Abstract
Bovine viral diarrhea virus (BVDV), a Pestivirus member of the Flaviviridae family, has a positive-stranded RNA genome which consists of a single open reading frame (ORF) and untranslated regions (UTRs) at the 5′ and 3′ ends. The 5′ UTR harbors extensive RNA structure motifs; most of them were shown to contribute to an internal ribosomal entry site (IRES), which mediates cap-independent translation of the ORF. The extreme 5′-terminal region of the BVDV genome had so far been believed not to be required for IRES function. By structure probing techniques, we initially verified the existence of a computer-predicted stem-loop motif at the 5′ end of the viral genome (hairpin Ia) as well as at the 3′ end of the complementary negative-strand replication intermediate [termed hairpin Ia (−)]. While the stem of this structure is mainly constituted of nucleotides that are conserved among pestiviruses, the loop region is predominantly composed of variable residues. Taking a reverse genetics approach to a subgenomic BVDV replicon RNA (DI9c) which could be equally employed in a translation as well as replication assay system based on BHK-21 cells, we obtained the following results. (i) Proper folding of the Ia stem was found to be crucial for efficient translation. Thus, in the context of an authentic replication-competent viral RNA, the 5′-terminal motif operates apparently as an integral functional part of the ribosome entry. (ii) An intact loop structure and a stretch of nucleotide residues that constitute a portion of the stem of the Ia or the Ia (−) motif, respectively, were defined to represent important determinants of the RNA replication pathway. (iii) Formation of the stem structure of the Ia (−) motif was determined to be not critical for RNA replication. In summary, our findings affirmed that the 5′-terminal region of the BVDV genome encodes a bifunctional secondary structure motif which may enable the viral RNA to switch from the translation to the replicative cycle and vice versa.
Bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and border disease virus of sheep are causative agents of economically important livestock diseases. They constitute the genus Pestivirus in the Flaviviridae family of RNA viral pathogens, which also includes the genera Flavivirus and Hepacivirus (human hepatitis C viruses [HCVs]) (12).
The pestivirus genome, a single-stranded, nonsegmented RNA molecule of positive polarity, has a size of approximately 12.5 kilobases and comprises a single open reading frame (ORF) that is flanked by untranslated regions (UTRs) at the 5′ and 3′ ends. Upon infection, the viral RNA functions as a messenger in the cytoplasm of the host cell and directs the synthesis of an unstable polyprotein, which is co- and posttranslationally processed to yield a range of different viral proteins. The array of genetic units along the ORF encoding the final products of polyprotein proteolysis has been determined to be 5′ Npro, C, Erns, E1, E2, p7, NS2-NS3, NS4A, NS4B, NS5A, and NS5B 3′ (reviewed in reference 30). Npro, a nonstructural autoprotease of unknown function, releases itself from the precursor (49). The activity of host signal peptidases gives rise to C, Erns, E1, and E2 which are destined to form the capsid and the envelope of the virion, respectively (reviewed in reference 44). The nonstructural (NS) proteins being encoded by the C-terminal part of the polyprotein are predominantly generated by the activity of a serine protease domain residing within the N terminus of NS3 (43, 50). Most NS proteins are presumed to act as catalytic components of the viral replication machinery (see below). Along this line, certain enzymatic functions which were demonstrated to be essentially involved in RNA replication, namely, a nucleoside triphosphatase/RNA helicase activity and an RNA-dependent RNA polymerase (RdRp) activity, were found to be associated with the NS3 and the NS5B protein, respectively (36, 40, 47, 52).
As with other positive-strand RNA viruses, replication of the pestivirus genome proceeds in an asymmetric manner along a two-step scheme. Supposedly in close functional linkage with the generation or maturation of the polyprotein, the nascent viral proteins and hypothetical cellular components associate with the 3′ terminus of the genomic RNA to form replication complexes. These catalyze the synthesis of a low copy number of complementary negative-strand RNA intermediates, which subsequently serve as templates for the transcription of an excess of progeny positive-strand RNA molecules (6).
cDNA copies of BVDV genomes (24, 25, 46) that are capable of producing infectious RNA transcripts in vitro permit detailed investigations of the virus life cycle by means of “reverse genetics”: i.e., mutagenesis of the viral RNA via the cDNA construct, introduction of the modified RNA into cultured cells, and monitoring of the effects of mutagenesis in vivo. In this way, genetic studies revealed that a subgenomic BVDV RNA (DI9c) consisting mainly of the 5′ and 3′ UTRs and the coding region of the nonstructural proteins NS3-NS5B replicates autonomously upon transfection into different host cells. BVDV DI9c RNA was hence verified to encode all factors and elements which—on the part of the virus—are necessary for the catalysis of both replication steps (6). The replicon system considerably facilitated probing the role of different functional features of the RNA replication pathway due to obvious experimental advantages: these concern a remarkable stability of the RNA, the inability of emerging revertants to spread, a high efficiency of multiplication, and, most importantly, the option to examine the replication process separately from events linked to RNA packaging and virion assembly. Sequence and structural motifs harbored in the 3′ terminus of the BVDV genome were thus defined to create a common cis-acting element, which is suspected to contribute to the negative-strand promoter of the initial replication complex (51). In a similar manner, the genomic 5′ terminus and the 3′ end of the negative-strand intermediate are expected to participate in the formation of a positive-strand promoter that initiates the second replication step. However, the situation is more complex, because the 5′ UTR of the Flaviviridae genome functions initially at the level of translation. In contrast to the flavivirus genome RNAs that contain a 5′ m7G cap structure (reviewed in reference 30) and ought to be translated in a cap-dependent manner, translation of the pestivirus and HCV ORFs is mediated by an internal ribosomal entry site (IRES), which involves internal attachment of the ribosomes to the initiation codon in a prokaryotic-like mode without scanning from the 5′ end (28, 29, 45). Reminiscent of similar features in picornaviruses such as encephalomyocarditis virus or poliovirus, the 5′ UTRs of the pestivirus and the HCV genomes are relatively long (BVDV, ca. 385 nucleotides; HCV, ca. 350 nucleotides), contain multiple cryptic AUG codons upstream of the authentic translation start, and share large conserved structure motifs as predicted by computer and refined by biochemical means (8, 11, 21, 22, 27). Although for HCV as well as for BVDV the exact boundaries of the functional IRES regions are still a matter of debate, they undoubtedly span the 3′ end of the 5′ UTR proximal to the translation initiation codon (13, 14, 19, 31, 32).
The 5′ UTR of poliovirus was shown to contain an additional functional RNA motif—a cloverleaf-like structure at the immediate 5′ terminus of the viral genome, which specifically associates with a viral and a cellular protein. This ternary ribonucleoprotein complex conceivably promotes catalysis of the second replication step in trans (1, 2, 15, 16). Although it was originally thought that the 5′-terminal region of the poliovirus RNA would operate independently of the IRES (1, 33), various pieces of evidence suggest a functional overlap between both RNA domains (7, 34, 35).
In this work, we demonstrate that a stem-loop structure motif encoded by the 5′-terminal region of the BVDV genome encompasses diverse cis-acting RNA elements which are critically implicated in the modulation of translation and RNA replication, respectively.
MATERIALS AND METHODS
Cells.
All types of cells utilized throughout this work were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and nonessential amino acids.
Construction of recombinant plasmids.
Restriction and subcloning procedures were performed according to standard protocols. Restriction enzymes were purchased from New England Biolabs (NEB) (Schwalbach, Germany), Pharmacia (Freiburg, Germany), and Roche Diagnostics (Mannheim, Germany). All primers used for mutagenesis, primer extension, and DNA sequencing (5′ IRD71-labeled oligonucleotides) were obtained from MWG Biotech GmbH (Ebersbach, Germany).
