All other steps of virus-like particle (VLP) isolation were performed at 4°C. After thawing of sporozoites-oocysts on ice, the cellular debris and nuclei were removed by centrifugation at 2,000 × g for 5 min and then twice at 10,000 × g for 10 min. Five milliliters of clarified supernatant (crude lysate) was layered on top of a 10 to 40% (wt/wt) linear sucrose gradient in buffer A (35 ml) and centrifuged at 100,000 × g for 3 h; fractions of 2 ml were collected. In other experiments, crude lysates were extracted twice with an equal volume of chloroform. The aqueous phase was collected and centrifuged at 10,000 × g for 10 min. A portion of the 4.5-ml clarified supernatant was pelleted through a cushion of 1 ml of 15% sucrose by centrifugation at 100,000 × g for 2 h. The pellet was resuspended by sonication in 1.2 ml of buffer A and loaded atop a preformed gradient of CsCl (1 ml with 1.5 g/ml, 1 ml with 1.4 g/ml, 1.5 ml with 1.3 g/ml, and 1 ml with 1.2 g/ml) and centrifuged at 100,000 × g for 16 h. Thirty fractions of 180 μl each were collected and dialyzed against buffer A overnight. Each fraction of the gradient was tested for the presence of (i) RNA polymerase activity; (ii) dsRNAs and single-stranded RNAs (ssRNAs), by Northern blot hybridization; and (iii) proteins encoded by ORFs of both segments of dsRNAs, by Western blot analysis.
Electron microscopy.
Aliquots of the CsCl gradient fractions were deposited onto carbon-coated 200-mesh copper grids, negatively stained with 2% uranyl acetate, and visualized with a Philips 201 transmission electron microscope for the presence of VLPs.
RESULTS
Stability of RNA polymerase activity.
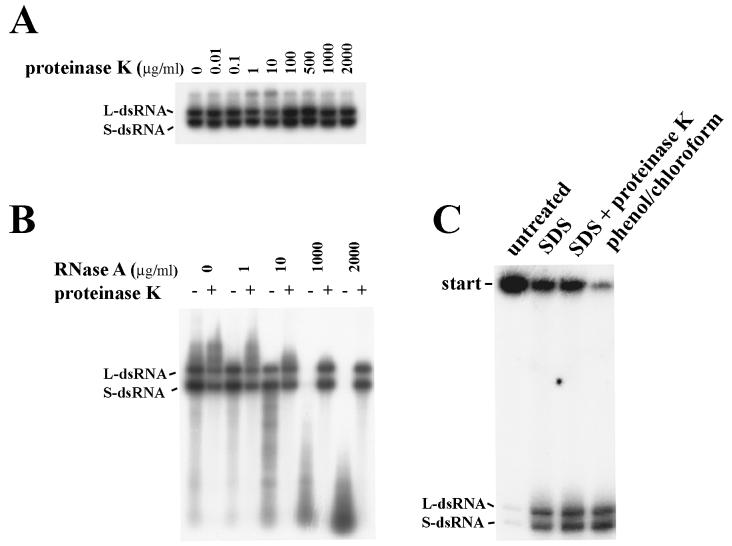
Incubation of crude oocyst lysates with proteinase K (up to 2 mg/ml) (Fig. 1A) did not have a significant effect on RNA polymerase activity, which suggests that the polymerase might be protected from digestion. When RNase A was eliminated by digestion with proteinase K before the RNA polymerase assay, treatment of lysate with RNase (up to 2 mg/ml) (Fig. 1B) did not change incorporation of 32P into nucleic acids. This suggests that templates for RNA polymerase are protected. When reactions were performed in the presence of 10 μg of RNase A per ml, 32P-labeled products of the polymerase assay were visible on autoradiograms (Fig. 1B). This suggests that the newly synthesized products of polymerase assay are also protected, as the purified dsRNAs and ssRNAs have been shown to be completely degraded at this RNase A concentration (17). Products of RNA polymerase activity are more likely to form complexes with the protein(s), as they do not migrate into the polyacrylamide gels if loaded directly from the polymerase mixture (Fig. 1C). However, 32P-labeled L- and S-dsRNAs were resolved by electrophoresis when complexes were destroyed before loading onto gels by incubating the polymerase mixture with 0.1% SDS, or SDS and 0.5 mg of proteinase K per ml, or extraction with phenol-chloroform (Fig. 1C). The extraction of crude lysate three times with organic solvents (butanol or chloroform) prior to the RDRP assay did not have any effect on RNA polymerase activity (data not shown). This may be explained by RNA polymerase activity not being associated with membranous structures.
