Abstract
The E1A gene products are required and sufficient for activation of adenovirus gene expression in cultured cells. The E4-6/7 gene product induces the binding of the cellular transcription factor E2F to the viral E2a promoter region. The induction of E2F binding to the E2a promoter in vitro is directly correlated with transcriptional activation of the E2a promoter in vivo. The E2 region encodes the viral replication proteins, yet adenoviruses lacking E4-6/7 function demonstrate no defective phenotype in infected cells. Here we show that the E4-6/7 protein can functionally compensate for E1A expression in virus infection. In the absence of the E1A gene products, expression of the E4-6/7 protein is sufficient to displace retinoblastoma protein family members from E2Fs, activate expression of early region 2 via induction of E2F DNA binding to the E2a promoter region, and significantly enhance replication of an E1A-defective adenovirus. These results have implications in the regulation of viral gene expression and for the development of recombinant adenovirus vectors.
Adenovirus (Ad) early gene expression is regulated by several viral gene products. E1A is the first transcription unit expressed during Ad infection, and the E1A proteins activate viral and cellular gene expression by multiple, independent mechanisms (8). Both the E1A 12S and 13S gene products bind to members of the retinoblastoma (Rb) protein tumor suppressor family (pRb, p107, and p130) (6). Among numerous cell binding partners, members of the Rb protein family interact with and repress the activity of members of the E2F transcription factor family (6). Sequestration of Rb protein family members by the E1A proteins releases free E2F, which transactivates both viral and cellular promoter regions containing E2F-responsive promoter elements. The E1A 13S gene product also transactivates viral and cellular promoters by different mechanisms involving interaction with cellular transcription factors such as the TATA binding protein, the activating transcription factor, and the suppressor of RNA polymerase B (SRB) mediator complex (2, 18, 19). The net effect of E1A gene expression early after infection is a significant increase in the activity of the other early Ad promoter regions. As expected, E1A mutants have dramatically reduced viral gene expression and, consequently, productive virus infection (16).
E2F was first described as a nuclear activity that bound to an inverted repeat in the Ad type 5 (Ad5) E2a promoter (17). The binding activity of E2F to these sites is stimulated by Ad infection dependent on the activities of two viral proteins. E2F transcriptional activity is positively regulated by the Ad E1A gene products, which dissociate Rb protein family members from E2Fs (4). Free E2F is then bound by the Ad E4-6/7 protein, which forms a complex with E2F and induces the cooperative and stable binding of E2F to the inverted binding sites in the Ad5 E2a promoter (12, 13, 15, 22, 30). The induction of E2F binding to the Ad5 E2a promoter in vitro is directly correlated with transcriptional activation of the E2a promoter as assayed using transient-expression assays and virus infection assays in vivo (24, 25, 26, 27, 32).
Ads lacking E4-6/7 function demonstrate no defective phenotype in the presence of the E1A gene products (11). Yet Ads of different serotypes encode E4-6/7 gene products capable of induction of E2F DNA binding activity and transactivation of promoter regions carrying inverted E2F binding sites (33). Further, the integrity of the E2F binding sites is important for E2a promoter activity in the context of Ad infection (21). The function of the E4-6/7 protein may be masked by or auxiliary to E1A during Ad infection of cultured cells. A role for the E4-6/7 protein cannot readily be addressed using viruses lacking the E1A transcription unit, since E1A proteins are required to express the E4 gene products. To ask if E4-6/7 could contribute to viral growth in the absence of the E1A proteins, wild-type E4-6/7 under control of the cytomegalovirus (CMV) promoter/enhancer was introduced in place of the E1A coding region in a virus background carrying a deletion that disrupts E4-6/7 activity (11). In the absence of the E1A gene products, we found that expression of the E4-6/7 protein was sufficient to displace Rb protein family members from E2Fs, activate expression of early region 2 via the induction of E2F DNA binding to the E2a promoter region, and significantly enhance replication of an E1A-defective Ad. These results suggest that the E1A and E4-6/7 proteins exhibit redundant functions for activation of the viral E2a promoter. Additionally, Ads that express all or part of early region 4 are being developed as gene therapy vectors for enhanced transgene expression (1, 3, 9, 20, 29, 37). The results presented here have important implications for the use of recombinant Ad vectors, since viruses that are deemed to be replication defective due to E1A deletion may in fact replicate and exhibit viability due to E4-6/7 protein expression.
MATERIALS AND METHODS
Viruses and infections.
