Abstract
The assembly and budding of lentiviruses, such as human immunodeficiency virus type 1 (HIV-1), are mediated by the Gag protein precursor, but the molecular details of these processes remain poorly defined. In this study, we have combined pulse-chase techniques with density gradient centrifugation to identify, isolate, and characterize sequential kinetic intermediates in the lentivirus assembly process. We show that newly synthesized HIV-1 Gag rapidly forms cytoplasmic protein complexes that are resistant to detergent treatment, sensitive to protease digestion, and degraded intracellularly. A subpopulation of newly synthesized Gag binds membranes within 5 to 10 min and over several hours assembles into membrane-bound complexes of increasing size and/or density that can be resolved on Optiprep density gradients. These complexes likely represent assembly intermediates because they are not observed with assembly-defective Gag mutants and can be chased into extracellular viruslike particles. At steady state, nearly all of the Gag is present as membrane-bound complexes in various stages of assembly. The identification of sequential assembly intermediates provides the first demonstration that HIV-1 particle assembly proceeds via an ordered process. Assembly intermediates should serve as attractive targets for the design of antiviral agents that interfere with the process of particle production.
The late phase in the human immunodeficiency virus type 1 (HIV-1) life cycle consists of the assembly of new virus particles and their release from the plasma membrane of the infected cell. Particle assembly is directed by the Gag protein (reviewed in references 8 to 10). Gag is produced as a polyprotein precursor and is cleaved near or at the time of budding (18) by the virus-encoded protease to produce mature virions. The major Gag cleavage products include p17MA, which coats the inner leaflet of the viral membrane; p24CA, which forms a cone-shaped structure that encapsulates the RNA virus genome; p7NC, which coats the genomic RNA; and p6. The Gag precursor, Pr55, plays a central role in the assembly of HIV-1 and other retroviruses (reviewed in references 8 and 9). Virionlike particles (VLPs) that bud from the plasma membrane can be produced when Gag is expressed in the absence of other viral proteins (12). In addition, Gag is involved in the recruitment of other viral proteins, including Pol and Env, to the budding virion (8, 9). The fact that Gag contains all the determinants for assembly makes retroviruses an excellent system with which to address fundamental mechanistic questions about the assembly and budding of viruses.
HIV-1 Gag, like all retroviral Gag precursors, is a peripheral membrane protein. Specific targeting of Gag to the plasma membrane is conferred by an N-terminal myristate-plus-basic motif (15, 35). Mutation of either the myristoylation site or the basic residue cluster within the N-terminal region of Gag inhibits Gag membrane binding and drastically reduces budding (13, 35). In addition, analyses of numerous other mutations within Gag have revealed a strong correlation between plasma membrane binding of Gag and budding of infectious virions (for example, references 23 and 24). Other domains in Gag necessary for budding (reviewed in reference 9) are the I domain, believed to mediate oligomerization and RNA binding (27), and the L domain (14), which plays a role in the pinching off of virions from the cell.
Although HIV-1 particles have been studied in structural and chemical detail, the processes of assembly and budding are just beginning to be characterized. Increasing progress has been made toward the assembly of capsidlike structures in vitro (4, 5, 21, 22, 31). These studies suggest several conclusions about the budding of HIV-1. First, Gag contains domains that are capable of self-assembly into distinct structures. Second, assembly requires posttranslational step(s) that involve ATP hydrolysis and multiple cellular factors. Third, RNA probably facilitates the assembly process. The relevance of these in vitro findings remains to be tested within in vivo systems of assembly.
Electron microscopy has also yielded many clues about the assembly process. For C-type retroviruses, and lentiviruses such as HIV-1, assembly intermediates can be visualized as electron-dense patches under the plasma membrane which deform the membrane outward as they grow (11). More advanced intermediates appear as spherical projections connected by a stalklike structure to the plasma membrane. After budding, the electron-dense layer is still visible under the membranes of immature virions. To date, assembly intermediates of HIV-1 have been observed at the plasma membrane only by electron microscopy. In contrast, several biochemical studies have reported the existence of cytosolic HIV-1 assembly complexes (19, 20, 23, 24). The roles of the putative membrane-bound and cytosolic complexes in the assembly process remain to be defined.