The DI9c cDNA construct pA/BVDV/D9, which is based on the low-copy-number plasmid pACYC177 (NEB), has been described previously (25). To obtain a more efficiently replicating DI9c cDNA construct, pA/BVDV/D9 was cut with SpeI and SmaI, and the resulting 8.0-kb fragment, which contained the multiple cloning site (MCS) of pACYC177, the promoter, and the entire coding region of DI9c cRNA, was ligated into a pBR322 derivative which had been cleaved previously with NheI and SmaI. The novel construct was termed pP/BVDV/D9.
To enable a straightforward mutagenesis of the BVDV Ia coding region, a BamHI-XhoI fragment of pP/BVDV/D9 (BamHI in the MCS, XhoI at position 208; nomenclature according to the full-length BVDV CP7 genome, see reference 42) was ligated into the corresponding restriction sites of pBluescript II KS+ (Stratagene, La Jolla, Calif.), forming subclone pBX. Mutant 1 (see Fig. 2) was created by digestion of pBX with AccI, removal of the 26-nucleotide fragment, and religation. Mutants 2 and 3 (Fig. 2) were originated by PCR primer-directed mutagenesis. For this purpose, pBX was used as a template, oligonucleotide Stu was applied as antisense primer, and oligonucleotides Hl and Hs were applied as sense primers, respectively (Stu, 5′-GATTTTTGTCCCCTAGGCCTAA-3′; Hl, 5′-CAGGAATTCAATACGACTCACTATAGTATACGCCCCCGATTGGGGGCGTATACATATTGG-3′; Hs, 5′-CAGGAATTCAATAC GACTCACTATAGCCAGCCCCCGATTGGGGGCGTATACATATTGG-3′). The PCR products were gel purified, digested with EcoRI and StuI, and cloned into pBX, which was previously cleaved with the same enzymes (EcoRI in the MCS and StuI at position 42). The residual mutations (mutants 4 to 18 in Fig. 2) were generated via “QuickChange” primer-directed mutagenesis. To introduce these mutations into the final pP/BVDV/D9 construct, a second cloning intermediate, pMsc, was created by cutting pP/BVDV/D9 with MscI, removal of the DNA fragment between two MscI sites at positions 7147 and 11903, and religation. The mutant BamHI-XhoI fragments of pBX were then used to replace the corresponding sequence in pMsc. Finally, 2.6-kb BglI-EagI fragments (BglI in the vector and EagI at position 5770) of the respective pMsc derivatives were introduced into pP/BVDV/D9 previously cut with the same enzymes. For inactivation of the NS5B RdRp, the GDD motif (amino acids 3635 to 3637 of BVDV CP7), which is highly conserved in most nucleic acid-polymerizing enzymes (3), was changed into GTR by applying QuickChange with appropriate oligonucleotide primers. All mutations were confirmed by dideoxy sequencing.
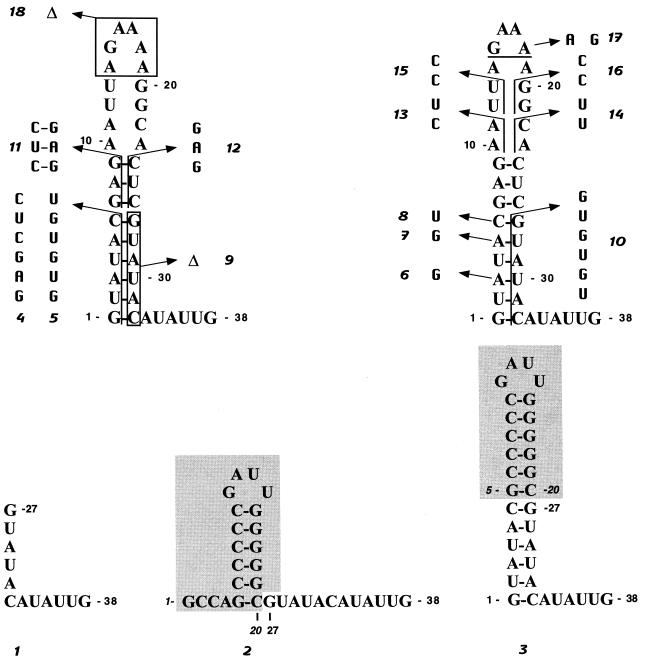
FIG. 2.
Mutagenesis of the 5′ terminus of BVDV DI9c RNA. Schematic drawing of the diverse mutations (denoted no. 1 to 18) introduced into the 5′-terminal stem-loop structure of the BVDV DI9c RNA via genetic means (see Materials and Methods). The substitutions and deletions are indicated by arrows. The numbers in standard type represent the corresponding positions in the DI9c wild-type RNA. The shaded regions in the schemes of mutations 2 and 3 indicate the sequence elements derived from the 5′ end of the HCV genome (39), and the italic numbers indicate their respective positions in the HCV RNA.
For RNA structure probing of the immediate 5′-terminal region of the BVDV RNA, several constructs were generated: for example, to obtain pΔBE, pA/BVDV/D9 was cut with BstEII and EcoRV; the resulting 7.1-kb fragment (position 453 to position 11888) was removed, blunted with T4 DNA polymerase (NEB), and religated. For in vitro transcription, all pP/BVDV/D9 derivatives and pΔBE were linearized with SmaI. The plasmid constructs used to generate the radiolabeled RNA probes for the RNase protection assay were described previously (6, 17).
DNA sequencing.
Dideoxy sequencing was carried out with 5′ IRD71-labeled primers and the Amersham cycle sequencing kit, applying DNA sequencer 4000L (Li-Cor; MWG) and the Genetics Computer Group software for computer-aided analysis of sequence data. To localize the modified nucleotides of chemical structure probing, the corresponding DNA plasmid was sequenced with a 5′ 32P-labeled primer (see below) and the T7 DNA polymerase dideoxy sequencing kit from Pharmacia.
Chemical and enzymatic probing of RNA secondary structure.
The 5′ end labeling of DNA oligonucleotides and of in vitro-transcribed RNA was performed as described previously (51), with T4 polynucleotide kinase (AGS GmbH, Heidelberg, Germany) and [γ-32P]ATP (5,000 Ci/mmol; Amersham). To determine the RNA secondary structure, enzymatic and chemical probing were performed according to the protocol of reference 51. Modified RNA molecules were analyzed by primer extension essentially as described previously and in the Results section by utilizing 5′-end-labeled primers (e.g., for Ia, primer Stu).
Transcription and transfection of RNA and RNase protection assay.
The mutant DI9c derivatives were generated by in vitro transcription according to a standard protocol. To remove residual amounts of DNA template, we digested the reaction mixtures with DNase and different restriction enzymes (for details, see reference 17). Unless indicated differently, the in vitro-transcribed RNAs were transfected into the target host cells via electroporation (the experimental setup is described in reference 6), and RNA replication was quantified by RNase protection at 9, 13, 24, and 48 h posttransfection. The quantification procedure is described in reference 17. RNase protection with two different probes (for details, see reference 17 and Fig. 3C) was applied to determine the stability of different BVDV DI9c derivatives.
