Abstract
F6A, a molecular clone of subgroup A feline leukemia virus (FeLV) is considered to be highly infectious but weakly pathogenic. In recent studies with a closely related subgroup A molecular clone, FRA, we demonstrated high pathogenicity and a strong propensity to undergo recombination with endogenous FeLV (enFeLV), leading to a high frequency of transition from subgroup A to A/B. The present study was undertaken to identify mechanisms of FeLV pathogenesis that might become evident by comparing the two closely related molecular clones. F6A was shown to have an infectivity similar to that of FRA when delivered as a provirus. Virus load and antibody responses were also similar, although F6A-infected cats consistently carried higher virus loads than FRA-infected cats. However, F6A-infected cats were slower to undergo de novo recombination with enFeLV and showed slower progression to disease than FRA-infected cats. Tumors collected from nine pF6A- or pFRA-inoculated cats expressed lymphocyte markers for T cells (seven tumors) and B cells (one tumor), and non-T/B cells (one tumor). One cat with an A-to-A/C conversion developed erythrocyte hypoplasia. Genomic mapping of recombinants from pF6A- and pFRA-inoculated cats revealed similar crossover sites, suggesting that the genomic makeup of the recombinants did not contribute to increased progression to neoplastic disease. From these studies, the mechanism most likely to account for the pathologic differences between F6A and FRA is the lower propensity for F6A to undergo de novo recombination with enFeLV in vivo. A lower recombination rate is predicted to slow the transition from subgroup A to A/B and slow the progression to disease.
Feline leukemia virus (FeLV) is a naturally occurring, horizontally transmissible viral infection of cats (9, 25) that was first isolated in 1964 (14). While FeLV causes a wide range of neoplastic and cytosuppressive diseases, it is unclear if the diversity of disease is related to disease-specific variants of FeLV or to the genomic instability of the virus (17, 29). FeLV is divided into three subgroups (A, B, and C) based on the apparent binding of the large external envelope glycoprotein gp70 to subgroup- specific receptors (13, 32, 33). The weakly pathogenic FeLV subgroup A (FeLV-A) is commonly transmitted in nature (9, 10) but rarely leads to disease (5) until new subgroups, FeLV-B or FeLV-C, arise de novo as a result of recombination and/or mutation. FeLV-B is derived through recombination of exogenous FeLV-A with endogenous FeLV sequences and is associated with lymphoma or other myeloproliferative diseases (2, 4, 7, 15, 22, 23, 24, 30, 31, 34, 35). The origin of FeLV-C is less clear but may also involve recombination and/or mutation (13, 20, 27, 30, 31). FeLV-C is capable of inducing erythroid hypoplasia and immunosuppression (1, 6, 16, 19, 27, 28).
In recent studies neonatal cats were inoculated with plasmid DNA containing a full-length molecular clone of FeLV derived from the Rickard strain of FeLV-A (pFRA) (4). Because the challenge was genetically homogeneous, high-fidelity mapping of genomic changes could be documented as in vivo recombinants arise de novo. The cats inoculated with pFRA developed classic FeLV infection with chronic lifelong viremia, and four out of five animals showed enhanced tumor induction in a period of 28 to 55 weeks postinfection (p.i.), while the fifth cat underwent a subgroup A-to-A/B-to-A/B/C transition and developed anemia at 65 weeks p.i. (4). Interestingly, genetic evidence of recombination between exogenous FeLV and endogenous FeLV-like viruses was detected in the first few weeks p.i., followed by the transition to FeLV-A/B in the plasma at 12 weeks p.i. (4). In comparison, FeLV-B was rarely detected in the terminal tissues (30 to 78 weeks p.i.) and was not detected in the plasma from three out of seven chronically viremic cats infected with cell-free FeLV-A (11). This observation suggested that FRA was more recombinogenic and perhaps more virulent than other FeLV-A isolates or that the unusual composition of the challenge (DNA) or the route of challenge enhanced recombination and the pathogenic process. To further understand the mechanisms of increased pathogenesis of the pFRA challenge, a widely studied and closely related molecular clone, pF6A, with 98% homology to FRA, was inoculated into neonatal cats by the same route and at the same dosage previously used (4). Tissue culture derived F6A as a prototypic cell-free whole virus inoculum has been widely used in FeLV studies and is generally considered to be highly infectious but marginally pathogenic (20, 21, 26, 30). When the results of several studies are combined, the frequency of tumor induction was 4 in 28 cats held for between 50 and 116 weeks p.i. (the mean tumor incubation period was 69 weeks) (20, 21, 26, 30).
The present study was undertaken to gather additional information on the mechanisms of FeLV pathogenesis and specifically to determine if inoculation of the FeLV provirus accounts for the more-severe pathogenic disease pattern. For this purpose, plasmid DNA from the FeLV molecular clones pF6A and pFRA was inoculated into neonatal cats and the cats were monitored for viremia, anti-FeLV antibody titer, recombinant phenotype, proviral genome stability, and disease pattern. The results of the study show F6A to be less pathogenic and to have a lower recombinational rate than FRA.
