Abstract
The human and simian immunodeficiency virus (HIV-1 and SIVmac) transmembrane proteins contain unusually long intracytoplasmic domains (ICD-TM). These domains are suggested to play a role in envelope fusogenicity, interaction with the viral matrix protein during assembly, viral infectivity, binding of intracellular calmodulin, disruption of membranes, and induction of apoptosis. Here we describe a novel mutant virus, SIVmac-M4, containing multiple mutations in the coding region for the ICD-TM of pathogenic molecular clone SIVmac239. Parental SIVmac239-Nef+ produces high-level persistent viremia and simian AIDS in both juvenile and newborn rhesus macaques. The ICD-TM region of SIVmac-M4 contains three stop codons, a +1 frameshift, and mutation of three highly conserved, charged residues in the conserved C-terminal alpha-helix referred to as lentivirus lytic peptide 1 (LLP-1). Overlapping reading frames for tat, rev, and nef are not affected by these changes. In this study, four juvenile macaques received SIVmac-M4 by intravenous injection. Plasma viremia, as measured by branched-DNA (bDNA) assay, reached a peak at 2 weeks postinoculation but dropped to below detectable levels by 12 weeks. At over 1.5 years postinoculation, all four juvenile macaques remain healthy and asymptomatic. In a subsequent experiment, four neonatal rhesus macaques were given SIVmac-M4 intravenously. These animals exhibited high levels of viremia in the acute phase (2 weeks postinoculation) but are showing a relatively low viral load in the chronic phase of infection, with no clinical signs of disease for 1 year. These findings demonstrated that the intracytoplasmic domain of the transmembrane Env (Env-TM) is a locus for attenuation in rhesus macaques.
Lentivirus transmembrane proteins (TM) share a conserved structural organization, characterized by an N-terminal hydrophobic fusion peptide, an extracellular domain, a hydrophobic membrane anchor domain, and a C-terminal intracytoplasmic domain (ICD-TM) (21, 41). In human and simian immunodeficiency viruses (HIV-1 and SIVmac), the ICD-TM is unusually long (150 to 200 residues) and contains two conserved amphipathic alpha-helices near the C terminus (38). The ICD-TM region of HIV-1 and SIVmac has been implicated in several virologic functions, notably induction of cytopathic effects (38, 55), interaction with the matrix (MA) protein during virion assembly (10, 17), modulation of fusogenicity (43, 51, 61), regulation of envelope (Env) expression at the cell surface (5, 29, 44), and binding of calmodulin (37, 54, 57, 58), possibly related to induction of apoptosis (37, 40). Several molecular clones of SIVmac and HIV-2 contain stop codons in the ICD-TM; these stop codons were introduced following adaptation to human cell lines (7, 19, 33). In some cases (i.e., SIVmac142 and SIVmac1A11), these molecular clones were nonpathogenic in rhesus macaques (7, 33). In the case of two other molecular clones, SIVmac239 and SIVmacBK28, single stop codons in TM were found to revert rapidly upon inoculation of rhesus macaques (19, 27). However, these studies did not address the potential role of the TM intracytoplasmic domain in viral pathogenesis.
We and others have demonstrated that individual point mutations, when introduced separately into the coding region for the SIVmac ICD-TM, reduce viral infectivity, in part by altering envelope stability, processing, and incorporation into virions (7, 24, 43, 50, 51). Based on these observations, we hypothesized that SIVmac ICD-TM mutants might be attenuated for disease induction in rhesus macaques. In an earlier study, we examined the reversion of ICD-TM stop codons in four rhesus macaques inoculated with a viral recombinant consisting of portions of pathogenic SIVmac239 and nonpathogenic SIVmac1A11 (32). Reversion of two ICD-TM stop codons derived from SIVmac1A11 was found to correlate with development of simian AIDS in the context of this recombinant virus. However, this report left several questions unanswered. First, the study was performed retrospectively and therefore did not include detailed longitudinal analysis of virus load and disease course. Second, the chimeric virus contained only simple stop codons in the ICD-TM coding region.
Removal of the entire ICD-TM domain by deletion mutagenesis is impractical for SIVmac because this domain overlaps with the second coding exons of tat and rev, genes that are essential for replication, and the nef gene. The phenotype of a truly “ICD-TM-minus” virus is therefore difficult to determine. To circumvent this difficulty, we constructed a virus, SIVmac-M4, containing multiple point mutations in conserved regions of ICD-TM, without changing sequences for tat, rev, or nef (50). This virus was constructed in the context of pathogenic molecular clone SIVmac239 (42), and contains three stop codons, a +1 frameshift, and three amino acid substitutions designed to disrupt the conserved C-terminal amphipathic alpha-helix of ICD-TM (50). In the present study, the pathogenic potential of SIVmac-M4 was tested in juvenile and neonatal rhesus macaques. Juvenile macaques intravenously inoculated with SIVmac-M4 developed transient viremia, which dropped below detectable levels by 8 to 12 weeks postinoculation, and these animals have remained asymptomatic for 1.5 years. Neonatal macaques that received SIVmac-M4 survived the acute phase of infection and are showing relatively low viral loads at 1 year postinoculation, with no disease signs. Taken together, these results demonstrate that the ICD-TM can be considered a locus for attenuation of SIVmac in rhesus macaques.
MATERIALS AND METHODS
Construction of mutants.
Mutants SIVmac-M1 and SIVmac-M4 were described in an earlier report as SIVmac239WT- stop and SIVmac239-M3 stop, respectively (50). The GenBank accession number for SIVmac239 is M33262 (42). Both mutants were created by oligonucleotide-directed mutagenesis of SIVmac239-Nef+; the oligonucleotide primers used are described in reference 50. SIVmac-M4 contains clusters of mutations at four sites: (i) a single stop codon at nucleotides (nt) 9056 to 9058 of env; (ii) two stop codons at nt 9197 to 9199 and 9203 to 9205; (iii) a single-base-pair deletion, nt 9318, creating a +1 frameshift mutation in the ICD-TM; and (iv) three conserved Arg codons changed to Gly codons in the C-terminal amphipathic alpha-helix of ICD-TM, encompassing nt 9459 to 9470. Taken individually, mutations ii, iii, and iv each reduce viral infectivity, as previously reported (50). SIVmac-M1 contains only the stop codon at nt 9056 to 9058 of env. Cassettes containing individual mutations were reintroduced into a plasmid encoding the 3′ half of the SIVmac239 genome (p239-3′-pSP72, obtained from D. Regier, New England Regional Primate Center). All mutations were verified by extensive restriction analysis followed by DNA sequencing of the entire mutated region. Nucleotide numbering in molecular clone SIVmac239 corresponds to that of Regier and Desrosiers (42).
Preparation of virus stocks.