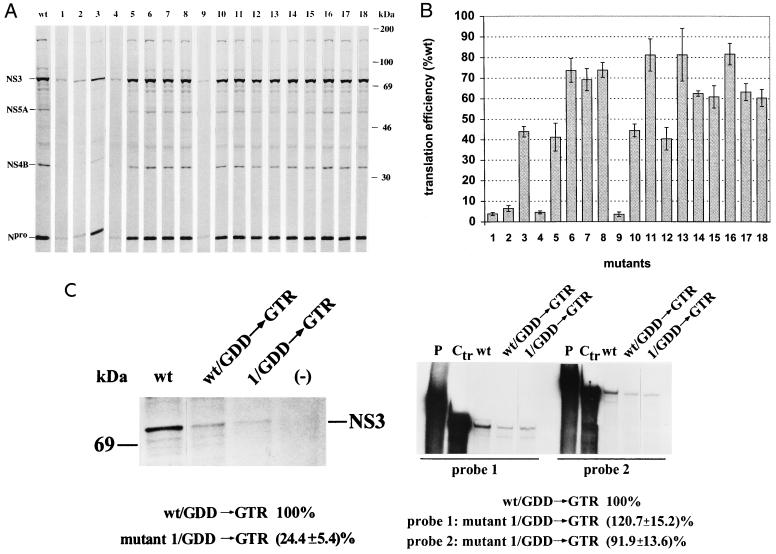
FIG. 3.
Measurement of the translation efficiency of mutant BVDV DI9c RNAs. (A) In vitro translation analysis. In vitro translation was performed with cytoplasmic extracts of BHK-21 cells as described in Materials and Methods by using the in vitro-transcribed wild-type RNA (wt) and the 18 mutant replicon RNAs as templates, respectively. A portion of the reaction mixture was solubilized in protein sample buffer, and the 35S-labeled proteins were analyzed by SDS-PAGE (10% polyacrylamide) (lanes wt and mutants 1 to 18; numbering scheme as described above). The positions of some mature NS proteins (see Results) are indicated on the left, with their molecular masses given on the right. As described in Results, no major differences were observed for the translation efficiency of the different DI9c Ia mutants, irrespective of whether the translation reaction was performed for shorter or longer intervals. The SDS-PAGE results shown in this figure were derived from a representative translation reaction carried out for 14 h. (B) Quantification of translation efficiency. The synthesis of Npro protein was measured with a phosphorimager system. Based on four in vitro translation experiments which were carried out with three independently prepared S10/eIF preparations of BHK-21 cells (see Materials and Methods), the relative translation efficiencies of the 18 mutants were calculated with respect to that of the wild-type (wt) replicon (calculated as 100% translation competent), as depicted (numbered as described above). The error bars indicate the mean deviations. (C) Measuring the translation efficiency and stability of mutant 1 (ΔIa) DI9c RNA in vivo. (Left panel) BHK cells were transfected with either mock RNA (−), the replication-competent DI9c wild-type replicon (wt) or DI9c RNA (wt/GDD→GTR) or mutant 1 RNA (1/GDD→GTR) encoding NS5B with a mutated GDD motif, respectively (see Materials and Methods and Results sections). De novo synthesis of NS3 was analyzed by growing the cells in the presence of [35S]methionine/cysteine (in the presented experiment, labeling was initiated at 2 h posttransfection and performed for a time period of 4 h). The viral protein was subsequently immunoprecipitated with a monoclonal anti-NS3 antibody, separated by SDS-PAGE, and quantitated by phosphorimaging (for details, see reference 17). The translation efficiency of the (nonreplicative) mutant 1 RNA was calculated from three independent transcription-translation experiments with respect to the nonreplicative wild-type replicon (calculated as 100% translation competent). (Right panel) RNase protection assay to compare the stability of wild-type and mutant 1 DI9c RNA in the course of an in vivo translation experiment. BHK cells were transfected with the same set of RNAs as above; i.e., replicative wild-type DI9c RNA (wt); nonreplicative DI9c RNA (wt/GDD→GTR) and nonreplicative mutant 1 (1/GDD→GTR) DI9c RNA, respectively. RNase protection was carried out at 6 h posttransfection (see above) as described in reference 17. Two different antisense probes were applied: probe 1, which hybridizes with the 5′-terminal NS5B-encoding region of the DI9c replicon RNA (nucleotides 10141 to 10421 in the nomenclature of BVDV CP7; see reference 42); probe 2, which hybridizes with nucleotides in the 5′-terminal Npro/5′-terminal NS3-encoding portion of the DI9c replicon RNA (nucleotides 635 to 1062 in the nomenclature of BVDV CP9; see reference 41). Lanes P represent the untreated probes, and lanes Ctr represent protection assays which were carried out with the probe on the original plasmid construct. The stability of the (nonreplicative) mutant 1 DI9c RNA (1/GDD→GTR) was compared with that of the (nonreplicative) DI9c RNA (wt/GDD→GTR), the latter estimated as 100% stable, by measuring the amounts of protected RNA fragments via phosphorimaging. The given values represent the averages (and mean deviations) of three independent protection experiments with each probe. The same experiments performed with a mock-translation assay yielded blank lanes (data not shown).
In vitro translation.
The preparation of BHK-21 S10 extract and BHK-21 cell eIFs (translation initiation factors) was carried out by following the protocol of Barton and Flanegan (4). In vitro translation reactions (50-μl total volume) were performed at 30°C for the indicated time intervals in the presence of 1 μg of in vitro-transcribed RNA template, 30% (vol/vol) BHK-21 S10 extract, 10% (vol/vol) BHK-21 cell eIFs, 40 U of RNaseOut (Gibco BRL), and 10 to 15 μCi of [35S]methionine, as described previously (17). Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) and monitored by autoradiography. Quantification was performed with a Fuji Bio Imaging Analyser and the corresponding software, TINA 2.09, by measuring the amounts of Npro and NS3, respectively.
Measuring the translation of BVDV replicon RNA in transfected cells.
Five micrograms of viral RNA transcript was transfected into BHK-21 cells via electroporation. At 2, 4, or 6 h posttransfection, the cells were suspended in Dulbecco's medium deficient in methionine and cysteine and supplemented with 100 μCi of ProMix (1,000 Ci/mmol; Amersham) and grown for 4 h in the presence of the 35S-labeled amino acids. The viral NS3 protein was immunoprecipitated from cytoplasmic extracts by the procedure published by Grassmann et al. (17), and the labeled protein was analyzed by SDS-PAGE (10% polyacrylamide) by standard procedures. Quantification of NS3 was performed via phosphoimaging (see above).
RT-PCR.
Reverse transcription (RT)-PCR to verify the stability of the diverse DI9c mutant RNAs under different assay conditions and/or to follow up the synthesis of the different replication products (see Results section) was performed essentially as described previously (6, 17).
RESULTS
The 5′-terminal region of BVDV DI9c RNA folds into a stable stem-loop structure, and a complementary motif is formed by the 3′ portion of the negative-strand replication intermediate.
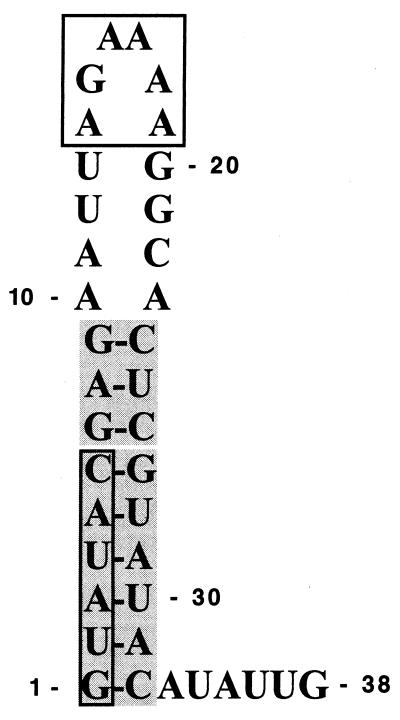
Alignment of different pestivirus genomes revealed that the 5′-terminal 29 to 32 residues of each viral RNA consist of two stretches of conserved nucleotides that are separated by an intervening sequence of rather variable composition (5, 11) (Fig. 1A). To gain a first idea as to the nature of the secondary structure of this part of the BVDV DI9c RNA, which, due to the cloning strategy of the original cDNA construct, corresponds to the 5′ portion of the BVDV NADL genome (25) (Fig. 1A), we applied the FOLD program of Zuker and Turner (23; http:\\mfold2.wustl.edu). Due to initial calculations which included all residues spanning the 5′ UTR as well as the 5′ extremity of the ORF (ca. 430 nucleotides), the 5′-most 32 nucleotides of DI9c should potentially form a pronounced stem-loop structure (ΔG = −9.2 kcal/mol at 37°C). This motif, which will be denoted as hairpin Ia (according to the nomenclature of references 8 and 32), was found to be thermodynamically favored as well if considerably longer or shorter parts of the RNA 5′ region were subjected to the folding (not shown). While the stem of Ia should be constituted of 18 nucleotides from which 4 GC and 4 AU base pairs are 100% conserved among all different virus strains, the loop part of Ia should comprise 14 nucleotides from which only 2 are conserved (Fig. 1A and C). Consistent with the computer-assisted prediction of Ia at the 5′ end of the positive-strand RNA genome, the 3′-terminal region of the negative-strand replication intermediate was calculated forming the complementary stem-loop motif (data not shown), which was consequently named Ia (−).