MATERIALS AND METHODS
Animals.
Seven specific-pathogen-free neonate kittens from a commercial source (Liberty Research, Waverly, N.Y.) were utilized in this study. All work was performed in accordance with the University Laboratory Animal Care and Use Committee and the U.S. Department of Health, Education, and Welfare (37a).
FeLV plasmid challenge and sample collection.
Seven 24- to 48-h-old kittens from three litters were inoculated intradermally with 50 μg of DNA (either pF6A or pFRA) combined with 0.50 mg of N-[1(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP; Roche Diagnostic Corp., Indianapolis, Ind.) in a final volume of 0.5 ml of HEPES-buffered saline (4, 38). The challenge DNA for the FRA group was from the same DNA preparation used in a previous study (4). The size of the DNA inoculum and the age of kittens were also identical to those of the previous study (4). The DNA-DOTAP mixture was inoculated intradermally into three sites over the dorsal thorax. The kittens remained with their natural queen until weaning at 10 weeks of age and were then separated by sex into one or two animals per cage. Once paired, cage mates were not changed throughout the remainder of the study. The FRA and F6A groups were maintained in contact isolation. Blood and bone marrow collections were performed at regular intervals using aseptic techniques and appropriate anesthesia.
FeLV Viremia.
FeLV viremia was measured by antigen capture enzyme-linked immunosorbent assay (Synbiotics, San Diego, Calif.) for the detection of p27 capsid protein in plasma. Optical density (OD) readings were normalized against kit standards using the following formula: relative OD = OD of unknown − OD of negative control/OD of positive control − OD of negative control
FeLV antibody.
Anti-FeLV antibody responses were detected by indirect immunofluorescence on the FL-74 cat T-lymphoma cell line chronically infected with FeLVs representing the A, B, and C subgroups (8).
Viral interference assay.
Viral interference assays to identify FeLV subgroups were performed as previously described (33). FeLV pseudotypes of murine sarcoma virus were generated with virus stocks FeLV-A/Glasgow-1 (36), FeLV-B/GA (7), and FeLV-C/Sarma (27), derived from the transfected plasmids pFGA, pBHM-1, and pFSC, respectively.
Immunophenotypic analysis of tumor cells.
Tumor tissue collected at necropsy from five cats in this study and four cats in a previous study (4) was phenotyped for lymphocyte surface markers by either flow cytometry or immunohistologic staining. For flow cytometry analysis, single cell suspensions were obtained from the solid tumors by passage through a tissue sieve (Cellector; Bellco Glass Co., Vineland, N.J.) and separated by centrifugation over a density gradient (Ficoll Paque Plus; Pharmacia Biotech AB, Uppsala, Sweden). Monoclonal anti-feline CD1a, CD21, and CD22 (obtained from the laboratory of Peter Moore, University of California, Davis); anti-feline CD4, CD5, and CD8 (Southern Biotechnology Associates, Inc., Birmingham, Ala.); anti-feline CD45, CD57, and major histocompatability complex class II (MHC-II) (Serotec, Washington, D.C.); and anti-feline MHC-I (VMRD Research, Pullman, Wash.) antibodies were diluted as directed and incubated with the cell suspensions. Anti-mouse immunoglobulin G- phycoerythrin (Sigma, St. Louis, Mo.) secondary antibody diluted at 1:200 was used to detect the unlabeled primary antibodies (CD1a, CD21, CD22, CD45, and MHC-II). Labeled cell suspensions were analyzed by flow cytometry (Coulter Electronics, Miami, Fla.).
For immunohistologic staining, tumor tissue collected at necropsy was embedded in OCT compound (Sakura Finetek, Torrance, Calif.) and frozen in liquid nitrogen. Frozen sections were adhered to charged slides, briefly air dried, and stored at −20° until immunostained. Tissue sections were fixed in acetone for 2 min and washed extensively in Tris-buffered saline. Monoclonal anti-feline CD1a, CD21, CD22, CD4, CD5, CD8, and MHC-II antibodies (obtained from the laboratory of Peter Moore, University of California, Davis) were diluted 1:10 and applied to the tissues. The secondary antibody was horseradish peroxidase-labeled anti-mouse immunoglobulin G (Sigma). Immunolabeled tissues were developed using diaminobenzidine (DAB kit; Sigma) and counterstained with methyl green. Frozen lymphoid tumor tissues from cats do not express endogenous peroxidase activity (unpublished observation). Percent positive staining of cells was estimated by counting cells in randomly selected fields within the tissue section.
PCR analysis.
The genomic DNA isolation and nested PCR were described previously (4). Briefly, genomic DNA was isolated from bone marrow and buffy coat samples at various p.i. time points with a tissue genomic DNA isolation kit (Qiagen, Santa Clarita, Calif.). Nested PCR was performed with 250 ng of genomic DNA in the first round of amplification (35 cycles), using primer set H18 and H20 (4). A 1-μl volume of PCR product from the first round of amplification was used in a second-round amplification with another 35 cycles and with primer set RB53 and RB17 for recombinant FeLVs (rFeLVs) and primer set RB59 and RB17 for FeLV-A (4). Reaction mixtures were then analyzed by gel electrophoresis.