To recover infectious virus, plasmids encoding the 5′ and 3′ halves of SIVmac239 (wild type or mutants) were linearized in equimolar amounts by digestion with SphI, ligated with bacteriophage T4 DNA ligase, precipitated in isopropanol-sodium acetate, and resuspended in sterile distilled water (50). COS-7 cells, grown in Dulbecco's minimal essential medium with 10% fetal calf serum, l-glutamine, and antibiotics, were transfected with 10 μg of DNA using the calcium phosphate coprecipitation method (47). At 48 h posttransfection, Dulbecco's minimal essential medium was removed and the cells were washed once in RPMI containing 10% fetal calf serum, l-glutamine, and antibiotics (R-10). Then 1 × 106 to 2 × 106 CEMx174 cells were added to each plate in 5 ml of R-10 medium. Coculture was continued overnight. The following day, CEMx174 cells were removed from virus-producing COS-7 cells and placed in individual T-25 flasks. These cultures were maintained for 10 to 15 days, and virus-containing supernatant was harvested on days 7, 10, and 14. Supernatants were evaluated for p27 content using an SIV p27gag enzyme-linked immunosorbent assay (ELISA) (Coulter Immunology, Hialeah, Fla.).
The 50% tissue culture infective dose (TCID50) of each virus stock was determined by serial dilution on replicate cultures of CEMx174 cells in a 96-well plate format. Calculations were performed by the method of Reed and Muench (23). To perform sequence verification, viral RNA was extracted from all stocks using the QIAmp viral RNA purification kit (Qiagen, Chatsworth, Calif.) and reverse transcribed using Superscript II (Gibco-BRL) primed with random hexamers (Pharmacia, Piscataway, N.J.). Amplification products were purified using Qiaquick (Qiagen) and sequenced using sense strand primers SIV-342, bases 8997 to 9019 (5′-TGCTAGCTAAGTTAAGGCAGGGG-3′), and SIV-337, bases 9265 to 9282 (5′-CCAGAGGCTCTCTGCGAC-3′).
In vitro growth kinetics.
Peripheral blood mononuclear cells (PBMC) were obtained from whole blood of uninfected rhesus macaques by Ficoll-Hypaque density centrifugation (Pharmacia). Cells were stimulated in R10 medium, supplemented with 0.5 μg of Staphylococcus enterotoxin A (Toxin Technology, Inc., Sarasota, Fla.) per ml, for 3 days before being infected. For each virus, duplicate infections were set up using 106 cells at a multiplicity of infection of 0.01. Infections were carried out for 2 h in a 37°C, 5% CO2 incubator. The cells were then washed twice in RPMI, placed in duplicate wells of a 24-well culture plate, and maintained for up to 3 weeks. Supernatants were withdrawn for p27 antigen testing every 3 to 5 days and replaced with fresh R10 medium containing 50 U of recombinant interleukin-2 per ml.
Inoculation of rhesus macaques and collection of samples.
The animals used in this study were colony-bred juvenile or newborn rhesus macaques (Macaca mulatta) housed at the California Regional Primate Research Center and determined to be free of simian type D retroviruses, SIV, and simian T-lymphotropic virus. These animals were maintained in accordance with the standards of the American Association for Accreditation of Laboratory Animal Care. Physical examinations were performed at regular intervals to detect lymphadenopathy, splenomegaly, and opportunistic infections. Clinical criteria for euthanasia consisted of three or more of the following: (i) greater than 10% weight loss within 2 weeks or greater than 20% within 2 months; (ii) chronic diarrhea unresponsive to treatment; (iii) infections unresponsive to antibiotic treatment; (iv) inability to maintain body heat or fluids without supplementation; and (v) persistent, marked hematological abnormalities or persistent, marked splenomegaly or hepatomegaly.
Juvenile rhesus macaques were inoculated with 103 or 104 TCID50 of cell-free virus by the intravenous route. From each juvenile macaque, 8 to 15 ml of blood was collected by venipuncture immediately prior to inoculation, at 1, 2, 3, 4, 6, and 8 weeks postinoculation, and at regular intervals thereafter. Plasma was stored for virus load and antibody determinations. Complete blood count and T-lymphocyte subset analysis were performed at each time point. Lymph node biopsies were performed at 2, 8, 16, 32, and 80 weeks postinoculation.
Neonatal rhesus macaques were inoculated intravenously at 2 days of age with 2 × 103 or 1 × 104 TCID50 of cell-free virus. Prior to inoculation, every 2 weeks through week 8 and at regular intervals thereafter, 1.5 ml of blood was collected from each macaque.
Virus load in plasma and mononuclear cells.
Viral p27gag antigen in longitudinal plasma samples was measured with an SIVmac p27gag ELISA kit (Coulter Immunology). Viral RNA in plasma samples was determined by the branched-DNA (bDNA) assay (J. Booth and P. Dailey, Bayer Diagnostics, Emeryville, Calif.) (6).
Cell-associated viremia was measured in peripheral blood and lymph node mononuclear cells (LNMC) by a limiting-dilution assay as previously described (34). Briefly, serial 10-fold dilutions of rhesus PBMC or LNMC were cocultivated with the SIV-susceptible cell line CEMx174 in replicate wells of a 24-well tissue culture plate. Cultures were maintained for 4 weeks. The presence of SIV in culture supernatants was assessed by a p27 antigen ELISA as described above. The cell-associated virus load was calculated using the method of Reed and Muench (34).
PCR amplification and sequencing of viral DNA.
Genomic DNA was isolated from either 107 PBMC or 107 CEMx174 cells cocultured with PBMC from infected macaques (Qiagen tissue kit). TM sequences were amplified in a nested PCR using first-round primers SIV-340 (nt 8901 to 8920) and SIV-341 (nt 9918 to 9940) and second-round primers SIV-342 (nt 8997 to 9019) and SIV-343 (nt 9816 to 9834) and sequenced on an ABI automated sequence analyzer (Applied Biosystems-Perkin Elmer, Foster City, Calif.).
Antibody titers.
Antibodies to SIV were assessed in longitudinal samples from experimentally inoculated macaques and control macaques by using an HIV-1/HIV-2 peptide ELISA (Genetic Systems Corp., Redmond, Wash.).
RESULTS
Rationale for ICD-TM mutations.
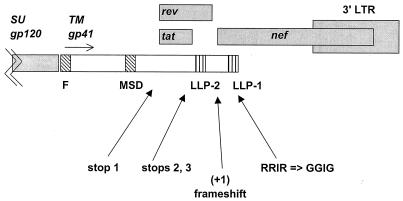
A single stop codon in the ICD-TM would be expected to revert rapidly in vivo in rhesus macaques (19, 25, 27). However, in previous studies we identified several point mutations in the ICD-TM that, taken individually, each reduced viral infectivity and/or delayed virus replication in vitro (50). The presence of stops 2 and 3 (Fig. 1) reduced the stability of Env, reduced Env incorporation into virions, and delayed viral replication by 2 to 3 weeks (50). The +1 frameshift dramatically reduced Env incorporation into virions and abrogated viral replication (50). The RRIR-to-GGIG mutation delayed viral replication by 2 to 3 weeks (50). We reasoned that the presence of all of these mutations downstream of a stop codon would minimize reversion to a full-length TM reading frame in vivo (Fig. 1). None of the mutations in SIVmac-M1 or SIVmac-M4 affected the translation frames for tat, rev, or nef.