FIG. 1.
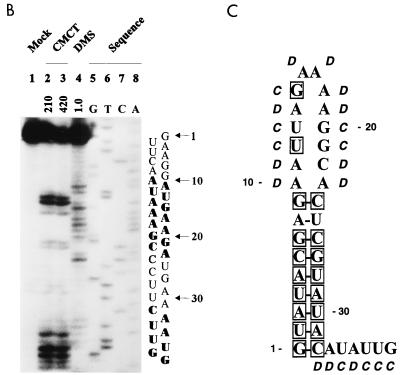
RNA primary and secondary structure of the BVDV DI9c 5′ terminus. (A) Alignment of 5′-terminal sequences of six representative pestivirus strains (BVDV-1 NADL, BVDV-1 SD-1, BVDV-1 Osloss, BVDV-2 890, CSFV Alfort-T, and BDV X818). The sequences were extracted from the GenBank/EMBL database. The numbers above the nucleotide sequence represent the coordinates utilized throughout this work. The 5′ terminus of the DI9c genome corresponds to the sequence of BVDV strain NADL shown in the top line, which was reconfirmed by sequencing of pA/BVDV/D9 (see Materials and Methods). Missing residues in the less-conserved intervening sequence are indicated by dashes. Nucleotides that are 100% conserved among all strains are boxed. (B) Chemical probing of the 5′-terminal RNA secondary structure. In vitro-transcribed pΔBE RNA (1 μg) was treated with single-strand-specific chemicals CMCT (G and U specific) and DMS (A and C specific) under native conditions. The products of the subsequent primer extension reaction, performed with 5′-end-labeled antisense primer Stu (see Materials and Methods), were electrophoretically analyzed on a 10% denaturing polyacrylamide gel. Lanes: 1, negative control performed in CMCT buffer; 2 and 3, CMCT modification (210 and 420 μg, respectively); 4, DMS modification (1.0 μl); 5 to 8, DNA sequence ladder obtained by dideoxy sequencing of the pΔBE plasmid, using the same radiolabeled primer, Stu, and T7 DNA polymerase. The corresponding sequence and the positions in the DI9c RNA are shown on the right, and the modified nucleotides are indicated in boldface. For comparison with the pattern obtained in the primer extension reaction, note that cDNA products synthesized from modified RNA are usually 1 nucleotide displaced with respect to the corresponding DNA sequencing products. This is due to the fact that RT extends the primer only to the position on the 3′ side of the modified nucleotide. (C) Secondary structure model of the BVDV DI9c 5′ terminus. The experimental data of chemical structure probing were superimposed on the computer prediction of the DI9c 5′ UTR which exhibited the lowest ΔGmax value of −9.2 kcal/mol (see Results). Nucleotides susceptible to the respective chemicals are denoted by italics (D, DMS; C, CMCT). The residues which are 100% conserved among all pestivirus strains are boxed. The numbering scheme is the same as in panels A and B.
Prior to commencing a study on the function of these RNA features, we decided to prove the predictions by biochemical means. To this end, we generated run-off transcripts that, for example, encompassed the entire BVDV DI9c 5′ UTR (see Materials and Methods). Under conditions which were established previously (51), the transcripts were treated with the chemicals dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT), respectively: DMS specifically modifies unpaired bases in the order of preference (most to least) G-N7, A-N1, and C-N3; CMCT modifies U-N3 and G-N2 in that order (10, 37). The sites of chemical modification were subsequently defined by a primer extension-termination assay in which cDNA was synthesized by reverse transcriptase from a 5′-end-labeled complementary oligonucleotide which was hybridized downstream of the region of interest (see Materials and Methods). Identification of modified nucleotides was achieved via side-by-side electrophoresis of a DNA sequencing reaction performed on the original plasmid construct with the same primer (Fig. 1B). Because methylation of the G-N7 position does not induce a specific stop of the reverse transcriptase, DMS was considered an AC-specific agent and CMCT was considered an UG-specific agent to detect nucleotides residing in single-stranded conformation within native RNA molecules.
The primer extension pattern unambiguously revealed the nucleotides at positions 1 to 9 and 24 to 32 of the DI9c RNA; i.e., those that were predicted to be involved in base pairs, as being almost entirely protected from chemical modification (the only exception is residue 32). In remarkable contrast, the residues at positions 10 to 23 were observed to be highly susceptible to DMS and CMCT, respectively (Fig. 1B). The former complementary nucleotide stretches were hence confirmed to fold into a stable stem helix, while the exposed nucleotides 10 to 23 were classified as constituents of a distinct loop structure. It should be noted that identical modification data were obtained, irrespective of whether shorter or longer RNA transcripts were examined in the assay. This finding implies that downstream sequences have negligible influence on the formation of Ia. Moreover, it is noteworthy that structure probing by partial digestion with RNases yielded congruent results (data not shown). In agreement with the computer model presented above, the native 5′ terminus of the DI9c RNA was thus defined to form a pronounced secondary structure motif, which is composed of a conserved stem and a variable loop. A final model summarizing the experimental data is drawn up in Fig. 1C. Using similar approaches, we confirmed that the 3′-terminal region of the DI9c negative-strand intermediate indeed forms the complement Ia (−) structure under native conditions (data not shown).
Mutagenesis of the 5′-terminal region of the BVDV DI9c RNA.
To analyze the function(s) of the above-defined RNA motifs in detail, we introduced a range of alterations into the Ia coding region of the BVDV DI9c RNA via mutagenesis of the respective cDNA construct (see above). An overview of this approach is given in Fig. 2; the rationale behind each intervention is summarized in Table 1. Below, we will focus more on presenting the experimental results which concern the genomic Ia motif rather than the Ia (−) structure. However, the consequences of each mutagenesis were also taken into account for the negative-strand intermediate and will be discussed later.
TABLE 1.