TA cloning and characterization of clone.
As described previously (4), the desired PCR product bands were purified and cloned into the TA cloning vector (pCR2.1) (Invitrogen, Carlsbad, Calif.). The sequences of the 3′ recombination crossover sites as well as a 600-bp sequence upstream of the crossover site were determined and compared to reported sequences of natural FeLV-B isolates.
Statistical analysis.
Survival times for cats inoculated with pFRA or pF6A were compared for statistical significance by the log-rank test using the PC-based program JMPIN (SAS Institute Inc., Cary, N.C.). Statistical analysis of mean antigenemia used one-way analysis of variance.
RESULTS
Inoculation of pF6A DNA resulted in the establishment of viremia, detectable anti-FeLV antibodies, and the de novo generation of FeLV-B and FeLV-C phenotypes.
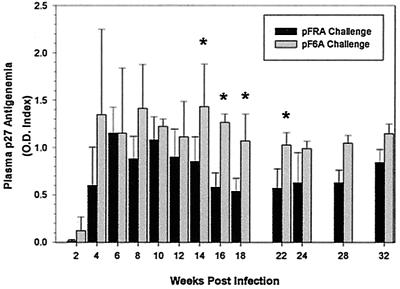
To determine its infectivity and pathogenicity, F6A plasmid DNA was inoculated intradermally into four neonate cats using a protocol identical to that previously described (4). Three additional age-matched cats were inoculated with FRA plasmid DNA as a challenge control. FeLV viremia, a presumptive test for virus load, was measured in serial plasma samples from the four pF6A-inoculated cats and a total of eight pFRA-inoculated neonatal cats (three cats from this study and five cats from a previous study given the same challenge material [4]). Mean FeLV plasma viremia levels were consistently higher for the pF6A-inoculated cats than the pFRA-inoculated cats. The differences were significant at weeks 14, 16, 18, and 22 p.i. (Fig. 1). No significant difference was found when the two groups of pFRA-challenged cats were compared (three from the current study and five from a previous study [4]).
FIG. 1.
Mean and standard deviation (error bars) of FeLV p27 plasma concentration from pF6A- and pFRA-challenged cats. p27 was determined by commercial enzyme-linked immunosorbent assay as described in Materials and Methods. Asterisks indicate mean values that were significantly different between pF6A- and pFRA-challenged cats.
Anti-FeLV antibody responses were detectable in the seven cats from our study by 7 weeks p.i. (data not shown). The peak antibody titers varied from 1:640 to 1:5,120. Antibody responses and peak titers were similar for the pFRA and pF6A groups.
Selected plasma samples were tested for the FeLV subgroup by interference assay. At 6 weeks p.i., all plasma samples from both groups of cats were positive for subgroup A, but negative for subgroup B or C (Table 1). By 28 weeks p.i., three of the four pF6A-inoculated and three of the three pFRA-inoculated cats had converted to an A/B or A/C subgroup phenotype. The conversion time in the plasma was comparable to that in our previous pFRA study (4). Cat 5037 (pF6A challenge) maintained the subgroup A phenotype alone to at least the 44-week-p.i. time point (last point tested). Cat 5051 (pF6A challenge) tested positive for subgroup A and B at 12 weeks p.i. but was positive for only subgroup A at 14 and 18 weeks p.i. At week 22 and later, all plasma samples from cat 5051 tested positive for subgroup A and B. Similarly, another cat (cat 5041 [pFRA group]) after exhibiting the A/B phenotype between 28 and 32 weeks p.i., became positive for subgroup A only at 36 to 43 weeks p.i. These patterns of either prolonged absence of subgroup B or vacillation between subgroup A and A/B phenotypes in plasma were observed previously in pFRA DNA-inoculated cats (4). Cat 5040, a pFRA-challenged cat, converted to an A/C subgroup phenotype at 18 weeks p.i., and replicating FeLV-C was consistently isolated from plasma until the cat was euthanatized due to severe anemia at 54 weeks p.i. This unusual conversion pattern (A to A/C) was analogous to another previous observation in pFRA-inoculated cats, in which the conversion was from the A to A/B to A/B/C phenotype (4).
TABLE 1.