FIG. 1.
Mutations in the ICD-TM. Mutants SIVmac-M1 and SIVmac-M4 are described in Materials and Methods and in an earlier report (50). Both mutants were created by oligonucleotide-directed mutagenesis of SIVmac239-Nef+ (GenBank accession number M33262) (42). Mutant SIVmac-M1 contains a single stop codon at nt 9056 to 9058 of SIVmac239-Nef+. Mutant SIVmac-M4 contains clusters of mutations at four sites: (i) a single stop codon at nt 9056 to 9058; (ii) two stop codons at nt 9197 to 9199 and 9203 to 9205; (iii) a single-base-pair deletion, nt 9318, creating a +1 frameshift mutation in ICD-TM; and (iv) three conserved Arg codons changed to Gly codons in the C-terminal amphipathic alpha-helix of ICD-TM, encompassing nt 9459 to 9470 (50). Abbreviations: SU, surface glycoprotein; TM, transmembrane glycoprotein; F, fusion domain; MSD, membrane-spanning domain; LLP, lentivirus lytic peptides 1 and 2.
In vitro replication kinetics of SIV mutants.
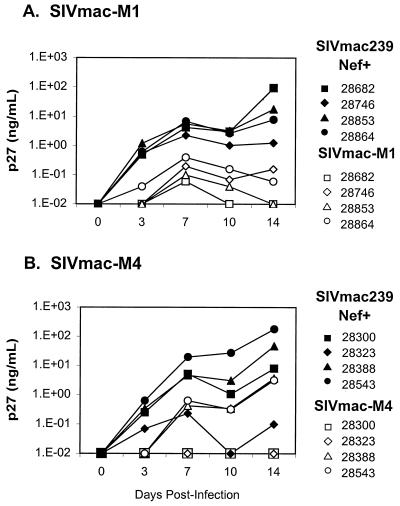
It has been reported that SIVmac encoding a truncated ICD-TM replicates more efficiently than wild-type SIVmac in established human T-cell lines such as HUT-78 (7, 19, 24, 27, 50). In an earlier study, we observed that both SIVmac-M1 and SIVmac-M4 showed more rapid replication than did wild-type SIVmac239-Nef+ in HUT-78 cells (50). In CEMx174 cells, SIVmac239-Nef+, SIVmac-M1, and SIVmac-M4 replicated with identical kinetics (50). It was unclear from these earlier studies whether the presence of ICD-TM mutations would impede the replication of SIVmac in primary mononuclear cells from rhesus macaques. To address this question directly, we prepared cell-free virus stocks of SIVmac-M1 and SIVmac-M4 on CEMx174 cells and used these to infect primary PBMC obtained prior to initiation of the study from each of the eight juvenile macaques that were later inoculated with infectious virus. The viral antigen content (p27) of the stocks was determined by ELISA, and the TCID50 was measured by serial dilution on CEMx174 cells. To rule out the possibility that mutations had reverted during expansion of the viruses in vitro, we performed reverse transcription-PCR, followed by sequence analysis, of the viral nucleic acid isolated from each stock (data not shown). Results of representative experiments are shown in Fig. 2; each experiment was performed in duplicate. For each infection, 106 PBMC were infected with 104 TCID50 of virus and supernatants were harvested every 3 to 5 days for SIV p27gag quantitation. Intriguingly, in primary PBMC from all eight rhesus macaques, SIVmac-M1 and SIVmac-M4 replicated less efficiently than SIVmac239-Nef+ did. The SIV-p27gag antigen level in these cultures was consistently 1 to 2 orders of magnitude lower than that observed in cultures of SIVmac239-Nef+. Taken together, these results demonstrate that SIVmac molecular clones containing a premature stop codon in ICD-TM replicate (i) more efficiently than wild-type SIV in the human T-cell line HUT-78, (ii) with a profile identical to wild-type SIVmac in human T/B hybrid cell line CEMx174, and (iii) less efficiently than wild-type SIVmac in primary rhesus PBMC.
FIG. 2.
Replication kinetics of ICD-TM mutants in PBMC from rhesus macaques. PBMC were obtained from whole blood of juvenile rhesus macaques prior to in vivo inoculation with SIVmac ICD-TM mutants. Staphylococcus enterotoxin A-stimulated PBMC were infected with SIVmac239-Nef+ or ICD-TM mutants at a multiplicity of infection of 0.01. Culture supernatants were withdrawn for p27 antigen testing every 3 to 5 days. All infections were performed in duplicate. (A) Results for PBMC infected with SIVmac-M1; (B) results for PBMC infected with SIVmac-M4.
Infection of juvenile macaques with SIVmac-M4.
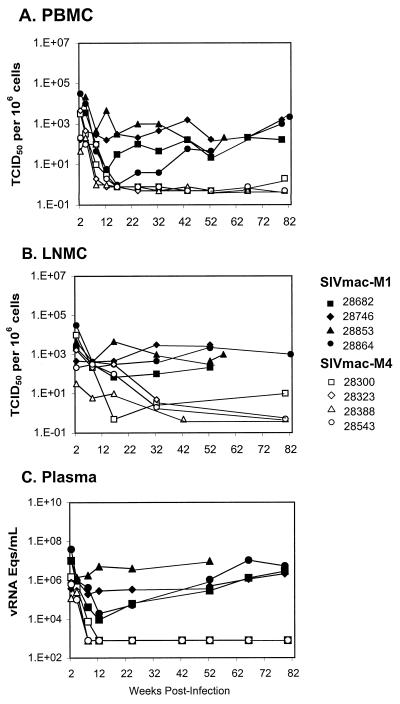
The goal of this study was to determine whether the SIVmac ICD-TM region is essential for high-level virus replication and pathogenesis in rhesus macaques. During the acute phase of infection, plasma viremia in all four juvenile macaques peaked 2 weeks after infection at 1.2 × 105 to 1.5 × 106 copies/ml (bDNA assay, Fig. 3C) and declined to undetectable levels (<1,500 copies/ml) by 8 to 12 weeks. Plasma viremia has remained below detectable limits in all four juveniles up to 80 weeks after infection.
FIG. 3.
Virus load in juvenile rhesus macaques infected with SIVmac ICD-TM mutants. (A and B) The cell-associated virus load was measured in PBMC (A) and LNMC (B) by the limiting-dilution assay (34). Briefly, serial 10-fold dilutions of rhesus PBMC or LNMC were cocultured with CEMx174 cells in replicate wells of a 24-well tissue culture plate. Cultures were maintained for 4 weeks. The presence of SIV in individual wells was assessed by a p27 antigen ELISA. The cell-associated virus load was calculated using the method of Reed and Muench (34). The sensitivity of this method is 1 TCID50 per 106 cells. (C) Viral RNA in plasma samples was determined by the bDNA assay; the sensitivity of the assay used in these experiments was 1,500 copy equivalents/ml of plasma. In all panels, animals inoculated with SIVmac-M1 are indicated by black symbols and animals inoculated with SIVmac-M4 are indicated by white symbols.