Intended effects of mutations initiated in the Ia coding region of BVDV DI9c RNA
| Mutationa | Intention |
|---|---|
| 1 | Deletion of nucleotides 1–26 of the BVDV DI9c 5′ terminus |
| 2 | Replacement of residues 1–26 of the BVDV DI9c 5′ terminus by 20 nucleotides of the genomic 5′ terminus of an HCV RNA (BK, 1b [see reference 39]), which also form a stable stem-loop structure (HCV Ia) |
| 3 | Replacement of the less-conserved (upper) region of the BVDV DI9c Ia motif by the upper part of HCV Ia, giving rise to an elongated stem and a smaller loop formed by non-BVDV sequences |
| 4 | Replacement of nucleotides 2–5 of the DI9c RNA by unrelated sequence elements; formation of the lower stem region abolished |
| 5* | Replacement of nucleotides 2–6 by sequence elements which maintain the stem of the Ia motif (compensatory exchanges); contrary effect on the complementary Ia structure (in this case, stem formation should be abolished) |
| 6, 7, 8* | Substitution of unrelated nucleotides for single residues, which in mutation 5 were replaced in a row; formation of the Ia stem should be only slightly affected; formation of the Ia (−) stem, however, expected to be weakened |
| 9 | Deletion of a major constituent part of the lower stem region |
| 10* | Replacement of nucleotides 27–32 of the DI9c RNA by different sequence elements, which maintains the stem of Ia, but abolishes stem formation in the Ia (−) motif (see mutation 5) |
| 11 | Conversion of nucleotide stretches which constitute the upper stem region of hairpin Ia |
| 12 | Destruction of the upper stem region and extension of the loop part of Ia |
| 13, 16, 17 | Substitution of unrelated residues for different parts of the loop region |
| 14, 15 | Modifications that either elongate the stem or create a bulge-stem-loop structure |
| 18 | Deletion of a considerable part of the Ia loop region |
All mutations except 5, 6, 7, 8 and 10 should have similar effects on the formation of the Ia and Ia (−) motifs, respectively. For details, see Fig. 2.
A first series of mutations was designed to obtain some initial idea of the general relevance of Ia by either deleting the structure (mutant 1) or replacing major parts of the motif (mutants 2 and 3) by similarly folded sequence stretches derived from the 5′-terminal region of an HCV 1B RNA (39; experimental data which defined the secondary structure of the HCV hairpin Ia structure as it is depicted in Fig. 2 are not shown). With a second set of mutations, we intended to explore the assignment of the “lower” (6 bp; highly conserved) and “upper” (3 bp; less conserved) parts of the stem structure of the DI9c Ia motif (Fig. 1) and to evaluate the importance of individual nucleotide constituents of the stem. In this way, parts of the Ia stem were either deleted (mutant 9) or replaced by unrelated sequence stretches (mutants 4 and 12) in order to entirely abolish double-strand formation. In the same context, one or more nucleotides were changed in such a way that the overall structure of the stem of hairpin Ia remained intact, but was made up by different sequence constituents (mutants 5, 6, 7, 8, 10, and 11). A last set of substitutions and deletions should affect either nucleotide components or the overall integrity of the loop structure of Ia (mutations 13 to 18; Fig. 2 and Table 1).
Significance of hairpin Ia for translation of BVDV DI9c RNA.
To evaluate the efficiency of IRES-mediated translation of each of the diverse hairpin Ia DI9c derivatives, the entire repertoire of mutant and wild-type RNAs was raised by in vitro transcription and initially tested via translation in vitro. Since commercial reticulocyte lysates yielded multiple aberrant translation products if programmed with the full-length replicon (data not shown), we employed freshly prepared cytoplasmic (S10) extracts and translation initiation factors from BHK-21 cells (see Materials and Methods). The latter reaction mixture—supplemented with [35S]methionine to visualize and/or to quantitate the process of protein synthesis—was recently demonstrated to promote efficient and accurate translation of the DI9c-encoded ORF as well as the processing of the nonstructural polyprotein (17). Consistent with these previous observations, subjecting the diverse replicon RNAs to the BHK S10-based translation reactions gave rise to a characteristic pattern of labeled bands on SDS-PAGE, which—by comigration experiments with individually translated proteins and mutagenesis of the different proteolytic cleavage sites—were identified as representing the mature viral NS proteins as well as some intermediates of the polyprotein processing pathway (Fig. 3A and data not shown). The general proteolysis profiles of the polyprotein (i.e., the efficiency of the NS3-mediated cis and trans cleavages) were determined to be virtually unaltered between the different DI9c Ia mutants and the wild-type replicon. However, measurement of the amounts of Npro or NS3 (the latter not shown) as the most prominent processing products revealed that the majority of alterations within the Ia region generally decreased the amounts of the diverse mature viral polypeptides with respect to the original DI9c RNA (Fig. 3A). Interestingly, the most drastic reduction of protein synthesis, to approximately 10% of the wild-type level, was observed if the hairpin Ia structure was either deleted (mutant 1) or substituted for by the corresponding HCV motif (mutant 2) or if formation of the lower portion of the stem region was severely affected (mutants 4 and 9). Abolishment of the upper stem (mutant 12), compensatory substitution of nucleotides at positions 3, 5, 6 or 30, 28, and 27 within the lower stem (mutants 5 and 10, respectively), and replacement of the upper stem and loop (mutant 3) by HCV sequence had a less drastic but still significant effect: in these cases, the amounts of proteins corresponded to roughly half of that of the wild-type RNA. Finally, changes of single nucleotides within the lower stem (mutants 6, 7, and 8), of parts of the loop region (mutants 13 to 18), and of nucleotides in the upper part of the stem (mutant 11) were found to have only minor consequences (Fig. 3A).
Certain mutations which were expected to affect formation and/or stability of hairpin Ia to a rather different extent (e.g., mutations 1, 2, and 4) nevertheless turned out to have a similar effect on the translation rate of the respective replicon RNA (Fig. 3A). It therefore appeared unlikely that the deviations in the accumulation of viral proteins listed above were caused by differences in the overall stability of the diverse RNA molecules in the course of the experimental approach. However, to minimize this possibility, we performed the in vitro translation reaction with the entire set of RNAs for different time periods. Thus, the relative degrees of inhibition of protein synthesis were confirmed to be mainly the same for each of the different DI9c Ia derivatives with regard to the wild-type replicon—independent of whether the assay was carried out for 1.5, 3, 10, or 14 h, respectively (data not shown). By RT-PCR and RNase protection before and at different time points during the translation reaction, we further ascertained the stability of all variant RNAs as being indistinguishable from that of the wild-type RNA under the chosen standards: both assays were performed on different regions of the respective RNA molecules and previously proven to indicate even slight variations in RNA stability (data not shown; see Fig. 3C for an example). We reasoned that the observed differences in translation efficiency could only be related to the mutagenized hairpin Ia motif (see also below).
It should be noted that identical translation values were measured with the various DI9c Ia RNAs, irrespective of whether they encoded an intact or an inactivated RdRp, respectively (data not shown, see also Fig. 3C). These results suggested that the level of RNA replication—whether it can occur at all in BHK S10 extracts has yet to be determined—should be so low that its effect on the in vitro translation process is negligible (also see Discussion).
Next, it was important to investigate whether the hairpin Ia motif has the same crucial function for translation of the BVDV DI9c RNA in vivo indicated by the in vitro data presented above. Several factors recommended particularly that the replicon derivative comprising mutation 1 be explored in this respect. (i) Deletion mutation 1 extends along almost the entire hairpin Ia-encoding region (Fig. 2), and it has a considerable negative impact on the translation of the BVDV DI9c RNA in vitro (Fig. 3A). (ii) Essentially the same mutation, analyzed in different pestivirus 5′ UTRs in the context of a flanking reporter gene, was measured to have no effect on the translation of the corresponding reporter enzyme (9, 32; H. Yu, unpublished results). Though the latter data are apparently inconsistent with the findings reported above (see Discussion), they imply mutation 1 has neither a destabilizing nor an interfering effect on the global folding of the remaining 5′ UTR.