FIG. 1.
Resistance of RNA polymerase activity in crude oocyst lysates. 32P-labeled products of RNA polymerase assays were resolved by electrophoresis on 0.8% agarose gels (A and B) or on a 6% polyacrylamide gel (C) under nondenaturing conditions and were then exposed to X-ray film. (A) Assays were performed in the presence of different concentrations of proteinase K. (B) Aliquots were incubated for 30 min at 37°C with different concentrations of RNase A, then RNase was destroyed by treatment with proteinase K (0.5 mg/ml, 30 min at 37°C), and finally [α-32P]UTP was added and the mixture was incubated for 1 h at 37°C. (C) RNA polymerase mixtures were untreated, incubated with 0.1% SDS or 0.1% SDS–0.5 mg of proteinase K per ml, or extracted once with phenol-chloroform and then loaded on the gel.
Some viral RNAs are associated with RNA polymerase activity, and others are not.
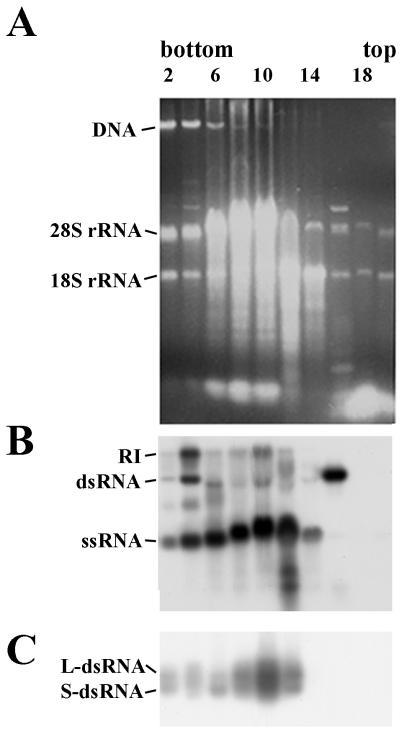
Results presented above suggest that viral RNA might be associated with RNA polymerase. Attempts were made to obtain evidence of copurification of viral RNAs and RNA polymerase. Crude oocyst lysates were fractionated by sucrose gradient centrifugation. The fractions were collected and analyzed for the presence of viral RNAs by staining with ethidium bromide (Fig. 2A), Northern blot hybridization (Fig. 2B), and also RNA polymerase activity (Fig. 2C). dsRNA and ssRNA (plus strands) were identified in fractions 2 to 16 (Fig. 2B); single-stranded minus strands were not found (data not shown). Free minus strands were not identified previously in crude oocyst extracts (17). RI and RNA polymerase activity were found only in fractions 2 to 12 and not in fractions 14 to 16 (Fig. 2B and C). These data suggest that at least two types of viral RNAs might be found in oocysts. Only one type was associated with polymerase activity. More likely, RNAs from fractions 14 to 16 are free, i.e., not bound to the protein(s), as they were localized in low-density fractions from the bulk of rRNAs derived from ribosomes (fractions 6 to 12) (Fig. 2A).
FIG. 2.
Presence of two distinct types of viral RNAs, with one of these associated with RNA polymerase activity. Crude lysates of oocysts were fractionated by sucrose gradient centrifugation, and the fractions were analyzed for the presence of viral RNA and RNA polymerase activity. (A) Ethidium bromide-stained 0.8% agarose gel with nucleic acids purified from even fractions of gradient. (B) Northern blot analysis, where nucleic acids were extracted from fractions and separated on agarose gels under nondenaturing conditions, then denatured in situ, and transferred to the filter. Hybridization was performed with 32P-labeled riboprobes complementary to the plus strand of the L-dsRNA. A similar pattern of hybridization was observed with riboprobes complementary to the plus strand of the S-dsRNA (data not shown). In experiments with 32P-labeled riboprobes complementary to the minus strands of the L- and S-dsRNAs, hybridization occurred only with RI and dsRNA (data not shown). (C) Autoradiogram of 32P-labeled products of RNA polymerase assay resolved by electrophoresis on an 0.8% agarose gel.
Association of RNA polymerase complexes with the VLPs.