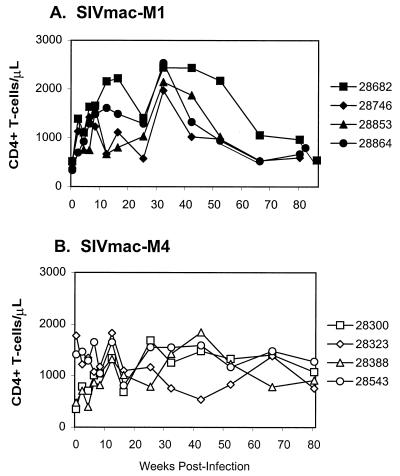
The viral load in PBMC exhibited a similar pattern, with a peak in cell-associated viremia at 2 weeks; subsequently, the PBMC-associated viral load declined to below detectable levels at 8 to 16 weeks and remained at or below the detection threshold thereafter (Fig. 3A). In LNMC, the viral load peaked at 2 weeks and dropped steadily during the first 6 months, reaching borderline-detectable levels (less than 10 TCID50 per 106 cells) at 32 weeks (Fig. 3B). At over 80 weeks postinoculation, all four SIVmac-M4 infected juveniles currently remain clinically healthy and asymptomatic, with CD4+ T-cell counts above 500 (Fig. 4B; Table 1).
FIG. 4.
CD4+-T-cell counts in peripheral blood of juvenile macaques. Lymphocyte subset analysis was performed by flow cytometry in a FACScan apparatus (Becton Dickinson, Mountain View, Calif.) after staining fresh whole blood with monoclonal antibodies recognizing CD4+ T cells (OKT4), CD8+ T cells (Leu2A), CD2+ T cells (Leu5b), and CD19+ B cells (Leu16). The figure shows CD4+-T-cell counts in macaques inoculated with SIVmac ICD-TM mutants SIVmac-M1 (A) or SIVmac-M4 (B).
TABLE 1.
Clinical status of rhesus macaques inoculated with SIVmac ICD-TM mutants
| Age | Virus | Animal | Inoculation dose (TCID50) | Clinical status |
|---|---|---|---|---|
| Juvenile | M1 | Mmu 28682 | 1 × 104 | Abdominal abscess; euthanized at 86 wk |
| Juvenile | M1 | Mmu 28746 | 1 × 103 | Lymphadenopathy, opportunistic infections; died of pneumonia at 88 wk |
| Juvenile | M1 | Mmu 28853 | 1 × 104 | Lymphadenopathy, splenomegaly, weight loss, CD4+ decline; euthanized at 57 wk |
| Juvenile | M1 | Mmu 28864 | 1 × 103 | Severe hemorrhagic enteritis, peritonitis; euthanized at 82 wk |
| Juvenile | M4 | Mmu 28300 | 1 × 104 | Healthy, asymptomatic through 80 wk |
| Juvenile | M4 | Mmu 28323 | 1 × 103 | Healthy, asymptomatic through 80 wk |
| Juvenile | M4 | Mmu 28388 | 1 × 103 | Healthy, asymptomatic through 80 wk |
| Juvenile | M4 | Mmu 28543 | 1 × 104 | Healthy, asymptomatic through 80 wk |
| Juvenile | 239-Nef+ | Mmu 26084a | 1 × 103 | Simian AIDS; euthanized at 51 wk |
| Juvenile | 239-Nef+ | Mmu 27098a | 1 × 103 | Simian AIDS, euthanized at 87 wk |
| Juvenile | 239-Δ152Nef | Mmu 26873a | 1 × 103 | Healthy through 105 wk |
| Juvenile | 239-Δ152Nef | Mmu 26939a | 1 × 103 | CD4+ T cells declined at 70 wk; euthanized at 105 wk |
| Neonate | M4 | Mmu 31342 | 2 × 103 | Unremarkable (acute phase), healthy (chronic phase)b |
| Neonate | M4 | Mmu 31345 | 2 × 103 | Treated for mild gastrointestinal disorders (acute phase), healthy (chronic phase) |
| Neonate | M4 | Mmu 31346 | 1 × 104 | Treated for gastrointestinal disorders; shigella (acute phase), healthy (chronic phase) |
| Neonate | M4 | Mmu 31348 | 1 × 104 | Unremarkable (acute phase), healthy (chronic phase) |
To determine whether the ICD-TM stop codons and other mutations of SIVmac-M4 had reverted to the wild-type coding sequence, DNA was isolated from PBMC or from PBMC-CEMx174 cocultures for all four juvenile macaques at time points ranging from 25 to 66 weeks. A portion of the SIV Env transmembrane protein (Env-TM) coding region was amplified by PCR, and the amplified DNA fragment was subcloned into a plasmid vector for DNA sequence analysis. Assessment of a total of 28 clones obtained from all four juvenile macaques revealed no reversion of any of the ICD-TM mutations (Table 2).
TABLE 2.
Reversion of ICD-TM mutations in rhesus macaquesa
| Age | Virus | Macaque | Time (wk) postinoculation | No. of clones showing reversion or no reversion |
|---|---|---|---|---|
| Juvenile | M1 | Mmu 28682 | 8 | 7 of 7 TAG → CAG |
| Juvenile | M1 | Mmu 28746 | 8 | 8 of 8 TAG → CAG |
| Juvenile | M1 | Mmu 28853 | 8 | 7 of 7 TAG → CAG |
| Juvenile | M1 | Mmu 28864 | 8 | 12 of 12 TAG → TGG |
| Juvenile | M4 | Mmu 28300 | 32 | 6 of 6 no reversion of TM mutations |
| 66 | 4 of 4 no reversion of TM mutations | |||
| Juvenile | M4 | Mmu 28323 | 32 | 6 of 6 no reversion of TM mutations |
| Juvenile | M4 | Mmu 28543 | 25 | 4 of 4 no reversion of TM mutations |
| 66 | 4 of 4 no reversion of TM mutations | |||
| Juvenile | M4 | Mmu 28388 | 42 | 4 of 4 no reversion of TM mutations |
| Neonate | M4 | Mmu 31342 | 24 | 5 of 5 no reversion of TM mutations |
| Neonate | M4 | Mmu 31345 | 24 | 10 of 10 no reversion of TM mutations |
| 32 | 5 of 5 no reversion of TM mutations | |||
| Neonate | M4 | Mmu 31346 | 24 | 5 of 5 no reversion of TM mutations |
| Neonate | M4 | Mmu 31348 | 24 | 6 of 6 no reversion of TM mutations |
Genomic DNA was isolated from either 107 PBMC or 107 CEMx174 cells cocultured with PBMC from infected macaques (Qiagen tissue kit). TM sequences were amplified in a nested PCR as describe in Materials and Methods. PCR products were sequenced on an ABI automated sequencer analyzer. Both strands were sequenced.