Accordingly, wild-type and mutant 1 DI9c transcripts, both of which encoded an inactivated NS5B protein to monitor intracellular translation independently from RNA replication, were transfected into BHK cells. The cells were subsequently grown in the presence of 35S-labeled amino acids, and the efficiency of translation of the input RNAs was determined by purification and quantification of the labeled NS3 protein (see Materials and Methods). As shown in Fig. 3C, which summarizes the results of three independent experiments, intracellular translation of NS3 was found to be generally low in the absence of RNA replication (compare the wt and wt/GDD lanes). However, in the case of the (nonreplicative) mutant 1 RNA, the amount of newly synthesized NS3 was found to be significantly reduced and to correspond to an average of approximately 25% of that of the (nonreplicative) wild-type RNA (Fig. 3C, compare lanes wt/GDD and 1/GDD). As in the previous series of experiments, we performed RT-PCR, RNase protection, and translation for different time intervals to confirm that this remarkable discrepancy in the accumulation of NS3 could not be explained by major differences in the decay of both RNAs within the cell (Fig. 3C, RT-PCR results and other translation data not shown). Taking in vitro and in vivo data together, we hence concluded that the hairpin Ia motif—particularly its constituent lower backbone stem structure—represents a sensitive molecular feature of the translation process of a genuine BVDV RNA (see summarizing model in Fig. 6 and Discussion).
FIG. 6.
The 5′-terminal stem-loop structure of the BVDV genome represents a bifunctional RNA signal involved in translation and RNA replication. The part of the Ia motif that has been identified as being essential for translation is shown as a bipartite shaded box (indicating the upper and lower parts of the stem structure); the elements which were demonstrated to be particularly important for RNA replication are depicted in open boxes.
Functional significance of sequence and structural elements encoded by the 5′-terminal region of the BVDV DI9c RNA for the RNA replication process.
In the subsequent set of experiments, we wanted to test the different Ia DI9c derivatives for their ability to replicate. For this purpose, the in vitro-transcribed RNAs were transfected into BHK-21 cells via electroporation, a procedure which was shown to yield reproducibly about 80 to 90% replicon-containing cells (reference 6 and data not shown). Amplification of each RNA was judged at different time points posttransfection by either monitoring the replication-associated synthesis of NS3 protein via immunofluorescence (IF) staining of the transfected cells or by quantifying the level of negative-strand intermediate and progeny positive-strand viral RNA in the cytoplasmic fraction via RNase protection and/or RT-PCR (RT-PCR and IF data not shown) (6). RNA molecules encoding an inactivated NS5B RdRp (see above) were applied as a negative control in all three assay systems. To exclude the possibility of cross-contamination and/or emerging revertants, we verified the identity of each RNA which was recovered as replication competent (see below) by RT-PCR and subsequent sequencing of the PCR products (data not shown). Figure 4 summarizes the data which were obtained in the course of five independent transcription-transfection experiments with each of the DI9c mutants. Like in previous works, RNase protection, RT-PCR, and IF analysis yielded corresponding results on the capability of the respective replicon derivatives to amplify within the host cells, irrespective of which time posttransfection the analysis was performed (some data not shown).
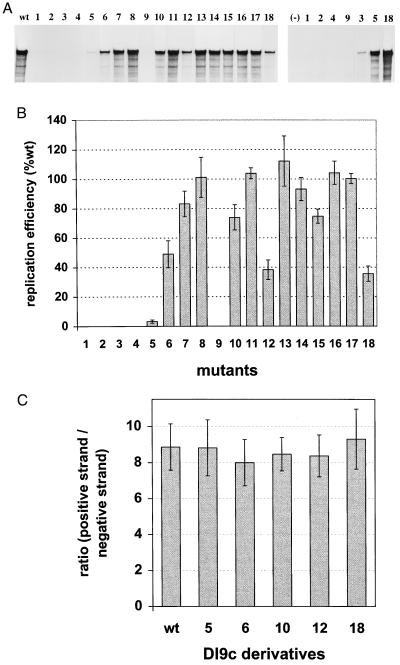
FIG. 4.
Replication capabilities of different BVDV mutant Ia DI9c RNAs. (A) Representative experiment of five independent transcription-transfection experiments of the different Ia replicon derivatives. RNA replication was monitored for novel synthesized positive-strand RNA with a 32P-labeled antisense probe after transfection into BHK-21 cells. The protected fragments were separated on a 5% denaturing polyacrylamide gel and analyzed by autoradiography. (Left panel) Lanes: wt, RNase protection of RNA derived from the cytoplasmic fraction of cells (24 h posttransfection) which had previously been transfected with the DI9c wild-type replicon RNA; 1 to 18, identical experiments carried out with each of the different Ia mutant RNAs (numbering scheme analogous to the one used in Fig. 2). (Right panel) Lanes: 1, 2, 4, 9, 3, 5, and 18, RNase protection assay performed 48 h posttransfection to measure the replication of mutants 1 to 5, 9, and 18; (−), negative control (BHK-21 cells transfected with a control replicon [see Results]). (B) Quantification of replication efficiency. The major protected band (A) of five independent experiments was quantified with a phosphorimaging analyzer system and normalized to a cotransfected control RNA (control of transfection efficiency [17]). The average replication ability was determined for each mutant with respect to the BVDV wild-type DI9c (estimated as 100% replication competent) as depicted. Error bars indicate the mean deviations. (C) Determination of the ratio of positive-strand RNA versus negative-strand RNA intermediate at 13 h posttransfection. Applying experimental protocols of references 6 and 17, five independent transfection/RNase protection experiments were performed with wild-type DI9c RNA and derivatives 5, 6, 10, 12 and 18, respectively (numbering scheme analogous to Fig. 2). The average ratios are depicted as a column diagram; error bars indicate the mean deviations. The general value was found to be slightly higher than previously published (6) and to vary around 8:1. Analogous experiments that were carried out at other time points yielded identical results (data not shown).
Interestingly, although the general amount of replication products that accumulated at different time points posttransfection in the cells varied dramatically between the different mutants (Fig. 4A and B), the ratio of newly synthesized positive-strand RNA to negative-strand intermediate (quantified by RNase protection; see also reference 17) was measured to be generally unchanged (Fig. 4C). From this result, we concluded that none of the alterations modifying the Ia or Ia (−) motif uncoupled both steps of the replication pathway (see Discussion).
As expected, provision of sufficient amounts of the viral proteins was found to be of crucial importance for efficient catalysis of RNA replication. Accordingly, alterations which abolished the architecture of the lower stem of Ia and which were previously shown to cause a drastic decline in the translation rate (mutants 1, 2, 4, and 9; see Fig. 3) led consequently to a complete block of the replication process. Mutations 5 and 10, which preserved the RNA double strand of the Ia motif [consequences for the Ia (−) motif are shown below] by essentially the same number of compensatory nucleotide exchanges within the 5′ or the 3′ constituent part of the lower stem, respectively, supported RNA replication to a strikingly different degree: while the multiplication capacity of mutant 5 was found to be rather deficient and to correspond a value of barely 5% of that of the wild-type RNA, replication of mutant 10 was, in contrast, observed to be only moderately diminished, giving rise to de novo synthesized RNA products at approximately 70% of the average wild-type level (Fig. 4B). Considering that both mutant RNAs were translated at a similar level (Fig. 3 and 5), these data suggested a sequence element in the 5′ constituent stretch of the stem of the Ia motif plays a pivotal role during RNA replication. The transition of each of the consecutive residues, which in the case of mutant 5 were replaced in concert, substantiated indeed the relevance of nucleotides A-3 (mutant 6) and A-5 (mutant 7). However, because the negative impact of individual nucleotide substitutions was observed to be not nearly as profound as if they were exchanged together, we concluded that these residues create a common replication signal. In keeping with this notion, modification of C-6 (mutant 8) had almost no influence on the replication behavior of the DI9c RNA (Fig. 4).
FIG. 5.
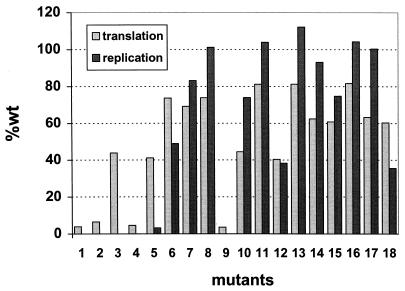
Side-by-side projection of the translation and replication capabilities of each of the DI9c derivatives used in this study (see also Fig. 3B and 4B).