Summary of results of antigenemia and subgroup analysis of pF6A- and pFRA-challenged cats
| Challenge and cat | Viremia status at the following time (wk p.i.)a:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 6 | 8 | 10 | 12 | 14 | 18 | 22 | 28 | 32 | 36 | 43 | 54 | 56 | 58 | 66 | 69 | >73 | |
| pF6A | ||||||||||||||||||
| 5051 | + A | + A | + A | + A | + A/B | + A | + A | + A/B | + A/B | + A/B | + A/B | + A/B | + | D-TLSA | ||||
| 5035 | + A | + A | + A | + A | + A | + A/B | + A/B | + A/B | + A/B | + A/B | + A/B | + | + | + | + | + | D-TLSA | |
| 5036 | + | + A | + A | + A | + A | + A | + A | + A/B | + A/B | + A/B | + | + A/B | + | + | + | D-TLSA | ||
| 5037 | − | + A | + A | + | + | + A | + A | + A | + A | + A | + | + A | + | + | + | + | + | Alive |
| pFRA | ||||||||||||||||||
| 5039 | + | + A | + | + A | + | + A/B | + A/B | + A/B | + A/B | + A/B | + | D-TLSA | ||||||
| 5040 | − | + A | + | + A | + | + A | + A/C | + A/C | + A/C | + A/C | + A/C | + A/C | D-AN | |||||
| 5041 | + | + A | + | + A | + | + A | + A | + A | + A/B | + A/B | + A | + A | + | + | D-BCL | |||
Viremia status is depicted as − (negative) or + (positive); subgroup determinations are based on interference assay, and the results are shown as A, A/B, or A/C where such tests were performed. Abbreviations: D, died; TLSA, thymic lymphosarcoma; AN, anemia; BCL, B-cell lymphoma.
Recombinant viral products were detected later in tissues of cats inoculated with pF6A than in those inoculated with pFRA.
The DNA extracted from bone marrow or buffy coat specimens collected from different infected cats at 8, 14, and 24 weeks p.i. was examined for the presence of FeLV-A or rFeLV exogenous proviruses by nested PCR. The results of the seven cats infected with pFRA or pF6A from this study combined with those of four cats infected with pFRA from our previous study are summarized in Table 2. For the pF6A-inoculated cats, two of eight samples (one bone marrow from cat 5035 and one buffy coat from cat 5037) were positive for rFeLV at 8 weeks p.i. By contrast 12 of 13 bone marrow or buffy coat samples were positive for rFeLV in the 8-week sample collection point for the pFRA group. By 14 or 24 weeks p.i. most samples from both groups were positive for rFeLV.
TABLE 2.
Detection of env gene recombinants in tissues of FRA- and F6A-infected cats
| FeLV inoculum and specimen | No. of recombinants detected/ no. of specimens tested at time point
|
|
|---|---|---|
| 8 wk p.i. | 14 or 24 wk p.i. | |
| pFRAa | ||
| Bone marrow | 6/7 | 7/7 |
| Buffy coat | 6/6 | 6/7 |
| pF6Ab | ||
| Bone marrow | 1/4 | 4/4 |
| Buffy coat | 1/4 | 4/4 |
Specimens collected from four cats from a previous study at 8 and 14 weeks p.i. (4) and three cats from the current study at 8 and 24 weeks p.i.
Specimens collected at 8 and 24 weeks p.i.
The structural features of env gene recombinants from the pF6A- and pFRA-inoculated cats were similar.
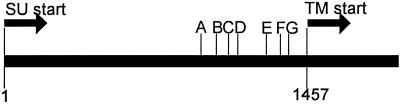
As previously done for pFRA (4), we analyzed the 3′ crossover sites of env gene recombinants from selected F6A-infected cats. The results of the analysis are summarized in Table 3. In bone marrow and buffy coat samples from cats 5036 and 5051 at 24 weeks p.i., all recombinants analyzed had 3′ crossover sites designated E, F, and G or 3′ to G (>G) (31). For samples at 8 weeks p.i., recombinant species were only detected from the buffy coat sample of cat 5037 and bone marrow sample of cat 5035. Six recombinant clones from each sample were evaluated. All clones except one from cat 5037 (5′ to A [<A]) were of crossover site E (Table 3).
TABLE 3.
Summary of 3′ recombination junction sites observed in env gene of in vivo-derived rFeLV clones from F6A-infected cats
| Cat | Specimen | Wk p.i. | No. of clones at crossover sitea
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| <A or A | B | C | D | E | F | G or >G | |||
| 5035 | Bone marrow | 8 | 6 | ||||||
| 5037 | Buffy coat | 8 | 1 | 5 | |||||
| 5036 | Bone marrow | 24 | 6 | ||||||
| 5036 | Buffy coat | 24 | 1 | 1 | 4 | ||||
| 5051 | Bone marrow | 24 | 6 | ||||||
| 5051 | Buffy coat | 24 | 4 | 2 | |||||
Crossover sites indicated in figure (above), A. through G. <A, 5′ to A; >G, 3′ to G.
When we aligned the deduced amino acid sequence of the region upstream of the crossover sites of these clones with sequences of other FeLVs (Fig. 2), we observed the same four amino acid differences as in our earlier report (4). This analysis included 11 clones from the two F6A samples at 8 weeks p.i., 23 clones from two F6A samples at 24 weeks p.i., and 1 exceptional clone from an 8-week sample for which the first amino acid difference (MGPNP to MGPNL) was not observed (Fig. 2). The crossover sites for F6A were similar to those reported for pFRA (4).