In this study, we present detailed biochemical and kinetic analyses of Gag membrane binding and particle assembly in HIV-1 provirus-transfected cells. Newly synthesized Gag is shown to be present mostly in cytoplasmic protein complexes that are rapidly degraded. We also demonstrate that pulse-chase analysis combined with velocity centrifugation can be used to separate distinct assembly intermediates. Moreover, we provide evidence for progressive assembly of membrane-bound intermediates, which can be chased into extracellular VLPs. These data provide the first demonstration that the process of HIV-1 particle assembly can be dissected into distinct temporal stages in a living cell.
MATERIALS AND METHODS
Plasmids and reagents.
Plasmid pHXB2gtpΔBal-D25S (pHXB2ΔBal) (35), a noninfectious HIV-1 proviral construct with a 2.2-kb deletion in Pol and a protease-inactivating point mutation, was a kind gift from L. Parent and J. Wills (Pennsylvania State University Medical School, Hershey, Pa.). p65-VALO (25) (a kind gift from Didier Picard) expresses β-galactosidase (β-Gal) under control of the α-1 globin promoter. p65-Gag-green fluorescent protein (GFP) chimeras have been previously described (36). pCMV5-Gag was created by amplification of p55 M 1-10 (28) (a kind gift from George Pavlakis, National Institutes of Health [NIH]), a Rev-independent form of Pr55gag, by PCR with oligonucleotide primers adding flanking KpnI and XbaI sites. The PCR product was digested with KpnI and XbaI and subcloned into pCMV5 (2), using the KpnI and XbaI sites in the polylinker. The plasmid expressing Raf–K-Ras was a kind gift of Xu Chen in our laboratory. Enhanced chemiluminescence and Western blotting reagents were purchased from Amersham-Pharmacia (Piscataway, N.J.). Secondary antibodies conjugated to horseradish peroxidase were obtained from Amersham-Pharmacia. Tran35S-label was obtained from ICN or NEN. Optiprep was obtained from Gibco Life Technologies (Rockville, Md.).
Antibodies.
Antibodies to Pr55gag and HIV-1 gp120env were obtained from the NIH AIDS Research and Reference Reagent Program. Rabbit anti-p24CA antiserum or human anti-HIV immune globulin was used to detect Gag. Sheep anti-gp120 antiserum was used to detect gp120env.
For detection of GFP, rabbit anti-GFP (Clontech, Palo Alto, Calif.) or monoclonal anti-GFP (Roche, Indianapolis, Ind.) was used. Monoclonal antibody to β-Gal was obtained from Promega (Madison, Wis.). Antibody to Na+,K+ ATPase was obtained from Biomol (Plymouth Meeting, Pa.).
Transfection, metabolic labeling, and cell fractionation.
COS-1 cells were maintained as previously described (33). The cells were seeded to about 50% confluency and transfected with pHXB2ΔBal using FuGene6 (Roche). The cells were passaged approximately 24 h later and were harvested 36 to 48 h after transfection. Metabolic labeling was performed as previously described (33), using 50 μCi of Tran35S-label/ml. The cells were pulse-labeled for 7 min at 37°C, followed by chases for various lengths of time in Dulbecco modified Eagle medium containing 10% fetal bovine serum and 100 μM cysteine and methionine. Jurkat T cells were maintained in RPMI plus 10% fetal bovine serum and were transfected with pHXB2ΔBal by electroporation.