Monolayer cultures of HeLa and A549 cells were maintained in Dulbecco's modified minimal essential medium containing 10% calf serum (HyClone); monolayer cultures of human embryonic lung (HEL) cells were maintained in Dulbecco's modified minimal essential medium containing 10% fetal bovine serum. Ad5 viruses WT300 and dl356 were described previously (11). dl356 contains a 2-bp deletion in E4 open reading frame 7 (ORF7) which disrupts the reading frame downstream of amino acid 91 and leads to expression of a truncated, nonfunctional E4-6/7 product. Ad-CMV-6/7-WT and Ad-CMV-6/7-F125P were constructed by inserting cDNAs corresponding to the Ad2 wild-type E4-6/7 protein or E4-6/7 mutant F125P (26) adjacent to the CMV promoter/enhancer in a plasmid containing the left 1,339 nucleotides (nt) of Ad5 (deletion of Ad5 nt 355 to 811). The E2-LS-CAT virus contains the XhoI-to-XbaI fragment of pE2a-E-CAT-LS-74/-85 (23) in place of the E1A region (nt 355 to 811). These plasmids were rebuilt into a dl356 virus background by the method of Stow (35) and propagated using 293 cells, an E1 complementing cell line (10). Recombinant viruses were plaque purified, virus stocks were amplified, and purified virus particles were prepared by CsCl equilibrium centrifugation following standard approaches (38). Virus particles were quantified by lysis of dilutions in buffer containing 0.1% sodium dodecyl sulfate (SDS) and absorbance at 260 nm was measured; 1 optical density unit at 260 nm equals 1012 particles/ml.
For viral growth curves, monolayer cultures of HeLa, A549, and HEL cells were grown to 75% confluency and infected with viruses at 100 particles per cell (corresponding to approximately 4 to 5 infectious viruses per cell) for 1 h at 37°C. The cell monolayers were then washed with phosphate-buffered saline solution, and fresh medium was added. Total cell lysates were prepared at different times after infection by freeze-thawing of infected cells in culture medium, and the virus yield was measured by serial dilution of infected cell lysates and plaque assay on 293 cells. Particle-PFU ratios were determined by serial dilution of purified virus particles and plaque assay on 293 cells.
Protein and DNA analyses.
For viral protein expression analyses, monolayer cultures of HeLa cells were grown to 75% confluency and infected with viruses at 200 particles per cell for 1 h at 37°C. At different times after infection, infected cells were washed with phosphate-buffered saline and harvested. Cell pellets were resuspended in SDS-lysis buffer (14), boiled, and sonicated, and the insoluble debris was removed by centrifugation at 12,000 × g for 15 min. For immunoprecipitation and Western blot analysis of E4-6/7 proteins, cleared cellular lysates were immunoprecipitated using anti-E4-6/7 monoclonal antibody M80 (MAb M80) (26), and the immune precipitates were analyzed by Western blotting using MAb M80 as the primary antibody. DNA-binding protein (DBP) expression was analyzed directly using cellular extracts by Western blotting with an anti-DBP monoclonal antibody (MAb 72-10) (31). Proteins were detected by enhanced chemiluminescence using ECL detection reagents (Amersham).
Viral DNA was isolated from purified particles by lysis with 0.1% SDS and proteinase K digestion. Following phenol-chloroform extraction and ethanol precipitation, DNA preparations were digested with XbaI to distinguish between the left-end fragments of different viral DNAs. Viral DNAs were analyzed by Southern blot hybridization using a fragment corresponding to the left 355 bp of Ad5 DNA, 32P labeled by the random primer method (7), as a probe.
Nuclear extract preparation and gel mobility shift assays.
Nuclear extracts were prepared according to the method of Dignam et al. (5). Nuclear fractions were dialyzed against dialysis buffer (DB; 20 mM HEPES [pH 7.5], 100 mM KCl, 10% glycerol, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride), and the dialysate was cleared by centrifugation at 25,000 × g. In vitro DNA binding assays were essentially performed as described previously (26, 27). Briefly, binding reaction mixtures (20 μl) contained 5 to 10 μg of nuclear extract, 2 μg of sonicated salmon sperm DNA, and 20,000 cpm of 32P-labeled Ad5 E2a promoter region probe DNA (1 to 2 fmol of DNA) in DB supplemented with Nonidet P-40 (final concentration, 0.1%). Binding reactions were performed for 1 h at room temperature, and then the products were electrophoresed in a 4% polyacrylamide nondenaturing gel at 4°C. The E2F double-site probe contains nt −30 to −73 from the E2a promoter plus additional vector sequences. Antibodies used for supershift analyses were anti-Rb (MAb C36) (Oncogene Science), anti-p107 (MAb SD9) (Santa Cruz Biotechnology), and anti-αE4-6/7 (MAb M45) (26).