Infection of juvenile macaques with SIVmac-M1.
Earlier studies revealed rapid in vivo reversion of single stop codons in the nef gene and in Env-TM of SIVmac (15, 19, 25, 27). Based on the results of these studies, we chose to use SIVmac-M1, which contains a single stop codon at position 733 of Env, as a control in our experiments. Juvenile rhesus macaques inoculated with SIVmac-M1 developed persistent plasma- and cell-associated viremia, as shown in Fig. 3. Plasma viremia, as measured by the bDNA assay, attained peak values at 2 to 4 weeks postinoculation (1 × 106 to 4 × 107 copies/ml) and remained above 0.9 × 104 copies/ml in all animals throughout the first year of infection (Fig. 3C). Virus was consistently rescued from PBMC and LNMC of these four animals at all time points from 2 to 52 weeks (Fig. 3A and B). One animal, Mmu 28853, had persistent viremia of over 106 viral RNA copies per ml of plasma; this macaque developed lymphadenopathy, splenomegaly, weight loss, and CD4+ T-cell decline and was euthanized at week 57 of the study (Table 1; Fig. 4). Two other animals in this group were euthanized due to deteriorating health: Mmu 28864 developed severe hemorrhagic enteritis and peritonitis and was euthanized at week 82, and Mmu 28682 developed an abdominal abscess and was euthanized at week 86. The fourth animal, Mmu 28746, succumbed to pneumonia at week 88; this animal had a history of lymphadenopathy and opportunistic infections prior to developing pneumonia.
The high viral loads measured in the four SIVmac-M1-infected animals suggested rapid reversion of the single ICD-TM stop codon in vivo. To evaluate potential reversions, genomic DNA was isolated from PBMC or from PBMC-CEMx174 cocultures for all four animals at 8 weeks postinoculation. TM sequences were amplified by PCR and cloned into a plasmid vector as described above. As shown in Table 2, all clones obtained from three of four animals (Mmu 28682, Mmu 28746, and Mmu 28853) inoculated with SIVmac-M1 showed reversion of the stop codon to the original CAG (Leu) codon. Sequences from the fourth animal (Mmu 28864) showed mutation of the stop codon to TGG (Trp) in all clones examined (Table 2).
Juvenile macaques infected with SIVmac239-Nef+ and SIVmac239Δ152Nef.
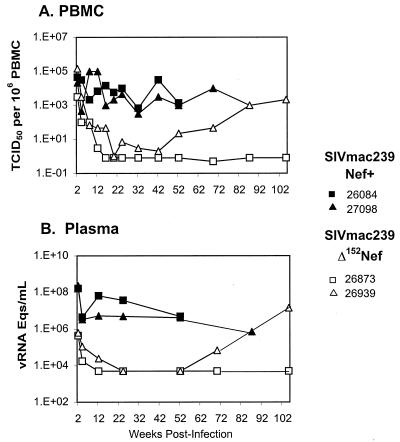
We previously described virologic and clinical data for two juvenile macaques intravenously inoculated with 103 TCID50 of SIVmac239-Nef+ (animals Mmu 26084 and Mmu 27098), and two juveniles (Mmu 26873 and Mmu 26939) inoculated in the same manner with SIVmac239Δ152Nef (26, 49). These data are briefly summarized here (Table 1; Fig. 5) to provide a basis for comparison with animals in the present study. The plasma viral load in animals Mmu 26084 and Mmu 27098 reached a peak at 1.6 × 108 to 2.3 × 108 copies/ml, 1 to 2 orders of magnitude higher than observed in the SIVmac-M1-inoculated animals during the acute phase, and remained above 3 × 106 copies/ml in both animals throughout the first year of infection (Fig. 5) (26). Both animals developed simian AIDS (Table 1). In contrast, plasma viremia in Mmu 26873 and Mmu 26939, which received SIVmacΔ152Nef, reached a peak at 4 × 105 to 6 × 105 copies/ml at 2 weeks postinoculation and dropped below detection limits by 12 weeks, similar to the pattern observed in SIVmac-M4-infected animals (Fig. 5B) (49). Mmu 26873 remained healthy and asymptomatic for 2 years, at which time it was euthanized. Mmu 26939 exhibited an increase in viral load after 70 weeks, concurrent with viral genetic changes leading to the expression of a novel truncated form of Nef, designated t-Nef (Fig. 5B) (49).
FIG. 5.
Virus load in juvenile rhesus macaques infected with SIVmac239-Nef+ or SIVmac239Δ152Nef. Juvenile macaques Mmu 26084 and Mmu 27098 were intravenously inoculated with 103 TCID50 of SIVmac239-Nef+ (black symbols) (26). Juvenile macaques Mmu 26873 and Mmu 26939 were intravenously inoculated with 103 TCID50 of SIVmac239Δ152Nef (white symbols) (49). (A) PBMC-associated viral load quantification was performed as described in the legend to Fig. 3. (B) Plasma viremia was determined by the bDNA assay. The sensitivity of the assay used in these experiments was 10,000 copy equivalents/ml of plasma.
Antibody titers in SIV-infected macaques.
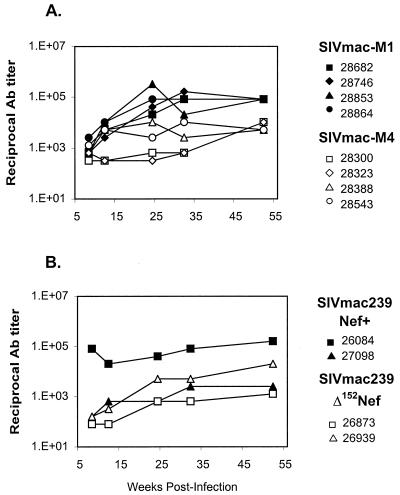
Antibodies to SIVmac were assessed in longitudinal samples from experimentally inoculated macaques and controls in an ELISA utilizing HIV-1 and HIV-2 peptides (Fig. 6). All animals exhibited detectable antibody responses by 8 weeks postinoculation. Peak reciprocal titers during the first 52 weeks ranged from 8 × 104 to 3 × 105 in the four animals that received SIVmac-M1 (Fig. 6A). Peak reciprocal titers were 2 to 3 orders of magnitude lower in animals that received SIVmac-M4, ranging from 6 × 102 to 1 × 104 (Fig. 6A). Of two macaques that received SIVmac239-Nef+, one had antibody titers comparable to those of animals that received SIVmac-M1; the titers in the other macaque were lower (Fig. 6B) (26). Antibody titers in juvenile macaques that received SIVmac-M4 were similar to those observed in animals that received SIVmac239Δ152Nef (Fig. 6B) (49).
FIG. 6.