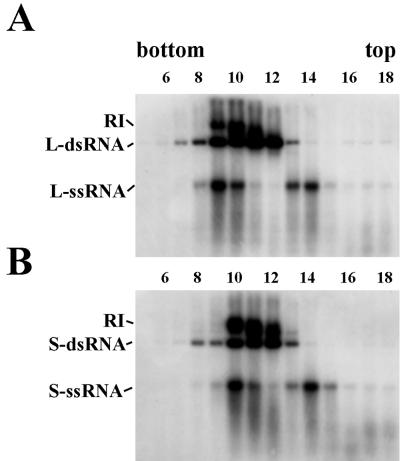
To further characterize the RNA polymerase complex, crude lysates of oocysts were twice extracted with chloroform, pelleted through a sucrose cushion, and fractionated by isopycnic centrifugation on CsCl density gradients. Northern blot analysis (Fig. 3) revealed that the lighter fractions (14 for L-probe and 14 and 15 for S-probe) contained mainly ssRNAs (plus strands) whereas the heaviest (7 for L-probe and 8 for S-probe) contained only dsRNA. Different types of RNA (dsRNA, ssRNA, and RI) were found together in fractions 9 to 11 for L-probe and 10 to 12 for S-probe. RI appeared to be growing in size, reaching maximum lengths in the heaviest fractions (9 for L-probe and 10 for S-probe). In these fractions, the highest amounts of ssRNA were detected (Fig. 3A, compare fraction 9 with fractions 10 and 11 for L-probe; Fig. 3B, compare fraction 10 with fractions 11 and 12 for S-probe). This suggests that, once synthesis of new strand is completed, one of the plus strands is released from the RI. More likely, the released strand still stays as a complex with the protein(s), as free ssRNA and dsRNA were removed (by centrifugation through a cushion of sucrose) prior to loading on CsCl gradients. RNA polymerase assays were performed on each fraction, and activity was found in fractions 8 to 14 with maximum activity in fractions 9 to 12 (see below). This suggests cosedimentation of viral RNAs with polymerase activity. All three kinds of viral RNA (dsRNA, ssRNA, and RI) were identified in the heaviest fractions when hybridization was performed with probes to L-dsRNA and then with probe to S-dsRNA (compare Fig. 3A and B). This indicates that large and small segments are associated with different polymerase complexes and that the complexes can be separated based on their densities on CsCl gradients.
FIG. 3.
Distribution of viral RNAs in CsCl fractions. Nucleic acids were purified from fractions from CsCl gradients, resolved on an 0.8% agarose gel under nondenaturing conditions, transferred to nitrocellulose filters, and hybridized with 32P-labeled riboprobe complementary to plus strands of L-dsRNAs (A) or S-dsRNAs (B). Only fractions 5 to 18 out of 30 total are shown.
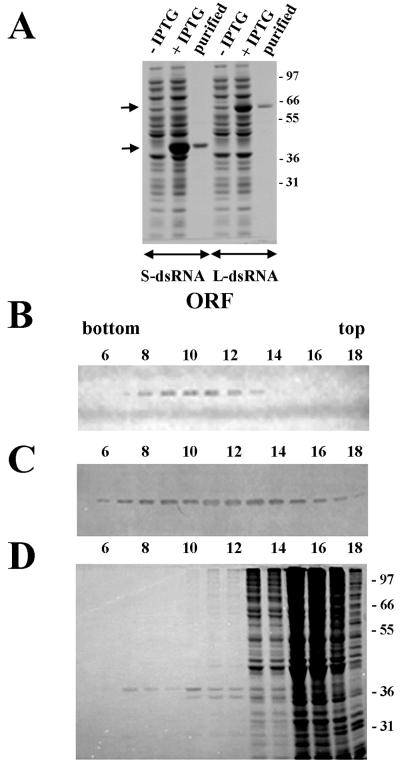
Because it was unknown whether the ORFs of the dsRNAs really encoded proteins, both ORFs were expressed in E. coli (Fig. 4A) and antiserum raised to the recombinant proteins was used to probe Western blots containing proteins from CsCl fractions. Antibodies to the protein encoded by the ORF of the L-dsRNA (putative RDRP) (17) recognized a band with a molecular size of 65 kDa in fractions 8 to 13 (Fig. 4B). Antisera to protein from the S-dsRNA (protein with unknown function) (17) identified a band with a molecular size of 37 kDa in fractions 6 to 17 (Fig. 4C). The sizes of both bands were similar to those predicted from amino acid sequences. This suggests that extensive modifications to the proteins are probably lacking. Coomassie blue staining of the SDS-polyacrylamide gel (Fig. 4D) showed that fractions 8 to 12 contained few proteins; one of them (37 kDa) was identified by Western blot analysis and the other proteins more likely had a host origin. The putative RDRP (65-kDa protein) was not visible, perhaps because this protein may be less abundant than the 37-kDa protein.