RESULTS
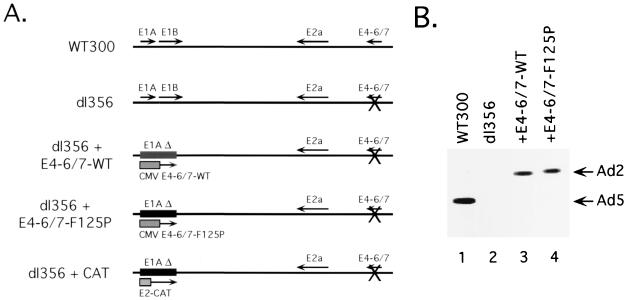
The function of the E4-6/7 protein during viral infection may be masked by or is auxiliary to E1A during Ad infection of cultured cells. A role for the E4-6/7 protein cannot readily be addressed using viruses lacking the E1A transcription unit, since E1A proteins are required to express the E4 gene products (8). To determine if E4-6/7 could contribute to viral growth in the absence of the E1A proteins, wild-type E4-6/7 under the control of the CMV promoter/enhancer was introduced in place of the E1A coding region in a virus background carrying a deletion that disrupts E4-6/7 activity (mutant virus dl356) (11). dl356 contains a 2-bp deletion in E4 ORF7 which disrupts the reading frame downstream of amino acid 91 and leads to expression of a truncated, nonfunctional E4-6/7 product. An E4-6/7 mutant (F125P) that binds E2F efficiently but contains a mutation that eliminates E4-6/7 dimerization, and thus the induction of E2F DNA binding to the inverted binding sites in the viral E2a promoter (26), was separately introduced into the same mutant virus background. These viruses, dl356+E4-6/7-WT and dl356+E4-6/7-F125P (Fig. 1A), expressed similar levels of E4-6/7 protein as wild-type Ad (WT300) in infected HeLa cells (Fig. 1B).
FIG. 1.
(A) Structural organization of recombinant Ads. The Ad genome is represented by a bold line. Mutant viruses described in the text are shown with the early proteins expressed (indicated with arrows). (B) E4-6/7 protein expression with recombinant adenoviruses. HeLa cells were infected at a multiplicity of infection of 100 virus particles per cell. Total cellular extracts were prepared 16 h after infection, and E4-6/7 proteins were immunoprecipitated and analyzed by Western blotting using MAb M80 (26). The Ad5 E4-6/7 protein migrates with faster mobility than the Ad2 E4-6/7 proteins produced by the recombinant viruses. The extracts were prepared from cells infected with the indicated viruses.
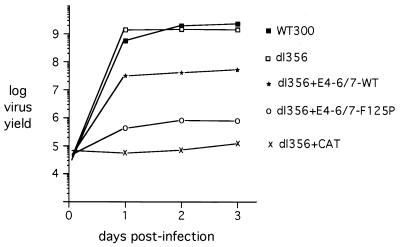
The ability of these recombinant viruses to undergo a lytic infection in HeLa cervical carcinoma cells was analyzed by a one-step growth curve. A virus that lacks the E1A coding region and carries the dl356 E4-6/7 mutation was used as a negative control (the E1 region was replaced by a chloramphenicol acetyltransferase [CAT] expression cassette; dl356+CAT) (28), and the E4-6/7 mutant virus, dl356, was tested as the parent of these recombinant viruses. Input virus was quantified at 6 h after infection and virus production was quantified at 1, 2, and 3 days after HeLa cell infection by a plaque assay using the E1 complementing cell line 293 (Fig. 2). As previously reported (11), mutation of the E4-6/7 reading frame in an otherwise wild-type Ad background (dl356) did not reduce virus growth in comparison to wild-type Ad5 (WT300). Deletion of the E1A region in a dl356 mutant background (dl356+CAT) reduced virus growth by more than 4 orders of magnitude, consistent with previous analyses of the growth of E1A mutant viruses (16). Surprisingly, constitutive expression of wild-type E4-6/7 in the absence of E1A (dl356+E4-6/7-WT) in part rescued the E1A defect. Levels of this recombinant virus were 50-fold lower than levels of WT300, an increase of 500-fold in comparison to the E1A/E4-6/7 double mutant virus, dl356+CAT. In contrast, a virus expressing the E4-6/7 protein deficient for E2F induction (dl356+E4-6/7-F125P) was increased only 10-fold in comparison to the E1A/E4-6/7 double mutant. The increase in growth of mutant dl356+E4-6/7-F125P in comparison to the E1A/E4-6/7 double mutant may reflect a limited activation of viral early gene expression by the CMV enhancer/promoter in the dl356+E4-6/7-F125P genome and/or the activation of E2F transcription factors by displacement of Rb protein family binding proteins (see Discussion). These results demonstrate that the E4-6/7 protein can contribute to viral growth in proliferating cultured cells in the absence of E1A expression.