Serum antibody titers to SIVmac in juvenile macaques. Antibodies to SIV were assessed in longitudinal samples from experimentally inoculated macaques, using an HIV-1/HIV-2 peptide ELISA. (A) Antibody titers in juvenile macaques inoculated with ICD-TM mutants SIVmac-M1 (black symbols) or SIVmac-M4 (white symbols); (B) titers in juvenile macaques inoculated with SIVmac239Nef+ (black symbols) or SIVmac239Δ152Nef (white symbols).
Infection of neonatal macaques with SIVmac-M4.
Recent reports suggested that multiply deleted viruses derived from SIVmac239, which may have an attenuated phenotype in adults, are pathogenic when administered orally or intravenously to newborn macaques (3, 4, 35, 49). It was therefore critical to assess the pathogenicity of SIVmac-M4 in rhesus neonates. Four neonatal macaques were inoculated intravenously with a cell-free preparation containing infectious SIVmac-M4, either 2 × 103 TCID50 (Mmu 31342 and Mmu 31345) or 1 × 104 TCID50 (Mmu 31346 and Mmu 31348) (Fig. 7). All four animals survived the acute phase of infection. Two of the four neonates (Mmu 31345 and Mmu 31346) were treated with antibiotics for gastrointestinal symptoms during the first 2 weeks; however, these animals responded to therapy. Plasma viral loads, as measured by bDNA, peaked at 1 × 106 to 5 × 107 copies/ml at 2 weeks and declined steadily thereafter, falling below the detection threshold in three of four animals (Fig. 7A). The PBMC-associated viral load also declined steadily during the first 16 weeks of infection (Fig. 7B). All four neonates had detectable antibodies to SIVmac (Fig. 7C). Currently, at 52 weeks postinoculation, all four neonates remain clinically healthy, with CD4+-T-cell counts of >1,500/μl (Fig. 7D), and will continue to be closely monitored.
FIG. 7.
Neonatal rhesus macaques infected with SIVmac-M4. Four neonatal macaques were intravenously inoculated with infectious SIVmac-M4. Macaques Mmu 31342 and Mmu 31345 received 2 × 103 TCID50 of infectious virus; macaques Mmu 31346 and Mmu 31348 received 1 × 104 TCID50. (A) Viral RNA in plasma samples was determined by the bDNA assay; the sensitivity of this method was 1,500 viral RNA copy equivalents. For neonatal macaques, the volume of plasma tested was 400 μl. (B) Cell-associated virus load was measured in PBMC by a limiting-dilution coculture assay as described in the legend to Fig. 3. (C) Antibody titers were assessed in serum from neonatal macaques as described in the legend to Fig. 6. (D) CD4+-T-cell counts were determined as described in the legend to Fig. 4.
To assess the stability of the ICD-TM mutations in newborn macaques, genomic DNA was isolated from PBMC or from cocultures of PBMC and CEMx174 cells for all four animals at 24 weeks and from one animal (Mmu 31345) at 32 weeks. PCR amplification followed by DNA sequence analysis of the TM region from a total of 31 clones from all four neonates revealed no reversion of any of the M4 mutations (Table 2).
DISCUSSION
ICD-TM and viral attenuation.
The major finding of this study, based on in vivo analysis of SIV clones with specific mutations, is that the ICD-TM domain of SIVmac is necessary for the development of persistent viremia and pathogenesis in rhesus macaques. The precise virologic functions of lentivirus ICD remain unresolved; however, in vitro studies have pointed to a role for these domains in viral replication kinetics, infectivity, host cell tropism, virion assembly, cytopathicity, intracellular trafficking, endocytosis, and signal transduction (5, 7, 10, 14, 17, 18, 24, 28, 44, 52, 61). The length of ICD-TM has been shown to influence envelope incorporation into virions, probably through interaction with the viral matrix protein (10, 17). The density of the Env glycoprotein is believed to be greater on viruses with a short ICD-TM (e.g., SIVmac239-M1 and SIVmac239-M4) than on those with a full-length TM (e.g., SIVmac239Nef+) (24). Envelope proteins with a short ICD-TM are also more fusogenic than those with a full-length TM (43, 51, 61). Although higher Env density and/or greater fusogenicity might be required for efficient viral entry in certain cell types (e.g., HUT-78), lower Env density and fusogenic capacity might permit more favorable virus-host cell interactions in other cases (e.g., rhesus PBMC) (24, 50). Another hypothesis can be drawn from studies that implicate conserved regions near the C terminus of ICD-TM in induction of cytopathic effects (lentivirus lytic peptides), calmodulin binding, and enhanced apoptosis (37, 38, 40, 58). According to this view, a full-length ICD-TM would be essential for gp160-enhanced apoptosis, mediated via calmodulin binding (37, 40). Calmodulin binding by ICD-TM might also be relevant to T-cell signaling events that play a role in viral replication (54). Further studies are required to identify the ICD-TM functions that are relevant to pathogenesis and to understand the virologic basis for attenuation of ICD-TM mutants.
In light of a recent report suggesting that genetic changes in the ICD-TM may compensate in vivo for deletion of nef sequences, it is intriguing to speculate on potential functional similarities between these proteins (L. Alexander, P. Ilyinski, X. Alvarez, R. Veazey, A. A. Lackner, and R. C. Desrosiers, Abstr. 7th Conf. Retroviruses Opportunistic Infect. abstr. 150, 2000). The SIVmac nef gene is dispensable for virus replication in cultured T-cell lines but enhances replication in unstimulated PBMC (1, 39, 53). Like ICD-TM, Nef interacts with cellular adaptor proteins, is membrane associated, and has been implicated in modulation of signal transduction in T cells (reviewed in references 16, 46, and 48). Because of these similarities, it will be important to monitor viral sequences isolated from macaques infected with SIVmac-M4 for potential compensatory changes in the nef coding region.
A hierarchy for live-attenuated SIVmac239 mutants.
A hierarchy for live-attenuated SIVmac239 mutants has been proposed, based on parameters such as disease induction, cell-associated virus load, plasma viremia, and level of antiviral antibody responses in rhesus macaques (13). The proposed ranking, from least attenuated to most highly attenuated, was as follows: SIVmac239 ΔVpr > ΔVpx > ΔVprΔVpx = ΔNef > Δ3 > Δ3x ≥ Δ4 > ΔVif > Δ5 (13). It is difficult to precisely situate SIVmac-M4 in this hierarchy because the methods for evaluating virus load and antibody titers differ among laboratories and because the Nef-deleted virus (SIVmac239Δ152Nef) constructed in our laboratory is not identical to the SIVmac239ΔNef mutant described elsewhere (25, 49). However, SIVmac-M4, which contains multiple mutations in ICD-TM, is clearly more strongly attenuated than SIVmac-M1, which contains a single stop codon in ICD-TM. Animals infected with SIVmac-M4 developed transient viremia in the range of 1 × 105 to 1.5 × 106 viral RNA equivalents/ml of plasma (mean, 7.5 × 105), 2 to 3 orders of magnitude below that detected in animals infected with SIVmac239-Nef+ but comparable to that observed in one juvenile macaque, Mmu 26873, infected with SIVmac239Δ152Nef (26, 49). In our laboratory, four juvenile macaques that received SIVmac-M4 remain healthy and asymptomatic with low viral loads at 80 weeks postinoculation whereas one of two macaques inoculated with SIVmac239Δ152Nef developed a high viral load and progressed to simian AIDS (49).