FIG. 2.
Deduced amino acid sequence comparison of the midregion of the SU gene of the in vivo-derived rFeLV clones with those of parent enFeLVs, various FeLV-B isolates, and F6A. Clones representative of recombination sites E, F/G, and >G (sites marked above the CFE-6 sequence) are shown. Numbers in parentheses indicate the total number of such clones examined. Amino acid sequences are presented relative to that of CFE-6, with dots indicating identity. Four consistent amino acid sequences differences observed in all in vivo-derived rFeLV clones as well as three isolates of FeLV-B, Gardner-Arnstein (GA), Snyder-Theilen (ST), and Rickard (R), are highlighted by boldface type (with the exception of the last position, for which only clones of recombinant site >G are highlighted) under the corresponding amino acid in CFE-6 (underlined). Numbers at both ends of the CFE-6 sequence depict the relative positions of these amino acids from the start point of the mature SU peptide. CFE-16 has a truncated SU peptide sequence because of a natural deletion. The reference F6A sequence is shown at the bottom, with gaps (dashes) introduced to maintain the sequence alignment.
The survival times for pFRA-inoculated cats were significantly shorter than for pF6A-inoculated cats.
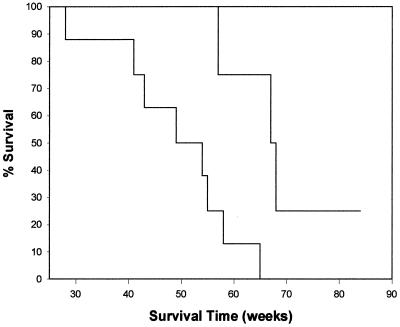
The average survival time of the eight pFRA-inoculated neonates (three from this study and five from the previous study) was 49 weeks p.i., with a range of 28 to 65 weeks p.i. By comparison, the average survival time for the four pF6A-inoculated neonates was >67 weeks, with tumor-related deaths at 57, 64, and 66 weeks p.i. and one cat alive and clinically healthy at >84 weeks p.i. (Fig. 3). Analysis of survival times using the log-rank test showed a significant difference between the pF6A and pFRA groups, with a P of 0.012. No significant difference in survival time was found between the two groups of cats inoculated with pFRA (three from this study and five from a previous study [4]).
FIG. 3.
Kaplan-Meier survival estimate showing survival times of cats challenged with pFRA (eight cats) and pF6A (four cats). All pFRA-challenged cats died from lymphoma (six cats) or anemia (two cats) prior to 65 weeks p.i. Three pF6A-challenged cats died from lymphoma, at weeks 57, 64, and 66 p.i. The remaining pF6A-inoculated cat was chronically FeLV viremic but clinically healthy at >75 weeks p.i. Kaplan-Meier plots and statistical analysis were performed using the PC-based program JMPIN (SAS Institute Inc.). The log-rank test showed a significant difference, with a P of 0.012.
Plasmid DNA inoculation with pFRA and pF6A resulted in the development of diverse FeLV-related neoplasias or hypoplastic anemia.
Analysis of stained tumor tissue by microscopy and flow cytometry revealed that of the eight neonates inoculated with pFRA from this study and from the previous study (4), five developed thymic lymphoma, two developed erythroid hypoplasia, and one developed multicentric B-cell lymphoma. By comparison, three of the four pF6A-inoculated cats developed thymic lymphoma during the study while the fourth cat remained viremic but clinically healthy. Phenotypic analysis of the lymphomas revealed a mixture of cell types; however, a predominant population could be identified. Thymic tumors from cats 5022, 5023, 5024, 5025, and 5035 consisted of predominantly CD5+ CD8+ CD22− cells, while thymic tumors from cats 5039, 5036, and 5051 were comprised of predominantly CD5+ CD4− CD8− CD22− cells (Table 4). CD1a+ cells could be detected but never comprised greater than 20 percent of the total population. MHC-II expression varied from <10 to 94% and did not appear to follow any specific pattern. Cat 5041 developed a multicentric B-cell lymphoma that was composed of predominantly CD21/22+ cells, with 90% of the tumor cells expressing MHC-II (Table 4). Thus, of the six tumors from cats inoculated with pFRA, four expressed T-cell markers (predominantly CD8+), one expressed neither T- nor B-cell markers, and one expressed B-cell markers. In addition, two cats developed erythroid hypoplasia. The diversity of disease manifestations in a relatively small group of outbred animals all receiving identical preparations of molecularly cloned FeLV DNA demonstrates that disease outcome is not dictated solely by the genotype of the initial virus challenge.
TABLE 4.