FIG. 2.
E1A-independent growth of Ad in HeLa cells. HeLa cells were infected at a multiplicity of infection of 100 virus particles per cell (∼4 to 5 PFU/cell), and cellular lysates were prepared at 6 h and 1, 2, and 3 days after infection. Virus production was quantified by plaque assay using 293 cells to complement the E1A defect with the recombinant viruses. The results are presented as log virus yield for each data point and represent the results of three independent experiments. The viruses used in the analysis are depicted in Fig. 1A.
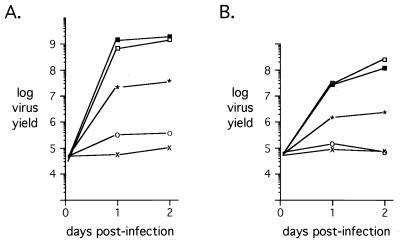
HeLa cells contain and express human papillovirus (HPV) DNA (34) and the E6 and E7 regulatory proteins that affect the p53 and pRb regulatory pathways (36). These functions are similar to those performed by Ad E1A and E1B 55-kDa proteins (6). To determine if HPV protein expression contributed to the growth of E1A-negative, E4-6/7-expressing recombinant Ad vectors, the growth properties of these viruses were analyzed in the human lung carcinoma cell line A549, which lacks endogenous HPV sequences, as well as in primary human HEL cells. The results of these analyses (Fig. 3A and B, respectively) were nearly indistinguishable from those obtained with HeLa cells. The recombinant virus that constitutively expressed wild-type E4-6/7 in the absence of E1A (dl356+E4-6/7-WT) was reduced 50-fold in yield in A549 cells and 100-fold in yield in primary HEL cells in comparison to WT300. In contrast, growth of a virus expressing the E4-6/7 protein deficient for E2F induction (dl356+E4-6/7-F125P) was increased only fivefold in A549 cells and less than twofold in HEL cells in comparison to that of the E1A/E4-6/7 double mutant dl356+CAT. These results confirm the interpretation made from experiments with HeLa cells that the E4-6/7 protein can complement the absence of the E1A gene expression in virus growth assays.
FIG. 3.
E1A-independent growth of Ad in A549 (A) and HEL (B) cells. A549 and HEL cells were infected at a multiplicity of infection of 100 virus particles per cell (∼4 to 5 PFU/cell), and cellular lysates were prepared at 6 h and 1 and 2 days after infection. Virus production was quantified as described in the legend to Fig. 2. Symbols: ■, WT300; □, dl356; ∗, dl356+E4-6/7-WT; ○, dl356+E4-6/7-F125P; ×, dl356+CAT.
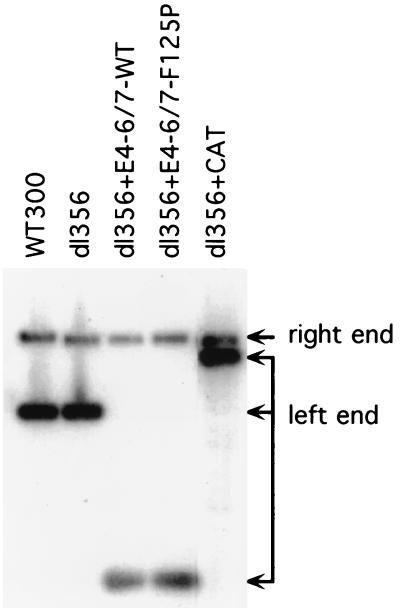
As further controls for these studies, the purity of the virus stocks was analyzed and the physical virus particle-to-infectious virus particle ratios (particle/PFU ratios) were determined. Figure 4 shows a Southern blot analysis of viral DNA isolated from the purified virus particle stocks used for these analyses. Distinct left-end DNA restriction fragments corresponding to the different viral genomes were observed, and no evidence of contamination of recombinant viruses with wild-type Ad was seen. The corresponding right-end fragment common to all of the viruses under study serves as an internal control for comparable DNA loading in this analysis. If the particle/PFU ratios of the recombinant Ads were significantly lower than those of the wild-type Ad5, then the growth properties of such viruses may be artificially increased due to a greater input of these viruses than of the wild type. While the 6-h time points in the growth curves did not indicate this to be the case, the results shown in Table 1 demonstrate that all viruses exhibited typical particle/PFU ratios for Ad5 (ranging from 14:1 to 33:1). We conclude that the growth properties of viruses that constitutively express the E4-6/7 protein do not reflect abnormalities in the virus stocks used for the studies.