Subcellular fractionations were performed as previously described (1). High-speed pellet (P100) and supernatant (S100) fractions were adjusted to 1× radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 1% Triton-X 100, 10 mM Tris, pH 7.4) and clarified at 100,000 × g for 15 min at 4°C. The fractions were then immunoprecipitated overnight at 4°C with 5 μl of rabbit anti-p25CA antiserum and 30 μl of agarose-protein A/G PLUS beads (Santa Cruz, Santa Cruz, Calif.). The beads were washed 3 times in RIPA buffer and were then adjusted to 1× SDS sample buffer (32 mM Tris [pH 6.8], 1% SDS, 10% glycerol, 0.05% bromphenol blue, 100 mM dithiothreitol). The samples were boiled for 2 min, and the beads were pelleted prior to loading the sample. SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting were performed as described previously (33). Analysis of radiolabeled Gag was performed by exposure to PhosphorImager screens, which were scanned using a Storm apparatus (Molecular Dynamics, Sunnyvale, Calif.). Quantitation and preparation of visual images was performed with ImageQuant software from the same company. Western blots were exposed to BioMax MR film (Kodak, New Haven, Conn.), scanned with an Epson scanner, and quantitated with MacBAS software.
For Optiprep gradient fractionations, the cell homogenate was centrifuged at 1,000 × g for 10 min to remove the nuclei. The pellet was resuspended by Dounce homogenization in isotonic buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris, pH 7.4) and recentrifuged, and the combined supernatants were centrifuged at 100,000 × g as described above. The pellet was resuspended by Dounce homogenization in 1 ml of isotonic buffer containing protease inhibitors and layered on top of a 0 to 18% Optiprep gradient containing 0.25 to 0.18 M sucrose, 1 mM EDTA, 10 mM Tris (pH 7.4), and protease inhibitors. Alternatively, NP-40 to 1% was added to the isotonic buffer after the P100 fraction was resuspended, and the sample was layered over an Optiprep gradient containing 0.1% NP-40. The gradients were centrifuged for 3 h at 37,000 rpm (100,000 × g) in an SW40 rotor (Beckman, Columbia, Md.) at 4°C, and 0.75-ml fractions were collected by puncturing the bottom of the tube. Aliquots from the gradient fractions and the gradient pellet were added to 1× RIPA buffer for immunoprecipitation or to SDS sample buffer for Western blotting or were used directly in enzymatic assays.
Equilibrium sucrose gradients were performed by a modification of a published protocol (20). Briefly, a postnuclear supernatant was prepared from transfected COS-1 cells as described above and was adjusted to 1% NP-40. The sample was then layered over a 16 to 60% continuous sucrose gradient made in TNE buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1 mM EDTA) and centrifuged at 28,000 rpm for 16 h in an SW40 rotor at 4°C.
Flotation assays were performed using slight modifications of a previously described protocol (30). Briefly, nucleus-depleted P100 fractions were prepared as described above and resuspended in 1 ml of 80% (wt/vol) sucrose by Dounce homogenization. The suspension was layered on the bottom of an SW55 tube and overlaid with 2 ml of 65% sucrose (wt/vol) and 1.5 ml of isotonic buffer. All solutions contained 1 mM EDTA, 10 mM Tris (pH 7.4), and protease inhibitors. Centrifugation was performed for 2 h at 200,000 × g and 4°C in an SW55 rotor. Identical results were obtained when centrifugation was performed for 16 h. Fractions were collected from the top of the tube.
Protease protection.
P1, S100, and P100 fractions were prepared as described above, except that the P100 fraction was resuspended in TNE buffer and no protease inhibitors were present at any stage of the cell fractionation. The samples were adjusted to 1× trypsin buffer (50 mM Tris [pH 8.0], 10 mM CaCl2, 1 mM dithiothreitol) with or without 0.2% NP-40 and were incubated briefly at room temperature. Freshly prepared trypsin was added to a concentration of 2 μg/ml, and the samples were incubated at room temperature for 20 min. A modified version of a previously described protease inhibitor cocktail (16) (the concentrations of all inhibitors were tripled, and 15 μg of soybean trypsin inhibitor/ml was also included) was added either before the trypsin or at the conclusion of the incubation, and the samples were incubated for an additional 5 min at room temperature. One milliliter of cold RIPA buffer containing the same protease inhibitor cocktail was added to the samples, and they were immediately placed on ice. Immunoprecipitations were performed with α-p25CA antibody. The immunoprecipitated protein was subjected to SDS-PAGE and autoradiography or Western blotting with human anti-HIV antiserum.