Phenotype analysis of tumor cells from FRA- and F6A-inoculated cats
| FeLV inoculum and cat | % Cells positive for:
|
|||||
|---|---|---|---|---|---|---|
| CD1a | CD4 | CD5 | CD8 | CD21/22 | MHC-II | |
| pFRA | ||||||
| 5022 | <10 | 53 | 90 | 84 | <10 | 75 |
| 5023 | <10 | 30 | 75 | 90 | <10 | 0 |
| 5024 | <10 | 50 | 90 | 90 | <10 | <10 |
| 5025 | <10 | 50 | 75 | 90 | <10 | 50 |
| 5039 | <10 | <10 | 47 | 14 | <10 | NDa |
| 5041 | <10 | <10 | 13 | 13 | 71 | 90 |
| pF6a | ||||||
| 5035 | 13 | <10 | 95 | 61 | <10 | 75 |
| 5036 | 20 | 38 | 99 | 18 | <10 | 33 |
| 5051 | 11 | 24 | 86 | 14 | <10 | 94 |
ND, not determined.
DISCUSSION
The diverse disease outcomes resulting from FeLV infection are likely to be due to both viral genetics and virus-host interactions. By using a model of in vivo transfection with molecularly cloned FeLV proviruses, we have begun to define the role of viral genetic determinants in pathogenesis. DNA inoculation with both pFRA and pF6A molecular clones, which have 98% nucleotide sequence identity, resulted in lifelong viremia. The virus replication kinetics for the two viruses were similar to each other and to that previously reported for cell-free virus inoculation with either the uncloned Rickard FeLV-A isolate (22) or cloned F6A FeLV-A (20, 21, 26, 30). Anti-FeLV antibody responses for both the pFRA- and pF6A-inoculated groups were comparable to the responses previously observed for cats inoculated with cell-free FeLV-A (24). Replicating FeLV-A was detected in the plasma of all seven DNA-inoculated cats prior to 12 weeks p.i. Beginning at 12 weeks p.i. replicating FeLV-B was also detected in plasma samples taken from the two pFRA- and three pF6A-inoculated cats which ultimately developed tumors during the study. When bone marrow or buffy coat samples, collected at 8 weeks p.i., were tested for recombinant provirus by PCR, 12 of 13 pFRA samples tested positive while only 2 of 8 pF6A samples were positive. However, when 24-week-p.i. bone marrow and buffy coat samples were tested, 13 of 14 FRA and 8 of 8 F6A samples were positive, suggesting the generation of rFeLV species was slower in pF6A- than in pFRA-infected cats. Analysis of the 3′ crossover sites of the rFeLV species revealed little difference between cats infected with pFRA and pF6A. Also, the recombinant species in F6A-infected cats, in general, bear the same env gene amino acid changes noted in FRA-infected cats, excluding one rFeLV clone in which the first amino acid difference was not observed (MGPNP to MGPNL). Thus, while the generation and selection of recombinant species in pFRA- and pF6A-infected animals seem to follow a similar pathway, the onset of the process for pFRA-infected cats occurs earlier. The delay in the recombinogenic events in pF6A- compared to pFRA-infected cats may be a factor in the ultimate disease outcome, in that the presence of rFeLVs may allow superinfection of target cells by pseudotype FeLV (A subgroup envelope and B subgroup virion RNA) and therefore more proviral integrations. In fact, a remarkable finding is that recombinants with structures similar to those found in experimental cats are also commonly present in naturally occurring feline lymphomas (35, 37). In addition to overcoming the interference barrier of FeLV-A infection, the recombinants may also utilize more than one cellular receptor to further increase the number of genetic “hits” (3).
Phenotypic analysis of tumors arising from pFRA or pF6A DNA inoculation revealed surprising tumor cell diversity, with T-cell, B-cell, and non-T-, non-B-cell lymphomas arising from the same clonal inoculum. What determines the cell origin of FeLV tumors is not known but may be related to the dominant rFeLV present or to the ontogenic stage of development of the target cells. The A-to-A/B recombination event is associated with lymphoid tumorigenesis (2, 4, 7, 15, 22, 23, 24, 30, 31, 34, 35), and the A-to-A/C subgroup transition is highly associated with red blood cell hypoplasia (1, 6, 16, 19, 27, 28). Perhaps the phenotypic shift in receptor utilization gives access to target cells not available to FeLV-A.
Finally, a number of questions are raised by this study. First, does the difference in recombination rate and disease progression between F6A- and FRA-inoculated cats relate to virus load? Overall, the results show similar virus load profiles for cats given the two provirus challenges but with F6A cats showing consistently higher mean virus loads throughout the observation period (Fig. 1). Thus, we conclude that higher virus load did not account for the increased recombinant activity observed in pFRA-inoculated cats. One possible model for retrovirus infection proposes a reverse relationship between virus load and recombination; that is, higher virus production reduces the frequency of recombination. This model is based on the premise that rapid spread of the parent virus tends to saturate the population of susceptible cells and therefore reduce the likelihood of recombination. In terms of growth kinetics, rFeLVs are at a select disadvantage because they appear only in the third round of infection (12).