FIG. 4.
Analysis of recombinant Ad DNAs. Viral DNAs from purified virions were isolated, digested with XbaI, and analyzed by Southern blotting using an Ad5 left-end radiolabeled probe (nt 1 to 355). The individual left-end DNA fragments of the different viruses and the common right-end fragment found with all viruses are indicated. The viral DNAs used in this analysis are indicated at the top of each lane.
TABLE 1.
Particle/PFU ratios of recombinant virusesa
| Virus | Particle/PFU ratio |
|---|---|
| WT300 | 20 |
| dl356 | 28 |
| dl356+E4-6/7-WT | 33 |
| dl356+E4-6/7-F125P | 14 |
| dl356+CAT | 21 |
Serial dilutions of each virus were used to infect monolayer cultures of 293 cells, and infectious virus concentration was determined by plaque assay. These values (PFU per milliliter) were compared to physical virus concentrations determined using absorbance at 260 nm where 1 optical density unit at 260 nm is equal to 1012 particles ml.
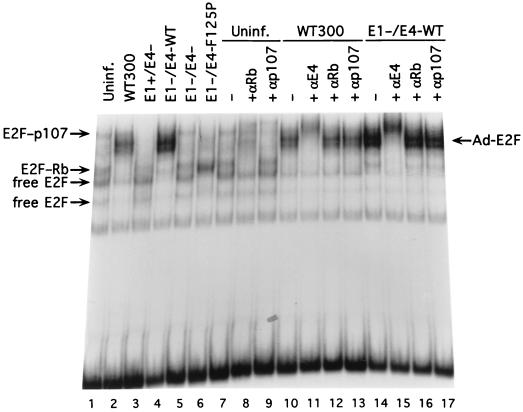
It is surprising that replacement of E1A with an E4-6/7 expression cassette would only cause a moderate defect in viral growth, since the E4-6/7 protein may not be sufficient to induce high levels of the E2 gene products required for viral DNA replication and since other early transcription units such as E4 would not be expected to be expressed. Since the E1A gene products normally are required in virus infection to liberate free E2F activity from repression by Rb protein family members to allow E2F to serve as a target for E4-6/7 binding, we analyzed E2F-specific complexes found after infection with the different recombinant viruses using a gel mobility shift assay with a radiolabeled probe corresponding to the inverted E2F sites of the Ad5 E2a promoter (Fig. 5). Different E2F binding complexes corresponding to both free E2F activity and E2F in complexes with pRb and p107 were observed using a nuclear extract from uninfected HeLa cells (Fig. 5, lane 1). The identification of higher-order E2F complexes is well documented in the literature and was confirmed using specific antibodies to E2F binding partners (pRb and p107) (Fig. 5, lanes 7 through 9). Infection with wild-type Ad5 (lane 2) resulted in the disappearance of E2F-Rb and E2F-p107 complexes due to E1A expression and the appearance of induced E2F activity containing the E4-6/7 protein. The loss of E2F-Rb and E2F-p107 complexes is evident visually and was confirmed using specific antibodies (lanes 10 through 13) where no change in binding pattern was seen in contrast to that observed with the antibodies and uninfected HeLa nuclear extract (lanes 7 through 9). E1A expression in the absence of E4-6/7 (E1+/E4− and dl356) (Fig. 5, lane 2) resulted in predominantly free E2F binding activity, while the absence of both the E1A and E4-6/7 gene products (E1−/E4− and dl356+CAT) (lane 5) yielded E2F complexes that were equivalent to uninfected HeLa nuclear extract. Note that expression of wild-type E4-6/7 in the absence of E1A (E1−/E4+ and dl356+E4-6/7-WT) (lane 4) gave rise to a binding pattern nearly identical to that found in the case of wild-type Ad5 (lane 2). That the major induced E2F complex with dl356+E4-6/7-WT contained the E4-6/7 protein was confirmed by using a monoclonal antibody against this product (26) (compare lanes 11 and 15). The minor new binding activity that was evident just above the free E2F activities corresponds to an E2F-E4-6/7 monomeric complex and also was recognized by the E4-6/7 antibody (lane 15). Additionally, expression of the E4-6/7 protein in the absence of E1A disrupted E2F-pRb and E2F-p107 complexes, since no evidence of these higher-order E2F complexes was observed when specific antibodies were used (compare lanes 12 and 13 to 16 and 17). Thus, expression of E4-6/7 in the absence of E1A displaced Rb protein family members from E2F and gave rise to induction of E2F activity in a manner identical to that observed with wild-type Ad.