Prior to initiating a detailed reverse genetics study, we substituted the homologous portion of the genome of other BVDV sources (for example strain Osloss) for the entire NADL-derived 5′-terminal part of the DI9c RNA (see above). Interestingly, the replication capability of the resulting RNAs, which deviated from the original replicon only in the loop part of the motif (Fig. 1), remained largely unaltered (data not shown). In the same context, subtle modifications of the loop sequence, such as in the cases of mutants 13, 14, 16, and 17 (Fig. 2), were detected not to have any effect or even a stimulatory effect on DI9c RNA replication (Fig. 4). On the other hand, several lesions in the loop region were found to interfere with the RNA replication pathway. These involved mutation 3, with which a huge portion of the motif had been converted into a HCV-BVDV chimera, and mutation 12, which besides abolishing the top part of the stem structure extended the loop region from 14 to 20 nucleotides. Because these mutations lowered the translation rate in a similar way to mutations 5 and 10 (see above), we explained their inhibitory influence by mixed effects on translation as well as replication. Other interventions, however, enabled translation to occur at reasonably high levels, but still affected replication (Fig. 3, 4, and 5): i.e., mutant 15, where the overall shape of the loop was modified into a bulge-stem-loop structure (chemical modification data confirming this assumption not shown), and mutant 18, which lacked a considerable part of the loop (nucleotides 14 to 19). We deduced from these results that Ia comprises an additional signal aside from the above-mentioned sequence element in the 5′ part of the stem, which is crucial for the RNA replication process; rather than its sequence composition, it apparently concerns the physical integrity of the loop of the motif (Fig. 6 [see also below]).
Formation of the stem structure of the Ia (−) motif is not needed for RNA replication.
It is important to state that the above-defined replication signals (the sequence motif as well as the loop region) may be active in either the Ia motif or the Ia (−) motif, respectively. Since correct folding of the stem structure of the positive-strand Ia motif was found to be an important prerequisite for the efficient initiation of translation, it is also impossible to differentiate its role during RNA replication. To determine on the other hand whether the negative-strand RNA requires formation of the stem helix, we could take advantage of a so-called asymmetric mutational approach, which is based on the particular property of G-U base pairs: incorporation of G-U pairs can replace A-U pairs in the positive-strand RNA, but on the complementary strand, the A-C base pairs cannot form. Thus, mutations 5, 6, 7, 8, and 10 kept the stem of Ia, but impaired or at least compromised stem formation in the Ia (−) motif (Fig. 2 and Table 1). In view of this scenario, our interpretation is that RNAs carrying mutations 7, 8, and 10 replicate at a reasonably high level in such a way that folding of the stem of the Ia (−) motif has only minor influence on the RNA replication pathway. Conversely, the replication debility of mutants 5 and 6 could evidently be traced back to the modification of the above-defined sequence element in the genomic or antigenomic RNA, respectively (see above).
DISCUSSION
The aim of this study was to gain evidence as to the assignment of cis-acting signals encoded by the extreme 5′ terminus of the BVDV genome during translation and replication of the viral RNA. To address these issues, we employed the BVDV DI9c replicon as a well-established experimental system, which during several recent studies considerably extended our knowledge of the role of virus-encoded determinants of the RNA replication pathway (6, 17, 51). In accord with some findings of these works, we investigated translation and replication of the BVDV replicon in the context of BHK-21 cells. (i) Besides MDBK cells, BHK cells support replication of BVDV RNA most efficiently. (ii) Due to constantly high transfection rates (see above) and an insignificant frequency of revertants or pseudorevertants that arise during the short-term experimental approaches making use of mutated replicons, in vivo replication studies were found to be most reproducible with BHK cells. (iii) Translation of the viral RNA, irrespective of whether it was performed in vivo or in vitro, was defined to be most efficient and reproducible in assay systems using BHK cells or cytoplasmic extracts of these cells, respectively (Fig. 3 [some data not shown]).
Before moving into functional studies of the 5′ terminus of the BVDV DI9c RNA, it was important to characterize its major RNA features. As demonstrated by biochemical structure probing, the immediate 5′-terminal 32 nucleotides and the complementary region at the 3′ end of the negative-strand replication intermediate unequivocally fold into a stable stem-loop secondary structure, which in accordance with previous computer predictions was designated as the Ia and Ia (−) motifs, respectively. Almost all nucleotides proposed to be involved in base pairing were shown to be inaccessible to single-strand-specific reagents (or enzymes), whereas all residues of the intervening (loop) sequence turned out to be highly reactive (Fig. 1). Therefore, intramolecular cross-talk such as of the distinctive loop region with distant parts of the RNA molecule (an attractive model considering the assumed regulatory function of the motif [see below]) was not recorded under the conditions of our analysis with naked RNA. As mirrored by the pattern of phylogenetically conserved nucleotide constituents, the stem-loop structure consists of several distinguishable elements: i.e., the lower backbone of the stem, which is identical in all pestiviruses; the less-conserved upper stem portion; and, finally, the loop region, which is only a little conserved. In the course of the reported experiments, this multipartite composition was validated as having a notable functional impact (Fig. 1C and 6).
Our data suggesting hairpin Ia as a crucial part of the IRES-mediated translation process of the BVDV ORF are predominantly based on a translation assay in vitro, with which we analyzed the genuine DI9c replicon and DI9c Ia derivatives in cytoplasmic-initiation factor fractions, which were prepared from authentic host cells (i.e., BHK cells). The applied experimental protocol followed essentially the example of reaction mixtures of HeLa cells, which were shown to allow translation as well as replication of poliovirus RNA up to the assembly of novel infectious virus particles in vitro (4, 26). Although we have no evidence that the BVDV replicon is capable of amplification under these in vitro conditions (see Results), the fact that translation initiates exclusively at the authentic start codon of the viral ORF and that the nascent polyprotein gets cleaved into the different viral proteins indicated that this in vitro system reflects the in vivo situation rather accurately. This hypothesis was clearly confirmed by our in vivo results applying the ΔIa (mutant 1) RNA: considering the differences between both experimental approaches, the data shown in Fig. 3A and C are surprisingly consistent and revealed a compelling relationship between the presence of an unaltered Ia motif on the one hand and efficient translation of the viral RNA on the other hand. Our analysis with a broad range of different Ia DI9c derivatives, which, within the technical limits, were all suggested to exhibit the same stability as the wild-type replicon (see Results), demonstrated particularly how the structure of the lower (conserved) stem region and, to a minor extent, the structure of the upper (less conserved) stem region account for the importance of hairpin Ia as a translation signal (Fig. 3). Thus, disintegration of translation as caused by disruption of the respective RNA double-strand region (mutants 4 and 12) could be mainly compensated for by restoration of the stem architecture with a different nucleotide sequence (mutants 5 and 11). Because the participation of single residues was found to be low (see mutants 6 to 8), we concluded that the overall sequence composition of the lower stem contributed to the function of Ia. In contrast, the (variable) loop region was shown as being only marginally involved in the translation process. Along this line, alterations that affected exclusively the loop were only slightly deleterious (Fig. 3), and substitution of HCV sequences for the entire upper part of the motif (mutant 3) lowered the translation efficiency, although not nearly as dramatically as certain alterations in the lower stem portion (for example, mutant 4).