FIG. 4.
Distribution of proteins encoded by ORFs of the L- and S-dsRNAs in CsCl gradient fractions. (A) Expression of the ORFs of the L- and S-dsRNAs in E. coli. Shown is a Coomassie blue-stained SDS–10% polyacrylamide gel with total proteins from uninduced (−IPTG) and induced (+IPTG) E. coli cells and affinity-purified recombinant proteins. Arrows show positions of recombinant proteins. Molecular size standards are shown in kilodaltons in the right margin. (B and C) Immunoblot analysis of the proteins from the CsCl fraction using antibodies raised against recombinant proteins of L-dsRNA (B) and S-dsRNA (C). (D) Coomassie blue-stained SDS–10% polyacrylamide gel with the proteins from CsCl fractions. Molecular size standards are shown in kilodaltons in the right margin. Only fractions 5 to 18 out of 30 total are shown.
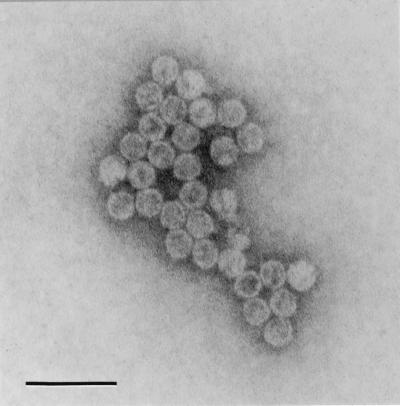
Electron microscopy analysis of CsCl gradient fractions revealed the presence of isometric VLPs with a diameter of about 31 nm in fractions 7 to 18. The concentration and purity of these particles were the highest in fractions 8 to 12 (density, 1.439 to 1.391 g/ml). Figure 5 shows particles from fraction 10. Observations of viral RNAs, RNA polymerase activity, proteins encoded by both dsRNAs, and the particles themselves in these same fractions of CsCl gradient strongly indicate their association and suggest that C. parvum harbors a potential dsRNA virus.
FIG. 5.
Electron micrograph of VLPs from fraction 10 (Fig. 3) of CsCl gradient. Bar = 100 nm.
Presence of transcriptase and replicase activities in identical fractions of CsCl gradient.
When RNA polymerase mixtures of the pulse experiment were resolved on nondenaturing agarose gels, several bands were visualized on autoradiograms (Fig. 6A). The heaviest fractions, from 8 to 12, synthesized products migrating as RI, dsRNA, and low-molecular-size material. Fraction 8 incorporated 32P predominantly into the band migrating as the L-dsRNA, and fraction 12 incorporated 32P mainly into the band migrating as the S-dsRNA. These data provide additional evidence that L- and S-dsRNAs form different complexes with polymerase and can be separated according to their buoyant density. Fractions 10 and 11 synthesized approximately similar amounts of L- and S-dsRNA. The lightest, fraction 14, incorporated 32P mainly in the bands migrating as ssRNA. In the pulse-chase experiment (Fig. 6B), most of the radioactivity was incorporated into dsRNA in all fractions. So far, it is difficult to find an explanation of why fraction 14 synthesizes ssRNA in pulse and double-stranded molecules in pulse-chase experiments. The amount of RI and the smaller-size products was decreased in fractions 8 to 12, compared to the amounts in the pulse experiment. The smaller-molecular-size molecules in fractions 8 to 12 most likely represent incomplete products synthesized at a limiting concentration of one of the nucleotides, as these small-size products were converted to full-size molecules at a higher concentration (1 mM) of all four nucleotides (pulse-chase experiment) (compare Fig. 6A and B). To confirm the chemical structure of the polymerase products, the 32P-labeled RNAs from fractions 10 and 14 were purified from the pulse experiment by phenol-chloroform extraction and subjected to RNase A digestion in low- and high-salt buffer. Figure 6C shows that the products of fraction 10 were mostly dsRNA, as they were not degraded in the presence of RNase A and 0.6 M NaCl, and that the products of fraction 14 were mostly ssRNA. To verify the polarity of the synthesized products, the 32P-labeled RNAs from fractions 10 and 14 were used as probes in hybridization experiments with unlabeled plus and minus transcripts obtained from cDNAs of the L- and S-dsRNAs (Fig. 6D). These experiments revealed that 32P-labeled plus and minus strands were present in fraction 10 and indicate that two polymerase activities exist in this fraction, transcriptase and replicase. The intensity of the hybridization signals suggests that fraction 10 synthesizes more plus strands than minus strands. Only replicase activity was observed in fraction 14, as the 32P-labeled products hybridized mostly with unlabeled transcripts, corresponding to plus strands of the L- and S-dsRNAs. The synthesis of minus strands most likely occurs on single-stranded plus strands, as only these kinds of molecules were identified in this fraction by Northern blot analysis (Fig. 3).