FIG. 5.
E2F complexes present in HeLa cells infected with recombinant viruses. HeLa cells were infected at a multiplicity of infection of 200 virus particles per cell (∼8 to 10 PFU/cell), and nuclear extracts were prepared 6 h after infection as previously described (20). E2F binding activities were assessed using a gel mobility shift assay with a radiolabeled probe corresponding to the E2F sites in the Ad5 E2a promoter region, as previously described (5). Cellular E2F complexes are indicated by arrows on the left, and the Ad-induced E2F complex is indicated on the right. The viruses used in the analysis are depicted in Fig. 1A and are indicated in the figure as WT300 (wild-type Ad5), E1+/E4− (dl356), E1−/E4-WT (dl356+E4-6/7-WT), E1−/E4− (dl356+CAT), and E1−/E4-F125P (dl356+E4-6/7-F125P). The antibodies used were anti-Rb (MAb C36) (Oncogene Science), anti-p107 (MAb SD9) (Santa Cruz Biotechnology), and anti-E4-6/7 (MAb M45) (26). Uninf.: uninfected.
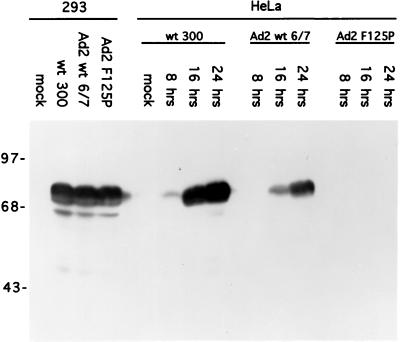
The simplest explanation for the efficient growth of recombinant virus dl356+E4-6/7-WT is that expression of the viral replication machinery encoded by the E2 transcription unit is an important rate-limiting event for a productive infection of cells in culture. To examine whether the growth properties of the different recombinant viruses were due to their level of E2 gene expression, accumulation of the E2 72-kDa DBP was measured by immunoblot analysis (31) following infection of HeLa cells. Protein lysates were prepared 8, 16, and 24 h after infection by wild-type Ad5 (WT300), dl356+E4-6/7-WT, and dl356-E4-6/7-F125P. As shown in Fig. 6, DBP expression levels with these viruses correlated very well with their growth properties in HeLa cells, where significant accumulation of DBP was evident with dl356+E4-6/7-WT in comparison to accumulation of wild-type Ad5, and no evidence of DBP expression was seen with dl356+E4-6/7-F125P. As a control, infection of 293 cells demonstrated that each of these viruses is equally capable of expressing DBP in the context of wild-type E1A.
FIG. 6.
DBP accumulation correlates with E2F induction and virus growth. HeLa cells and 293 cells were infected at a multiplicity of infection of 200 virus particles per cell (∼8 to 10 PFU/cell), and cellular lysates were prepared 8, 16, and 24 h after infection for HeLa cells and 16 h after infection for 293 cells. Fifty micrograms of cellular extract was loaded per lane on an SDS–10% polyacrylamide gel, electrophoresed, transferred to nitrocellulose, and analyzed for DBP expression by Western blotting using an anti-DBP MAb (31). The viruses used in the analysis were WT300, dl356+E4-6/7-WT (Ad2 WT 6/7), and dl356+E4-6/7-F125P (Ad2 F125P).