In a study of three highly attenuated mutants (SIVmacΔ3, SIVmacΔ3× and SIVmacΔ4) in juvenile macaques, peak plasma viral loads for SIVmacΔ3 were comparable to those we detected in animals infected with SIVmac-M4 (13). Peak levels of PBMC-associated virus for SIVmacΔ3 showed a 13.5-fold reduction relative to those of wild-type SIVmac239-Nef+, compared to a 16.8-fold reduction for SIVmac-M4 relative to SIVmac239-Nef+ in the present study (Fig. 3) (13). Taken together, these results suggest that the attenuation level of SIVmac-M4 is similar to or slightly greater than that of SIVmac239Δ152Nef or SIVmacΔ3.
Correlation of acute-phase virus load with clinical outcome.
Several recent reports suggested that events occurring during the first 6 weeks of infection determine the eventual disease course (20, 30, 36, 56, 59). In the present study, sequence analysis of viral nucleic acid obtained from SIVmac-M1-inoculated animals at week 8 postinoculation indicated that the single ICD-TM stop codon had mutated to a coding triplet in all clones obtained from all four macaques. In contrast, viral genomes obtained from SIVmac-M4-inoculated animals as late as 66 weeks showed no reversion of any of the ICD-TM mutations (Table 2). This result strongly suggests that early restoration of the wild-type ICD-TM in the SIVmac-M1 animals correlated with a high viral load during the acute phase. High acute-phase viremia, in turn, was predictive of persistent viremia and progression to disease, as previously reported (20, 30, 36, 56, 59).
Attenuation threshold.
The “threshold hypothesis” was proposed to account for the in vivo attenuation of SIVmac clones containing mutations in accessory genes (45). According to this view, mutations in individual viral genes contribute to attenuation not by abrogating essential virologic functions but by modulating virus replication efficiency in vivo. The reduced replication observed for SIVmac-M1 and SIVmac-M4 in primary macaque PBMC (Fig. 2) supports the notion that a full-length ICD-TM is required for optimal replication in primary macaque mononuclear cells. Therefore, decreased replication efficiency alone may be sufficient to account for the observed in vivo attenuation of SIVmac-M4. Additional studies are required to establish whether the ICD-TM domain also performs an essential virologic function (i.e., cellular activation, modulation of protein trafficking, or facilitation of viral assembly) in primary cells.
Implications for vaccine development.
Live-attenuated viruses derived from SIVmac molecular clones have shown promising levels of protection in macaques (2, 9, 11, 60). However, the ability of an attenuated viral vaccine to induce protective immunity is dependent on the ability of the virus to replicate in the host (12, 22, 31) and the length of time between vaccination and pathogenic challenge (8, 9). The most highly attenuated viral strains, which replicate poorly in vivo, are unlikely to induce protection; additionally, an inverse relationship was demonstrated between the level of attenuation and the degree of protection from challenge with virulent virus (13, 22, 31). Enthusiasm for this approach has also been somewhat moderated by the observation that live, attenuated mutants can induce disease in newborn (3, 4) and juvenile (4, 49) macaques. Additional investigations are required to determine to what extent the macaques that received SIVmac-M4 can be protected from challenge with pathogenic viruses. However, used in combination with mutations in other viral genes, site-directed mutations in the ICD-TM may serve to expand the repertoire of attenuated SIVmac molecular clones that will be useful in vaccine development.
ACKNOWLEDGMENTS
We thank the veterinary and Colony Services staffs of the California Regional Primate Research Center, especially Linda Hirst, David Bennett, and Wilhelm von Morgenland, for their help with the rhesus macaques. We thank J. Booth and P. Dailey (Bayer Diagnostics) for bDNA analysis. Lou Adamson, Elmer Mortel, Joseph Oh, Kimberli Schmidt, Mike Stout, and Joann Yee (UC Davis) and Bertrand Boson (Institut Cochin) are acknowledged for expert technical assistance. We also thank Marta Marthas for retrospective data on SIVmac1A11-infected animals, and helpful discussions.
This research was supported by a National Institutes of Health (NIH) grant (RO1-A139415) to M.B.G. and the base grant to the California Regional Primate Research Center, CRPRC (NIH-RR00169). B.L.S. was a Research Fellow of SIDAction, Paris, France, and received a Pediatric AIDS Foundation short-term scientific award (PS-22046-22).
REFERENCES
- 1.Alexander L, Du Z, Rosenzweig M, Jung J U, Desrosiers R C. A role for natural simian immunodeficiency virus and human immunodeficiency virus type 1 nef alleles in lymphocyte activation. J Virol. 1997;71:6094–6099. doi: 10.1128/jvi.71.8.6094-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almond N, Kent K, Cranage M, Rud E, Clarke B, Stott E J. Protection by attenuated simian immunodeficiency virus in macaques against challenge with virus-infected cells. Lancet. 1995;345:1342–1344. doi: 10.1016/s0140-6736(95)92540-6. [DOI] [PubMed] [Google Scholar]
- 3.Baba T W, Jeong Y S, Pennick D, Bronson R, Greene M F, Ruprecht R M. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science. 1995;267:1820–1825. doi: 10.1126/science.7892606. [DOI] [PubMed] [Google Scholar]
- 4.Baba T W, Liska V, Khimani A H, Ray N B, Dailey P J, Penninck D, Bronson R, Greene M F, McClure H M, Martin L N, Ruprecht R M. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 5.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Y, Ho D D, Todd J, Kokka R, Urdea M, Lifson J D, Piatak M, Jr, Chen S, Hahn B H, Saag M S, Shaw G M. Clinical evaluation of branched DNA signal amplification for quantifying HIV type 1 in human plasma. AIDS Res Hum Retroviruses. 1995;11:353–361. doi: 10.1089/aid.1995.11.353. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole K S, Rowles J L, Jagerski B A, Murphey-Corb M, Unangst T, Clements J E, Robinson J, Wyand M S, Desrosiers R C, Montelaro R C. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor R, Montefiori D, Binley J, Moore J, Bonhoeffer S, Gettie A, Fenamore E, Sheridan K, Ho D, Dailey P, Marx P. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Virol. 1998;72:7501–7509. doi: 10.1128/jvi.72.9.7501-7509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 12.Denesvre C, Le Grand R, Boissin-Cans F, Chakrabarti L, Hurtrel B, Vaslin B, Dormont D, Sonigo P. Highly attenuated SIVmac142 is immunogenic but does not protect against SIVmac251 challenge. AIDS Res Hum Retroviruses. 1995;11:1397–1406. doi: 10.1089/aid.1995.11.1397. [DOI] [PubMed] [Google Scholar]