A second question is whether the route of challenge affects pathogenesis. In almost all experimental studies with FeLV, cell-free virus has been administered by the intravenous, oronasal, intraperitoneal, intraosseous, and subcutaneous routes. The high infection efficiency observed with intradermal provirus inoculation, presumed to be less infectious than whole virus, suggests that the intradermal route may provide some advantage. It is possible that a subset of cells in the skin, such as Langerhans cells, may be highly permissive to FeLV. The spread of infection following intradermal DNA challenge is about 1 to 2 weeks slower than that seen with intravenous inoculation with cell-free virus (24). It is assumed that the lower rate of systemic virus spread reflects a smaller initial virus burden at a systemically remote site. The low initial virus burden may permit more rounds of infection and increase the probability of copackaging of endogenous FeLV RNA and subsequent recombination. Supporting this view is the observation that F6A-inoculated cats carry a higher virus load and have a reduced frequency of recombination. This issue will be resolved with additional challenge studies using whole cell-free virus administered intradermally.
Third, with 98% homology between the two subgroup A viruses, what genetic differences might explain the differences in recombinogenic rate. Of 22 predicted amino acid differences between F6A and FRA, there are at least 2 in the long terminal repeat (LTR) (4), 16 in the envelope region (4), and 4 in the product encoded by pol (Roy-Burman, unpublished data). Any one or more of these changes might affect pathogenecity. The four predicted amino acid differences in the reverse transcriptase could alter processivity and allow a greater frequency of recombination. Likewise, changes in the LTR could affect gene promotion and virus production which, by one model, would alter the likelihood of recombination. Env changes may also affect infection rate. Future gene substitution studies, where pol, env, or the LTR are switched between the two viruses should help define which genotypes affect pathogenesis.
ACKNOWLEDGMENTS
Andrew J. Phipps and Hang Chen contributed equally to this work.
We acknowledge the support of the Center for Retrovirus Research, the OSU Comprehensive Cancer Center, the Arthur G. James Cancer Hospital, and the Solove Research Institute, all of The Ohio State University.
This project was funded in part by Public Health Service grant R01 CA51485 and P30 CA16058 from the National Cancer Institute.
REFERENCES
- 1.Abkowitz J L. Retrovirus-induced feline pure red blood cell aplasia: pathogenesis and response to suramin. Blood. 1991;77:1442–1451. [PubMed] [Google Scholar]
- 2.Boomer S, Gasper P, Whalen L R, Overbaugh J. Isolation of a novel subgroup B feline leukemia virus from a cat infected with FeLV-A. Virology. 1994;204:805–810. doi: 10.1006/viro.1994.1597. [DOI] [PubMed] [Google Scholar]
- 3.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Bechtel M K, Shi Y, Phipps A, Mathes L E, Hayes K A, Roy-Burman P. Pathogenicity induced by feline leukemia virus, Rickard strain, subgroup A plasmid DNA (pFRA) J Virol. 1998;72:7048–7056. doi: 10.1128/jvi.72.9.7048-7056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donahue P R, Hoover E A, Beltz G A, Riedel N, Hirsch V M, Overbaugh J, Mullins J I. Strong sequence conservation among horizontally transmitted, minimally pathogenic feline leukemia viruses. J Virol. 1988;62:722–731. doi: 10.1128/jvi.62.3.722-731.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dornsife R E, Gasper P W, Mullins J I, Hoover E A. Induction of aplastic anemia by intra-bone marrow inoculation of a molecularly cloned feline retrovirus. Leukemia Res. 1989;13:745–755. doi: 10.1016/0145-2126(89)90087-8. [DOI] [PubMed] [Google Scholar]
- 7.Elder J H, Mullins J I. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol. 1983;46:871–880. doi: 10.1128/jvi.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essex M, Klein G, Snyder S P, Harrold J B. Feline sarcoma virus (FeSV)-induced tumors: correlation between humoral antibody and tumor regression. Nature (London) 1971;233:195–197. doi: 10.1038/233195a0. [DOI] [PubMed] [Google Scholar]
- 9.Hardy W D, Jr, Old L J, Hess P W, Essex M, Cotter S. Horizontal transmission of feline leukemia virus. Nature (London) 1973;244:266–269. doi: 10.1038/244266a0. [DOI] [PubMed] [Google Scholar]
- 10.Hardy W D, Jr, Hess P W, MacEwen E G, McClelland A J, Zuckerman E E, Essex M, Cotter S M, Jarrett O. Biology of feline leukemia virus in the natural environment. Cancer Res. 1976;36:582–588. [PubMed] [Google Scholar]
- 11.Hayes K A. Studies in the pathogenesis of FeLV infection in cats. M.S. thesis. Columbus: The Ohio State University; 1989. [Google Scholar]
- 12.Hu W-S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 13.Jarrett O, Hardy W D, Jr, Golder M C, Hay D. The frequency of occurrence of feline leukemia virus subgroups in cats. Int J Cancer. 1978;21:334–337. doi: 10.1002/ijc.2910210314. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett W F H, Crawford E M, Martin W B, Davie F A. A virus-like particle associated with leukemia (lymphosarcoma) Nature (London) 1964;202:567–569. doi: 10.1038/202567a0. [DOI] [PubMed] [Google Scholar]