DISCUSSION
Previously it has been difficult to reconcile the conservation of functional E4-6/7 proteins among Ads of evolutionarily divergent subgroups with the fact that this protein is not essential for virus infection in cultured cells (33). The results presented in this report directly correlate the ability of the E4-6/7 protein to induce stable E2F DNA binding to the viral E2a promoter with increased expression of E2 gene products and the production of infectious virus. Furthermore, they indicate the importance that E2F induction by E4-6/7 might have an Ad infection under more stringent conditions of natural infection. For example, the relative levels of viral proteins observed in infections of cultured cells may not accurately reflect the levels found in natural infection. If E1A levels were lower and E4 levels were higher due to the relative activities of their respective promoter regions, the ability of E4-6/7 to displace Rb protein family members from E2Fs may be more essential for virus growth. Additionally, the composition of E2F complexes in normal versus cultured cells may be different: higher levels of higher-order E2F complexes may be found in natural infection, whereas in cultured cells where free E2F activity may be more abundant, reducing the need for Rb protein family displacement from E2Fs. The ability of the E4-6/7 protein to displace pRb and p107 from E2Fs in HeLa cells is entirely consistent with our previous observation that the E4-6/7 protein and pRb bind to E2F-1 in a mutually exclusive manner in vitro, since both proteins require the conserved marked box region of E2F-1 for stable protein interaction (28).
The fact that the levels of recombinant virus dl356-E4-6/7-WT, which does not express functional E1A proteins in infected cells, were not more than 50-fold lower than those of wild-type Ad5 has several implications. First, the cumulative effect of all other viral early gene products on productive infection in cells in culture does not enhance virus growth more than 50-fold. One could argue that the CMV promoter/enhancer driving expression of the E4-6/7 protein with this recombinant virus might allow expression of other early viral genes, but this phenomenon does not appear to have had a major effect in these assays. If it had, then one would anticipate that activation of other early genes also would enhance growth of dl356+E4-6/7-F125P (CMV enhancer positive) in comparison to dl356+CAT (CMV enhancer negative), yet this was not the case in HEL cells (Fig. 3B), and early gene activation at most contributed a 5- to 10-fold increase in virus growth in A549 (Fig. 3A) and HeLa (Fig. 2) cells, respectively. The 5- to 10-fold effect of expression of the E4-6/7-F125P mutant protein on virus growth in HeLa and A549 cells may reflect the ability of this protein to displace Rb protein family members from E2Fs as was seen with the wild-type E4-6/7 product (Fig. 5). Thus, the Ad DNA replication proteins appear to play the major rate-limiting role for Ad infection in cultured cells.
Finally, several recent reports have noted that Ad E4 expression enhances the duration of transgene expression in infected animals and promotes viability of primary endothelial cells in culture (1, 3, 9, 20, 29, 37). It was noted that for these reasons, E4 expression may be beneficial for improving the utility of Ad gene therapy vectors. The results presented here provide a caveat to possible benefit, since the enforced expression of E4 in E1A-negative recombinant Ad vectors may lead to unintended viral DNA replication due to E4-6/7 expression. Thus, there is clearly the need to anticipate that the expression of specific E4 gene products in recombinant Ad vectors to enhance transgene expression may also have undesired consequences for viral infection.
ACKNOWLEDGMENTS
We thank our colleagues for many helpful discussions, and we thank Gia Feeney and Tina Philipsberg for excellent technical help. We are grateful to David Spector for providing human embryonic lung cells.
This research was supported by Public Health Service grants CA28146 and AI41636 from the National Institutes of Health to P.H. R.J.O. was supported by NIH Training Grant CA09176.
REFERENCES
- 1.Armentano D, Zabner J, Sacks C, Sookdeo C C, Smith M P, St. George J A, Wadsworth S C, Smith A E, Gregory R J. Effect of the E4 region on the persistence of transgene expression from adenovirus vectors. J Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 3.Brough D E, Hsu C, Kulesa V A, Lee G M, Cantolupo L J, Lizonova A, Kovesdi I. Activation of transgene expression by early region 4 is responsible for a high level of persistent transgene expression from adenovirus vectors in vivo. J Virol. 1997;71:9206–9213. doi: 10.1128/jvi.71.12.9206-9213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cress W D, Nevins J R. Use of the E2F transcription factor by DNA tumor virus regulatory proteins. Curr Top Microbiol Immunol. 1996;208:63–78. doi: 10.1007/978-3-642-79910-5_3. [DOI] [PubMed] [Google Scholar]
- 5.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 8.Flint J, Shenk T. Adenovirus E1A protein: paradigm viral transactivator. Annu Rev Genet. 1989;23:141–161. doi: 10.1146/annurev.ge.23.120189.001041. [DOI] [PubMed] [Google Scholar]