Preparations of VLPs.
VLPs were purified according to standard protocols (3). Briefly, medium from pHXB2ΔBal-transfected COS-1 cells was clarified at 500 × g for 30 min at 4°C and filtered through a 0.2-μm-pore-size filter. The filtrate was layered on top of a 20% sucrose cushion and spun for 2 h at 28,000 rpm in an SW40 rotor. To assess the purity of the VLP preparation, the pellet was resuspended in 1 ml of phosphate-buffered saline, layered over a 0 to 18% Optiprep gradient, and centrifuged for 3 h at 100,000 × g (a variation of a published protocol [7]). The Optiprep fractions were then analyzed as described below.
RESULTS
Identification of HIV-1 Gag assembly domains in Optiprep gradients.
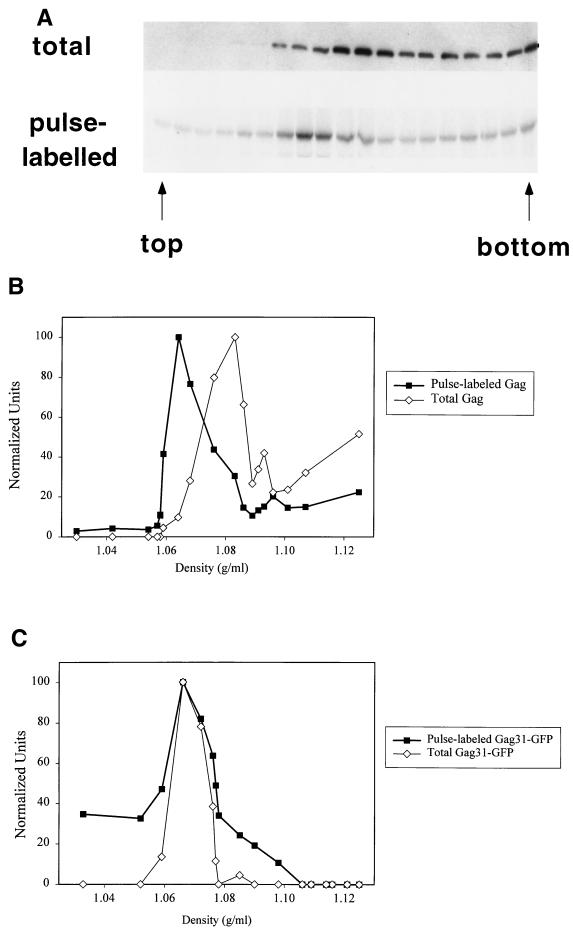
We wished to characterize the assembly and budding of HIV-1 and identify potential intermediates in these processes. Our studies focus on the Gag polyprotein precursor, since it plays a central role in directing the budding of HIV-1 and other retroviruses (9) and is capable of forming VLPs in the absence of other viral proteins. The first set of experiments was designed to follow various stages in Gag assembly by using density gradient centrifugation. Pr55gag was expressed from pHXB2ΔBal (35), a noninfectious proviral construct with an inactivated HIV-1 protease. Transfected cells were pulse-labeled with [35S]cysteine-methionine (Tran35S-label) and hypotonically lysed, and the homogenate was centrifuged at 1,000 × g for 10 min to deplete the preparation of nuclei. The resulting S1 (postnuclear supernatant) fraction was centrifuged at 100,000 × g, and the P100 fraction (26) was fractionated through an Optiprep density gradient. Aliquots from each gradient fraction were immunoprecipitated with anti-p24CA, followed by SDS-PAGE and phosphorimaging to detect newly synthesized Gag or by Western blotting to monitor the total Gag population. Figure 1A and B reveals that newly synthesized Gag and total Gag display strikingly different mobilities on Optiprep gradients, with the newly synthesized Gag migrating in a sharp peak and the total Gag exhibiting a broad distribution throughout much of the gradient. This difference between pulse-labeled and total Gag was reproducibly observed in over five different experiments. A similar result was obtained when Gag expressed in Jurkat T cells was analyzed on Optiprep gradients. Newly synthesized Gag migrated in a sharp peak, whereas the total Gag was broadly distributed throughout the gradient (M. Tritel and M. D. Resh, unpublished data). The Gag-containing structures do not reach their equilibrium densities in these velocity gradients; thus, the difference in mobility could correspond to a difference in the sizes and/or the densities of the structures.