- 15.Kumar D V, Berry B T, Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989;63:2379–2384. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathes L E, Pandey R, Chakrabarti R, Hofman F M, Hayes K A, Stromberg P, Roy-Burman P. Pathogenicity of subgroup C feline leukemia virus (FeLV) is augmented when administered in association with certain FeLV recombinants. Virology. 1994;198:185–195. doi: 10.1006/viro.1994.1021. [DOI] [PubMed] [Google Scholar]
- 17.Mullins J I, Hoover E A, Quackenbush S L, Donahue P R. Disease progression and viral genome variants in experimental feline leukemia virus-induced immunodeficiency syndrome. J Acquir Immune Defic Syndr. 1991;4:547–557. [PubMed] [Google Scholar]
- 18.Neil J C, Fulton R, Rigby M, Stewart M. Feline leukemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol. 1991;171:68–92. doi: 10.1007/978-3-642-76524-7_4. [DOI] [PubMed] [Google Scholar]
- 19.Onions D, Jarrett O, Testa N, Frassoni F, Toth S. Selective effect of feline leukaemia virus on early erythroid precursors. Nature. 1982;296:156–158. doi: 10.1038/296156a0. [DOI] [PubMed] [Google Scholar]
- 20.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 21.Overbaugh J, Hoover E A, Mullins J I, Burns D P W, Rudensey L, Quackenbush S L, Stallard V, Donahue P R. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology. 1992;188:558–569. doi: 10.1016/0042-6822(92)90510-v. [DOI] [PubMed] [Google Scholar]
- 22.Overbaugh J, Reidel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukemia virus in vitro. Nature (London) 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 23.Pandey R, Ghosh A K, Kumar D V, Bachman B A, Shibata D, Roy-Burman P. Recombination between feline leukemia virus subgroup B or C and endogenous env elements alters the in vitro biological activities of the viruses. J Virol. 1991;65:6495–6508. doi: 10.1128/jvi.65.12.6495-6508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey R, Bechtel M K, Su Y, Ghosh A K, Hayes K A, Mathes L E, Roy-Burman P. Feline leukemia virus variants in experimentally induced thymic lymphosarcomas. Virology. 1995;214:584–592. doi: 10.1006/viro.1995.0069. [DOI] [PubMed] [Google Scholar]
- 25.Pedersen N C, Theilen G, Keane M A, Fairbanks L, Mason T, Orser B, Chen C-H, Allison C. Studies of naturally transmitted feline leukemia virus infection. Am J Vet Res. 1977;38:1523–1531. [PubMed] [Google Scholar]
- 26.Quackenbush S L, Donahue P R, Dean G A, Myles M H, Ackley C D, Cooper M D, Mullins J I, Hoover E A. Lymphocyte subset alterations and viral determinants of immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1990;64:5465–5474. doi: 10.1128/jvi.64.11.5465-5474.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riedel N, Hoover E A, Gasper P W, Nicolson M O, Mullins J I. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus-C Sarma. J Virol. 1986;60:242–260. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riedel N, Hoover E A, Dornsife R E, Mullins J I. Pathogenic and host range determinants of the feline aplastic anemia retrovirus. Proc Natl Acad Sci USA. 1988;85:2758–2762. doi: 10.1073/pnas.85.8.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezanka L J, Rojko J L, Neil J C. Feline leukemia virus: mechanisms of neoplastic disease. Cancer Investig. 1992;10:367–385. doi: 10.3109/07357909209024796. [DOI] [PubMed] [Google Scholar]
- 30.Rohn J L, Linenberger M L, Hoover E A, Overbaugh J. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol. 1994;68:2458–2467. doi: 10.1128/jvi.68.4.2458-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy-Burman P. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Genes. 1996;11:147–161. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- 32.Sarma P S, Log T. Viral interference in feline leukemia-sarcoma complex. Virology. 1971;44:352–358. [PubMed] [Google Scholar]
- 33.Sarma P S, Log T. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization test. Virology. 1973;54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 34.Sheets R L, Pandey R, Klement V, Grant C K, Roy-Burman P. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and an endogenous FeLV element. Virology. 1992;190:849–855. doi: 10.1016/0042-6822(92)90924-e. [DOI] [PubMed] [Google Scholar]
- 35.Sheets R L, Pandey R, Jen W-C, Roy-Burman P. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol. 1993;67:3118–3125. doi: 10.1128/jvi.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsatsanis C, Fulton R, Nishigaki K, Tsujimoto H, Levy L, Terry A, Spandidos D, Onions D, Neil J C. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J Virol. 1994;68:8296–8303. doi: 10.1128/jvi.68.12.8296-8303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.U.S. Department of Health, Education, and Welfare. Guide for the care and use of laboratory animals. Washington, D.C.: National Academy Press; 1996. [Google Scholar]
- 38.Willems L, Kettmann R, Dequiedt F, Portetelle D, Voneche V, Cornil I, Kerkofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]