FIG. 6.
Association of transcriptase and replicase activities with fractions of CsCl gradients. (A and B) 32P-labeled products from pulse (A) and pulse-chase (B) RNA polymerase assays resolved by electrophoresis on 0.8% agarose and exposed to X-ray film. (C) Effect of RNase on products of RNA polymerase reactions from fractions 10 and 14. Purified products were treated with RNase A (10 μg/ml, for 30 min at 37°C) in the absence (−) or presence (+) of 0.6 M NaCl. After treatment, RNA was separated on an 0.8% agarose gel and detected by autoradiography. (D) Identification of the polarity of the RNA polymerase products from fractions 10 and 14. Unlabeled transcripts corresponding to minus and plus strands of the L-dsRNA (L) and S-dsRNA (S) were synthesized on cDNA clones using T7 and T3 RNA polymerases. Usually, two bands were identified by ethidium bromide staining of the products of the reaction, as was observed previously (18). Then unlabeled products were electrophoresed on 1.2% agarose, transferred to filters, and probed with 32P-labeled products of polymerase reactions synthesized by fractions 10 and 14.
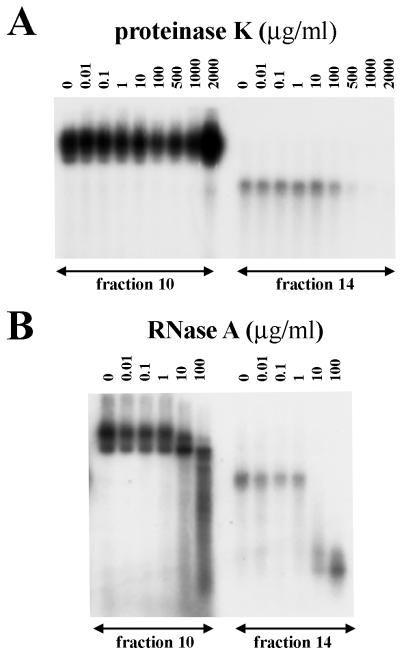
RNA polymerase assays were performed on fractions 10 and 14 in the presence of different concentrations of proteinase K (Fig. 7A) and RNase A (Fig. 7B). RNA polymerase activity in fraction 10 was more resistant to both treatments than was activity in fraction 14. The products of polymerase activity were still observed in fraction 14 after treatment with 1 μg of RNase A per ml. These results indicate that newly synthesized ssRNAs are protected in this fraction, as this concentration of RNase A is known to completely degrade purified ssRNA (17). The protection assays provide evidence of encapsidation of viral RNAs and RNA polymerase into the particles.
FIG. 7.
Resistance of viral RNA polymerase activity in fractions 10 and 14 of the CsCl gradient. 32P-labeled products of RNA polymerase assays were resolved by electrophoresis on an 0.8% agarose gel and exposed to X-ray film. Assays were performed in the presence of different concentrations of proteinase K (A) or RNase A (B).
DISCUSSION
This study demonstrates association of enzymatically active RNA polymerase complexes with VLPs. The population of complexes can be divided into two groups. The first type, with a higher density, contains a large segment, and the second, with the lower density, contains a small segment of viral RNA. Each type represents a heterogeneous system and can be separated into three classes according to the type of copurified nucleic acids: complexes with only dsRNA, complexes with only ssRNA, and complexes with both dsRNA and ssRNA. The proteins encoded by the ORFs for both dsRNAs were identified and found to be associated with RNA polymerase complexes. This is direct evidence for the expression of dsRNA genes in the oocysts of C. parvum. The study of RNA polymerase activity suggests encapsidation of viral RNAs and RNA polymerase into the particles. If the RNAs are encapsidated into VLPs, then the functions of both proteins can be explained. The protein product of the L-dsRNA is most likely a polymerase and has significant similarity with RDRP from viruses of the family Partitiviridae (17). The product of the S-dsRNA, which is more abundant than polymerase, most likely represents the capsid protein. It is in agreement with the hypothesis that all dsRNA viruses share a common capsid architecture and have a 120-capsid subunit and one or fewer molecules of polymerases (5, 14).