FIG. 1.
Distribution of total and newly synthesized HIV-1 Gag on Optiprep velocity gradients. Transfected COS-1 cells were pulse-labeled for 7 min with Tran35S-label, and a denucleated P100 fraction was centrifuged over a 0 to 18% Optiprep gradient. The fractions were analyzed for newly synthesized and total Gag. (A) Top, distribution of total Gag in a representative gradient. Gradient fractions were analyzed directly by anti-p24CA Western blotting. Bottom, newly synthesized Gag distribution in the same gradient. Gradient samples were immunoprecipitated with anti-CA antiserum, followed by SDS-PAGE and phosphorimaging. (B) Graphic depiction of the gradient in panel A. Bands of total and newly synthesized Gag were quantitated, and the values were normalized to the highest value in each gradient (arbitrarily set at 100) and plotted against the density of the fraction. (C) Graphic depiction of distribution of total and newly synthesized Gag31-GFP on an Optiprep gradient. Optiprep gradient fractionation was also performed with Gag31-eGFP and Gag69-eGFP, which express protein at levels comparable to pHXB2ΔBal, yielding the same results (not shown).
Optiprep gradient fractionation was next performed on Gag31-GFP, a construct that contains the N-terminal membrane-binding motif of Gag (35) fused to GFP. This construct lacks downstream oligomerization domains. No difference between the gradient profiles of newly synthesized and total Gag31-GFP proteins was observed (Fig. 1C), implying that the mobility shift seen with full-length Gag was due to the presence of downstream Gag domains.
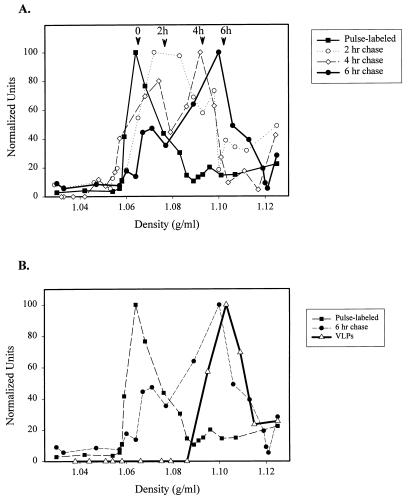
We next combined pulse-chase labeling with Optiprep gradient analysis in order to resolve multiple assembly intermediates. Transfected cells were pulse-labeled for 7 min and chased for various lengths of time. The P100 pellets were resuspended and fractionated on Optiprep gradients. The total amount of radiolabeled Gag decreased with increasing chase time (see below). For comparison, the amount of Gag in each fraction was quantitated and normalized to the levels detected in the peak fraction. As depicted in Fig. 2A, a clear progression of radiolabeled Gag towards heavier gradient fractions was apparent with increasing chase times. By 6 h, migration through the gradient was largely complete, as evidenced by comparison with the steady-state Gag distribution. To compare the migration of the chased material with that of VLPs, VLPs from COS-1 cells were prepared by pelleting the media through a sucrose cushion, as described in Materials and Methods. The Gag protein in the virion pellet was confirmed to be present in bona fide VLPs by fractionation on a velocity gradient that separates cellular contaminants from VLPs (7). Gag was found in the heavier fractions of the gradient, while most cellular proteins were in the lighter fractions (not shown). When the VLPs were fractionated on the Optiprep gradient, their migration was similar to that of Gag after a 6-h chase (Fig. 2B), suggesting that after 6 h, Gag may be present in cellular domains that resemble immature virions. It is also possible that a portion of the 6-h chase sample is derived from VLPs that have not yet detached from the cell plasma membrane.