Our results seemingly contradict those of previous studies which suggested that the 5′ termini of the pestivirus as well as of the HCV 5′ UTR were dispensable for IRES-mediated translation (9, 31, 32 [see also above]). The authors of these works, however, investigated the 5′ UTR separately from the residual viral genome in reporter gene-based assay systems. In fact, constructs in which the wild-type and the mutant 1 (ΔIa) DI9c 5′ UTRs were fused with a reporter gene, respectively, yielded almost identical reporter enzyme activities (H. Yu, unpublished data), a result which is in striking contrast to the negative effect of mutation 1 on the translation of the complete replicative RNA (Fig. 3). Efficient translation of the BVDV DI9c RNA has therefore to be considered as an intricate process, which, besides the 5′ UTR, may demand additional parts of the viral RNA and/or intramolecular communication of different parts of the viral RNA (see also below). The data presented here suggest a correctly folded Ia structure represents an essential prerequisite in this functional framework. Hence, as an intriguing topic of further experiments, it has to be proven whether these observations are also valid for the translation processes of other pestiviruses or even other members of the family Flaviviridae.
Correlation of the translation data of different BVDV replicon derivatives with their capacity to replicate, as is drawn in Fig. 5, revealed a translation rate of 60 to 80% of that of the wild-type RNA, providing apparently sufficient amounts of the NS proteins to catalyze RNA replication at the wild-type level (see, for example, mutant 17). This side-by-side projection allowed us to discriminate two elements of the Ia motif that do not seriously affect translation, but function as signals for an efficient replication of the RNA: i.e., a sequence element consisting of a highly conserved stretch of cooperating nucleotide residues in the lower stem region of the Ia or Ia (−) motif and, more unexpectedly, a variable RNA structure motif encompassing the loop region (Fig. 6). Interestingly, only severe interventions, such as a reduction in the number of constituting residues of the loop from 14 to 8 (mutant 18), were found to be accompanied by an evident negative effect on RNA replication, and even distantly related RNA features, such as similarly folded HCV sequences, could (inefficiently) compensate for the BVDV Ia loop (see mutant 3). Thus, in contrast with the terminal sequence element, the loop signal turned out to be surprisingly tolerant of variations—a fact which may explain the evolutionary drift of this region among different pestiviruses.
Our results are largely compatible with data from previous infection studies with BVDV-HCV or BVDV-encephalomyocarditis virus chimeric viruses which defined the initial four nucleotides of the BVDV genome to represent important replication signals (13). The observation of these authors that (low titers of) hybrid virus particles could be recovered even from RNAs, the 5′ UTR of which contained only these four residues of BVDV origin, might be explained by the fact that the long-term infection experiments bear multiple possibilities for the viral RNA to improve. This may occur either by restoration of a hairpin-like structure via base pairing of the tetranucleotide with other complementary sequence stretches or by second-site mutations, events that are difficult to control and that were apparently not detected under the stringent conditions of the replicon approach presented here (see above).
In the poliovirus system, Andino et al. were able to characterize mutations in the 5′ cloverleaf structure that caused a selective defect in positive-strand RNA synthesis and hence implied that this motif is an essential functional part of a positive-strand promoter (1). Accordingly, the replication signals of BVDV defined here were originally envisaged to operate preferentially in the second step of the replication pathway. However, our finding that none of the introduced Ia or Ia (−) mutations could uncouple both replication steps clearly contradicts this supposition (Fig. 4C [some data not shown]). At the current stage of knowledge, it is difficult to decide whether technical reasons (caused, for example, by a lower replication rate of BVDV) may explain this discrepancy between BVDV and poliovirus. More likely, the data reflect some fundamental differences in the multiplication strategies of both viruses. Similarly as was determined for the BVDV-encoded ATPase/RNA helicase function (17), it is tempting to assume that the Ia motif and/or the Ia (−) motif is recruited at a very early step of the replication pathway, possibly even during formation of the initial replication complex. Considering the bifunctional organization of Ia, it seems plausible that, besides mediating the formation of the proximal IRES structure, the motif may also be involved in other RNA-RNA interactions, such as direct or indirect long-distance cross-talking of the genomic 5′ and 3′ termini. The latter interaction, which has been proposed for several other positive-strand RNA viruses (18, 20, 38, 48), may be necessary for the mutually exclusive initiation of either translation or replication. The elucidation of the exact mode of action of the Ia motif as a potential regulator or modulator of both processes and the characterization of viral and/or cellular factors promoting such higher-order RNA conformations will be a fascinating task of future investigations.
ACKNOWLEDGMENTS
H.Y. and O.I. contributed equally to this work.
C.W.G. and O.I. were supported by SFB 535 “Invasionsmechanismen und Replikationsstrategien von Krankheitserregern,” and H.Y. was supported by the Graduiertenkolleg “Biochemie von Nukleoproteinkomplexen” from the Deutsche Forschungsgemeinschaft at the Justus-Liebig-Universität Giessen. S.-E.B. was partly supported by the Infektionsforschung-Stipendienprogramm (2131) of the BMBF (Bundesministerium Bildung und Forschung) administrated by the Deutsches Krebsforschungszentrum (DKFZ).
We thank N. Tautz for critically reading the manuscript and H.-J. Thiel for generous support.
REFERENCES
- 1.Andino R, Rieckhof G E, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′ end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton D J, Flanegan J B. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becher P, Orlich M, Thiel H-J. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A M, Deliat F G, Kean K M. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown E A, Zhang H, Ping L, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chon S K, Perez D R, Donis R O. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhea virus. Virology. 1998;251:370–382. doi: 10.1006/viro.1998.9425. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen J, Garrett R. Enzymatic and chemical probing of ribosomal RNA-protein interactions. Methods Enzymol. 1988;164:456–468. doi: 10.1016/s0076-6879(88)64061-4. [DOI] [PubMed] [Google Scholar]
- 11.Deng R, Brock K V. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 1993;21:1949–1957. doi: 10.1093/nar/21.8.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francki R I, Fauquet D L, Knudson D L, Brown F. Classification and nomenclature of viruses. Arch Virol Suppl. 1991;2:223–233. [Google Scholar]
- 13.Frolov I, McBride M S, Rice C M. Cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 15.Garmanik A V, Andino R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 16.Garmanik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 17.Grassmann C W, Isken O, Behrens S-E. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol. 1999;73:9196–9205. doi: 10.1128/jvi.73.11.9196-9205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn C S, Hahn Y S, Rice C M, Lee E, Dalgarno L, Strauss E G, Strauss J H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol. 1987;198:33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- 19.Honda M, Ping L-H, Rijnbrand R C A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry site within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Lai M C. An internal polypyrimidine-tract binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated region. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- 21.Jang S K, Pestova T V, Hellen C U T, Witherell G W, Wimmer E. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- 22.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 24.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed from a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 28.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of hepatitis C virus and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 30.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 931–959. [Google Scholar]
- 31.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 32.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohll J B, Percy N, Ley R, Evans D J, Almond J W, Barclay W S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J Virol. 1994;68:4384–4391. doi: 10.1128/jvi.68.7.4384-4391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoes E A, Sarnow P. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steffens S, Thiel H-J, Behrens S-E. The RNA-dependent RNA polymerases of different members of the Flaviviridae family exhibit similar properties under in vitro conditions. J Gen Virol. 1999;80:2583–2590. doi: 10.1099/0022-1317-80-10-2583. [DOI] [PubMed] [Google Scholar]
- 37.Stern S, Moazed D, Noller H F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 38.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura J K, Warrener P, Collett M S. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 41.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H-J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 1059–1074. [Google Scholar]
- 45.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:471–478. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wimmer E, Hellen C U T, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 49.Wiskerchen M, Belzer S K, Collett M S. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J Virol. 1991;65:4508–4514. doi: 10.1128/jvi.65.8.4508-4514.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine protease: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu H, Grassmann C W, Behrens S-E. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J Virol. 1999;73:3638–3648. doi: 10.1128/jvi.73.5.3638-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]