Using a taxonomic key for the placement of viruses in taxa (22), it is possible to unambiguously classify the virus of C. parvum as a new member of the family Partitiviridae. Table 1 shows a comparison of the main features of viruses from this family with those of the C. parvum virus (CPV). The partitiviruses include four genera, Partitivirus, Chrysovirus, Alphacryptovirus, and Betacryptovirus. Up until this time, all members of the Partitiviridae have been found in only fungi and plants. It appears that CPV is the first member of the family in a protozoan host. The other dsRNA viruses of protozoa belong to the family Totiviridae (13).
TABLE 1.
Comparison of the main characteristics of viruses of the family Partitiviridae and a CPV
| Property | Partitiviridaea | CPVb |
|---|---|---|
| Localization in the host cell | Cytoplasm | Cytoplasm |
| Particle diameter (nm) | 30–40 | 31 |
| Shape of virion | Isometric, nonenveloped | Isometric, nonenveloped |
| Density in CsCl (g/ml) | 1.34–1.39 | 1.35–1.46 |
| Stability of virion | Stable in butanol and chloroform | Stable in butanol and chloroform |
| Capsid protein | Single capsid protein, 42–73 or 125 kDa | May be a single capsid protein, 37 kDa |
| Nucleic acids | Two monocistronic dsRNA segments, 1.4–3.0 kbp, each segment separately encapsidated | Two monocistronic dsRNA segments, 1.4 and 1.7 kbp, each segment separately encapsidated |
| Satellite and defective dsRNAs | Some viruses contain additional dsRNA segments, which are considered satellite or defective | Not found |
| Large dsRNA segment | Encodes RDRP | Encodes RDRP |
| Small dsRNA segment | Encodes capsid protein | May encode capsid protein |
| Mode of transcription | Semiconservative | Semiconservative |
| RNA polymerase activity | Associated with virion | Associated with virion |
| Particles with RI | Particles with ssRNA and particles with both dsRNA and ssRNA | Particles with ssRNA and particles with both dsRNA and ssRNA |
| Mode of transmission | Do not have extracellular phase; transmitted vertically | Unknown |
| Host | Fungi, plants | Protozoan parasite C. parvum, both genotype 1 and genotype 2 |
| Genera | Partitivirus, Chrysovirus, Alphacryptovirus, Betacryptovirus | May represent new genus according to protozoan host |
| Effect on host | Many with no effect | Unknown |
Analysis of viral RNAs by Northern blot analysis and products of RNA polymerase assays in the CsCl gradient fractions allow us to identify intermediates of the multiplication cycle and propose an in vitro model of virus replication. For simplicity, only replication of the L-dsRNA will be discussed below. The RNA polymerase from the particles with lighter density (1.348 g/ml), containing mainly single-stranded plus strands (fraction 14, Fig. 3A), synthesizes a minus strand using plus strand as template (replication). Newly synthesized minus strand anneals with plus strand and forms dsRNA, as was observed in pulse-chase experiments (Fig. 6B). One molecule of dsRNA per particle would be a result of this first step, and these particles should have higher density. Only dsRNA was found in fraction 12 with a density of 1.391 g/ml (Fig. 3A). These dsRNAs more likely appeared from the particles with only one molecule of dsRNA. The majority (70%) of polymerase activity was found in fractions 9 to 11 (Fig. 6A and B) containing dsRNA, ssRNA, and RI (Fig. 3). Transcriptase and replicase activities were observed in these same fractions. The polymerase synthesizes a new plus strand (transcription) by a semiconservative mechanism, as was shown previously (18). Thus, the synthesis of a new strand occurs through RI in fractions 9 to 11 (Fig. 3A) using the dsRNA as template. When synthesis is completed (fraction 9), parental plus strand is released from the RI but still stays inside the particle. This ssRNA most likely serves as a template for synthesis of the complementary minus strand (replication), forming a new dsRNA molecule. As a result of transcriptase and replicase activities, one particle could have two identical molecules of dsRNA. These particles should have the highest buoyant density in the CsCl gradient. Indeed, only dsRNAs were detected in fraction 7 (density, 1.460 g/ml) by Northern blot hybridization (Fig. 3A). These dsRNAs could be derived from particles with two molecules of dsRNA.