- 9.Gorziglia M I, Lapcevich C, Roy S, Kang Q, Kadan M, Wu V, Pechan P, Kaleko M. Generation of an adenovirus vector lacking E1, E2a, E3, and all of E4 except open reading frame 3. J Virol. 1999;73:6048–6055. doi: 10.1128/jvi.73.7.6048-6055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 11.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy S, Engel D A, Shenk T. An adenovirus early region 4 gene product is required for induction of the infection-specific form of cellular E2F activity. Genes Dev. 1989;3:1062–1074. doi: 10.1101/gad.3.7.1062. [DOI] [PubMed] [Google Scholar]
- 13.Hardy S, Shenk T. E2F from adenovirus-infected cells binds cooperatively to DNA containing two properly oriented and spaced recognition sites. Mol Cell Biol. 1989;9:4495–4506. doi: 10.1128/mcb.9.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 15.Huang M-M, Hearing P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 16.Jones N, Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovesdi I, Reichel R, Nevins J R. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 18.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 19.Liu F, Green M R. Promoter targeting by adenovirus E1A through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 20.Lusky M, Grave L, Dieterlé A, Dreyer D, Christ M, Ziller C, Furstenberger P, Kintz J, Ali Hadji D, Pavirani A, Mehtali M. Regulation of adenovirus-mediated transgene expression by the viral E4 gene products: requirement for E4 ORF3. J Virol. 1999;73:8308–8319. doi: 10.1128/jvi.73.10.8308-8319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manohar C F, Kratochvil J, Thimmapaya B. The adenovirus EII early promoter has multiple E1A-sensitive elements, two of which function cooperatively in basal and virus-induced transcription. J Virol. 1990;64:2457–2466. doi: 10.1128/jvi.64.6.2457-2466.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marton M J, Baim S B, Ornelles D A, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy S C S, Bhat G P, Thimmappaya B. Adenovirus EIIA early promoter: transcriptional control elements and induction by the viral pre-early E1A gene, which appears to be sequence independent. Proc Natl Acad Sci USA. 1985;82:2230–2234. doi: 10.1073/pnas.82.8.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neill S D, Hemstrom C, Virtanen A, Nevins J R. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neill S D, Nevins J R. Genetic analysis of the adenovirus E4 6/7 trans activator: interaction with E2F and induction of a stable DNA-protein complex are critical for activity. J Virol. 1991;65:5364–5373. doi: 10.1128/jvi.65.10.5364-5373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obert S, O'Connor R J, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Connor R J, Hearing P. The C-terminal 70 amino acids of the adenovirus E4-ORF6/7 protein are essential and sufficient for E2F complex formation. Nucleic Acids Res. 1991;19:6579–6586. doi: 10.1093/nar/19.23.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor R J, Hearing P. Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1. J Virol. 1994;68:6848–6862. doi: 10.1128/jvi.68.11.6848-6862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramalingam R, Rafii S, Worgall S, Brough D E, Crystal R G. E1(−)E4(+) adenoviral gene transfer vectors function as a “pro-life” signal to promote survival of primary human endothelial cells. Blood. 1999;93:2936–2944. [PubMed] [Google Scholar]
- 30.Raychaudhuri P, Bagchi S D, Neill S, Nevins J R. Activation of the E2F transcription factor in adenovirus-infected cells involves E1A-dependent stimulation of DNA-binding activity and induction of cooperative binding mediated by an E4 gene product. J Virol. 1990;64:2702–2710. doi: 10.1128/jvi.64.6.2702-2710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 32.Reichel R, Neill S D, Kovesdi I, Simon M C, Raychaudhuri P, Nevins J R. The adenovirus E4 gene, in addition to the E1A gene, is important for trans-activation of E2 transcription and for E2F activation. J Virol. 1989;63:3643–3650. doi: 10.1128/jvi.63.9.3643-3650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaley J, O'Connor R J, Taylor L J, Bar-Sagi D, Hearing P. Induction of the cellular E2F-1 promoter by the adenovirus E4-6/7 protein. J Virol. 2000;74:2084–2093. doi: 10.1128/jvi.74.5.2084-2093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 35.Stow N D. Cloning of a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J Virol. 1981;37:171–180. doi: 10.1128/jvi.37.1.171-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tommasino M, Crawford L. Human papillomavirus E6 and E7: proteins which deregulate the cell cycle. Bioessays. 1995;17:509–518. doi: 10.1002/bies.950170607. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Greenburg G, Bunch D, Farson D, Finer M H. Persistent transgene expression in mouse liver following in vivo gene transfer with a ΔE1/ΔE4 adenovirus vector. Gene Ther. 1997;4:393–400. doi: 10.1038/sj.gt.3300404. [DOI] [PubMed] [Google Scholar]
- 38.Wold W S M. Adenovirus methods and protocols. Totowa, N.J: Humana Press; 1999. [Google Scholar]