- 13.Desrosiers R C, Lifson J D, Gibbs J S, Czajak S C, Howe A Y, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edmonson P, Murphey-Corb M, Martin L N, Delahunty C, Heeney J, Kornfeld H, Donahue P R, Learn G H, Hood L, Mullins J I. Evolution of a simian immunodeficiency virus pathogen. J Virol. 1998;72:405–414. doi: 10.1128/jvi.72.1.405-414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerman M, Malim M H. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science. 1998;280:1880–1884. doi: 10.1126/science.280.5371.1880. [DOI] [PubMed] [Google Scholar]
- 17.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J I. SIV adaptation to human cells. Nature. 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R P, Lifson J D, Czajak S C, Cole K S, Manson K H, Glickman R, Yang J, Montefiori D C, Montelaro R, Wyand M S, Desrosiers R C. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson V, Byington R. Quantitative assays for virus infectivity. In: Aldovini A, Walker B, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. pp. 71–76. [Google Scholar]
- 24.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 26.Khan I H, Sawai E T, Antonio E, Weber C J, Mandell C P, Montbriand P, Luciw P A. Role of the SH3-ligand domain of simian immunodeficiency virus Nef in interaction with Nef-associated kinase and simian AIDS in rhesus macaques. J Virol. 1998;72:5820–5830. doi: 10.1128/jvi.72.7.5820-5830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodama T, Wooley D P, Naidu Y M, Kestler H W, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Hoxie J A. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J Virol. 1994;68:7665–7667. doi: 10.1128/jvi.68.11.7665-7667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohman B L, McChesney M B, Miller C J, McGowan E, Joye S M, Van Rompay K K, Reay E, Antipa L, Pedersen N C, Marthas M L. A partially attenuated simian immunodeficiency virus induces host immunity that correlates with resistance to pathogenic virus challenge. J Virol. 1994;68:7021–7029. doi: 10.1128/jvi.68.11.7021-7029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luciw P A, Shaw K E, Shacklett B L, Marthas M L. Importance of the intracytoplasmic domain of the simian immunodeficiency virus (SIV) envelope glycoprotein for pathogenesis. Virology. 1998;252:9–16. doi: 10.1006/viro.1998.9467. [DOI] [PubMed] [Google Scholar]
- 33.Luciw P A, Shaw K E, Unger R E, Planelles V, Stout M W, Lackner J E, Pratt-Lowe E, Leung N J, Banapour B, Marthas M L. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res Hum Retroviruses. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- 34.Marthas M, Ramos R A, Lohman B L, Van Rompay K K A, Unger R E, Miller C J, Banapour B, Pedersen N C, Luciw P A. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marthas M L, van Rompay K K, Otsyula M, Miller C J, Canfield D R, Pedersen N C, McChesney M B. Viral factors determine progression to AIDS in simian immunodeficiency virus-infected newborn rhesus macaques. J Virol. 1995;69:4198–4205. doi: 10.1128/jvi.69.7.4198-4205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 37.Micoli K J, Pan G, Wu Y, Williams J P, Cook W J, McDonald J M. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J Biol Chem. 2000;275:1233–1240. doi: 10.1074/jbc.275.2.1233. [DOI] [PubMed] [Google Scholar]
- 38.Miller M A, Cloyd M W, Liebmann J, Rinaldo C R, Jr, Islam K R, Wang S Z, Mietzner T A, Montelaro R C. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 39.Miller M D, Feinberg M B, Greene W C. The HIV-1 nef gene acts as a positive viral infectivity factor. Trends Microbiol. 1994;2:294–298. doi: 10.1016/0966-842x(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 40.Pan Z, Radding W, Zhou T, Hunter E, Mountz J, McDonald J M. Role of calmodulin in HIV-potentiated Fas-mediated apoptosis. Am J Pathol. 1996;149:903–910. [PMC free article] [PubMed] [Google Scholar]
- 41.Pancino G, Ellerbrok H, Sitbon M, Sonigo P. Conserved framework of envelope glycoproteins among lentiviruses. Curr Top Microbiol Immunol. 1994;188:77–105. doi: 10.1007/978-3-642-78536-8_5. [DOI] [PubMed] [Google Scholar]
- 42.Regier D A, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 43.Ritter G D, Jr, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 44.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 45.Ruprecht R M, Baba T W, Rasmussen R, Hu Y, Sharma P L. Murine and simian retrovirus models: the threshold hypothesis. AIDS. 1996;10:S33–S40. doi: 10.1097/00002030-199601001-00005. [DOI] [PubMed] [Google Scholar]
- 46.Saksela K. HIV-1 Nef and host cell protein kinases. Front Biosci. 1997;2:606–618. doi: 10.2741/a217. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sawai E T, Cheng-Mayer C, Luciw P A. Nef and the Nef-associated kinase. Res Virol. 1997;148:47–52. doi: 10.1016/s0923-2516(97)81913-9. [DOI] [PubMed] [Google Scholar]
- 49.Sawai E T, Hamza M S, Ye M, Shaw K E S, Luciw P A. Pathogenic conversion of live, attenuated simian immunodeficiency virus vaccines is associated with expression of truncated Nef. J Virol. 2000;74:854–899. doi: 10.1128/jvi.74.4.2038-2045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shacklett B L, Denesvre C, Boson B, Sonigo P. Features of the SIVmac transmembrane glycoprotein cytoplasmic domain that are important for Env functions. AIDS Res Hum Retroviruses. 1998;14:373–383. doi: 10.1089/aid.1998.14.373. [DOI] [PubMed] [Google Scholar]
- 51.Spies C P, Compans R W. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology. 1994;203:8–19. doi: 10.1006/viro.1994.1449. [DOI] [PubMed] [Google Scholar]
- 52.Spies C P, Ritter G D, Jr, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:585–591. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivas S K, Srinivas R V, Anantharamaiah G M, Compans R W, Segrest J P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J Biol Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 55.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 56.Staprans S I, Dailey P J, Rosenthal A, Horton C, Grant R M, Lerche N, Feinberg M B. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J Virol. 1999;73:4829–4839. doi: 10.1128/jvi.73.6.4829-4839.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tencza S B, Mietzner T A, Montelaro R C. Calmodulin-binding function of LLP segments from the HIV type 1 transmembrane protein is conserved among natural sequence variants. AIDS Res Hum Retroviruses. 1997;13:263–269. doi: 10.1089/aid.1997.13.263. [DOI] [PubMed] [Google Scholar]
- 58.Tencza S B, Miller M A, Islam K, Mietzner T A, Montelaro R C. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J Virol. 1995;69:5199–5202. doi: 10.1128/jvi.69.8.5199-5202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyand M, Manson K, Garcia M, Montefiori D, Desrosiers R. Vaccine protection by a triple deletion mutant of SIV. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]