Two identical molecules of dsRNA in the same particle have been found previously in viruses of the family Partitiviridae, for example, from Penicillium stoloniferum (1) and Aspergillus foetidus (3). It was proposed previously that these particles could be uncoated in vivo, followed by RNA polymerase activity to produce copies of the mRNA using uncoated dsRNA as a template (4). The presence of free viral dsRNA and ssRNA in crude lysates of oocysts of C. parvum may offer some support for this model, but RNA polymerase activity was not found to be associated with these RNAs in vitro. An alternative possibility is that particles with two molecules of dsRNA synthesize and release mRNA, as was shown previously for VLPs from yeast with M1 dsRNA encoding a killer toxin (6). These authors proposed a head-full type of replication mechanism. The M1 virus is a satellite of L-A virus from the family Totiviridae, and the M1 dsRNA is packaged and replicates in L-A particles. L-A particles have a structure primarily adapted to encapsidate only one molecule of L-A dsRNA (10); however, because the M1 dsRNA (1.8 kbp) is less than half the size of L-A dsRNA (4.6 kbp), the L-A particles can comfortably encapsidate one or two molecules of M1 dsRNA as well (6).
The dsRNA virus of C. parvum has features most similar to viruses of the family Partitiviridae (Table 1). However, the head-full replication mechanism of satellite viruses of the family Totiviridae seems to be the most likely mechanism for the replication of both the C. parvum dsRNA virus and other partitiviruses. Since partitiviruses are thought have evolved from the totiviruses by dividing their genome between two separately encapsidated dsRNA segments (11), then the head-full mechanism of replication is plausible. The capsids of the partitiviruses might have an architecture and size similar to those of the ancestral totiviruses, which may allow them to encapsidate one or two identical molecules of dsRNA, as the C. parvum dsRNAs are around one-half the size of the putative ancestral RNA.
Several important issues about the CPV still remain unknown. Both the route of transmission and the relative importance of this virus for parasite survival or pathogenicity are areas of research yet to be explored. However, based on our data and what is known for other related viruses, it is likely that extracellular, horizontal transmission of the virus does not occur and the virus are passed only during cell division and gamete fusion. Thus, it is likely that the virus will be shown to have little or no negative effect on the host, as in the case for partitiviruses in general.
ACKNOWLEDGMENTS
We acknowledge A. Q. Pauson for help with electron microscopy. We also thank R. A. Consigli for critically reading the manuscript and for helpful advice during experiments.
This work was supported by NIH grant 1RO1AI/DK42545-01A1 to S.J.U. and N.V.K. and by EPA grant R825148-01-0 to S.J.U.
Footnotes
Kansas Agricultural Experiment Station contribution no. 00-301-J.
REFERENCES
- 1.Buck K W. Replication of double-stranded RNA in particles of Penicillium stoloniferum virus S. Nucleic Acids Res. 1975;2:1889–1902. doi: 10.1093/nar/2.10.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck K W. Molecular variability of viruses of fungi. In: Bridge P, Couteaudier Y, Clarkson J, editors. Molecular variability of fungal pathogens. Oxford, United Kingdom: CAB International; 1998. pp. 53–72. [Google Scholar]
- 3.Buck K W, Ratti G. Biophysical and biochemical properties of two viruses isolated from Aspergillus foetidus. J Gen Virol. 1975;27:211–224. doi: 10.1099/0022-1317-27-2-211. [DOI] [PubMed] [Google Scholar]
- 4.Buck K W, Ratti G. A model for the replication of double-stranded ribonucleic acid mycoviruses. Biochem Soc Trans. 1975;3:542–544. doi: 10.1042/bst0030542. [DOI] [PubMed] [Google Scholar]
- 5.Cheng R H, Caston J R, Wang G J, Gu F, Smith T J, Baker T S, Bozarth R F, Trus B L, Cheng N, Wickner R B, Steven A C. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol. 1994;244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 6.Esteban R, Wickner R B. Three different M1 RNA-containing viruslike particle types in Saccharomyces cerevisiae: in vitro M1 double-stranded RNA synthesis. Mol Cell Biol. 1986;6:1552–1561. doi: 10.1128/mcb.6.5.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteban R, Rodriguez-Cousino N, Esteban L M. Genomic organization of T and W, a new family of double-stranded RNAs from Saccharomyces cerevisiae. Prog Nucleic Acids Res Mol Biol. 1993;46:155–182. doi: 10.1016/s0079-6603(08)61021-1. [DOI] [PubMed] [Google Scholar]
- 8.Fayer R, Speer C A, Dubbey J P. The general biology of Cryptosporidium. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press, Inc.; 1997. pp. 1–41. [Google Scholar]
- 9.Fichera M E, Roos D S. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 10.Fujimura T, Esteban R, Esteban L M, Wickner R B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 11.Ghabrial S A. Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes. 1998;16:119–131. doi: 10.1023/A:1007966229595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghabrial S A, Bozarth R F, Buck K W, Yamashita S, Martelli G P, Milne R G. Family Partitiviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy, classification and nomenclature of viruses, sixth report of the international committee on taxonomy of viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 253–260. [Google Scholar]
- 13.Ghabrial S A, Bruenn J A, Buck K W, Wickner R B, Patterson J L, Stuart K D, Wang A L, Wang C C. Family Totiviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy, classification and nomenclature of viruses, sixth report of the international committee on taxonomy of viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 245–252. [Google Scholar]
- 14.Hill C L, Booth T F, Prasad B V V, Grimes J M, Mertens P P C, Sutton G C, Stuart D I. The structure of a cypovirus and the functional organization of dsRNA viruses. Nat Struct Biol. 1999;6:565–568. doi: 10.1038/9347. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins M C, Fayer R, Tilley M, Upton S J. Cloning and expression of a cDNA encoding epitopes shared by 15- and 60-kilodalton proteins of Cryptosporidium parvum sporozoites. Infect Immun. 1993;61:2377–2382. doi: 10.1128/iai.61.6.2377-2382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khramtsov N V, Oppert B, Montelone B A, Upton S J. Sequencing, analysis and expression in Escherichia coli of a gene encoding a 15 kDa Cryptosporidium parvum protein. Biochem Biophys Res Commun. 1997;230:164–166. doi: 10.1006/bbrc.1996.5877. [DOI] [PubMed] [Google Scholar]
- 17.Khramtsov N V, Woods K M, Nesterenko M V, Dykstra C C, Upton S J. Virus-like, double-stranded RNAs in the parasitic protozoan, Cryptosporidium parvum. Mol Microbiol. 1997;26:289–300. doi: 10.1046/j.1365-2958.1997.5721933.x. [DOI] [PubMed] [Google Scholar]
- 18.Khramtsov N V, Upton S J. High-temperature inducible cell-free transcription and replication of double-stranded RNAs within the parasitic protozoan Cryptosporidium parvum. Virology. 1998;245:331–337. doi: 10.1006/viro.1998.9181. [DOI] [PubMed] [Google Scholar]
- 19.Khramtsov N V, Chung P A, Dykstra C C, Griffiths J K, Morgan U M, Arrowood M J, Upton S J. Presence of double-stranded RNAs in human and calf isolates of Cryptosporidium parvum. J Parasitol. 2000;86:275–282. doi: 10.1645/0022-3395(2000)086[0275:PODSRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.McCabe P M, Pfeiffer P, Van Alfen N K. The influence of dsRNA viruses on the biology of plant pathogenic fungi. Trends Microbiol. 1999;7:377–381. doi: 10.1016/s0966-842x(99)01568-1. [DOI] [PubMed] [Google Scholar]
- 21.Milne R G, Natsuaki T. Cryptoviruses. In: Singh R P, Singh U, Kohmoto K, editors. Pathogens and host specificity in plant disease. Vol. 3. Oxford, United Kingdom: Pergamon; 1994. pp. 239–247. [Google Scholar]
- 22.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy, classification and nomenclature of viruses, sixth report of the international committee on taxonomy of viruses. Vienna, Austria: Springer-Verlag; 1995. [Google Scholar]
- 23.Nuss D L, Koltin Y. Significance of dsRNA genetic elements in plant pathogenic fungi. Annu Rev Phytopathol. 1990;28:37–58. doi: 10.1146/annurev.py.28.090190.000345. [DOI] [PubMed] [Google Scholar]
- 24.Patterson J L. Viruses of protozoan parasites. Exp Parasitol. 1990;70:111–113. doi: 10.1016/0014-4894(90)90091-p. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schmitt M J, Neuhausen F. Killer toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J Virol. 1994;68:1765–1772. doi: 10.1128/jvi.68.3.1765-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Upton S J, Tilley M, Nesterenko M V, Brillhart D B. A simple and reliable method of producing in vitro infections of Cryptosporidium parvum (Apicomplexa) FEMS Microbiol Lett. 1994;118:45–49. doi: 10.1111/j.1574-6968.1994.tb06801.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang A L, Wang C C. Viruses of the protozoa. Annu Rev Microbiol. 1991;45:251–263. doi: 10.1146/annurev.mi.45.100191.001343. [DOI] [PubMed] [Google Scholar]
- 29.Wang A L, Wang C C. Viruses of parasitic protozoa. Parasitol Today. 1991;7:76–80. doi: 10.1016/0169-4758(91)90198-w. [DOI] [PubMed] [Google Scholar]
- 30.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]