FIG. 2.
Optiprep gradient migration of intracellular Gag at various times after synthesis and comparison to that of VLPs. (A) Migration of intracellular Gag. Transfected cells were pulse-labeled and incubated in chase medium for 0, 2, 4, or 6 h, and denucleated P100 fractions were prepared and fractionated on Optiprep gradients as described in the legend to Fig. 1. The total counts per minute of the Gag bands decreased dramatically by the first chase point (not shown). The counts per minute of radiolabeled Gag in each fraction was therefore normalized to the densest band in the gradient and plotted against density. A representative experiment is depicted. (B) Migration of Gag in VLPs. VLPs were isolated from the medium of transfected COS-1 cells as described in Materials and Methods and fractionated on an Optiprep gradient. The distribution of Gag in the gradient was determined by Western blotting and is shown superimposed upon the profiles of the 0- and 6-h chase points in the graph in panel A. Each experiment was performed twice.
Newly synthesized Gag rapidly targets to the P100 fraction.
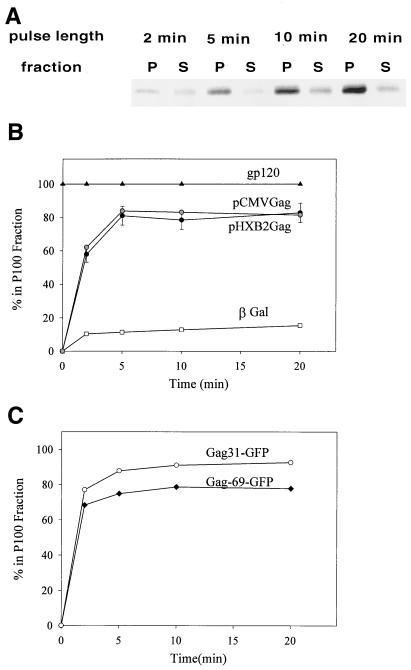
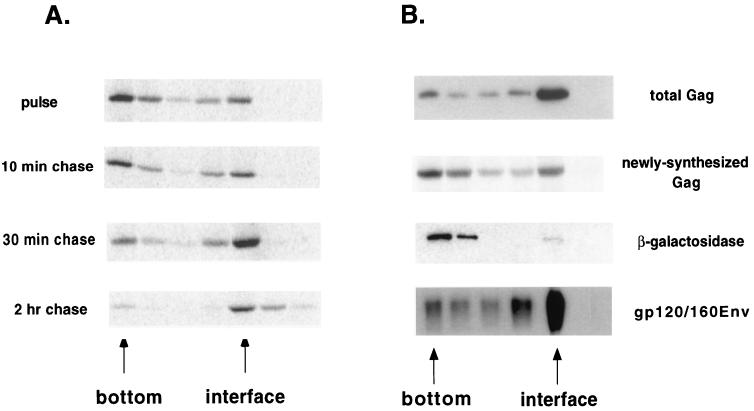
The next set of experiments was designed to determine the kinetics of appearance of the early assembly intermediates and their subcellular localization. Transfected COS-1 cells were pulse-labeled for short times and lysed in hypotonic buffer, and S100 and P100 fractions were generated. Samples were immunoprecipitated with anti p24CA antibody, and SDS-PAGE and autoradiography were performed to determine the distribution of the labeled Gag protein. Fig. 3A shows a representative experiment. At the 2-min time point, approximately 50% of the Gag protein was in the cytosolic fraction. At 5-min and later time points, approximately 80% of the Gag was found in the P100 fraction, equivalent to the steady-state distribution (36). A graph of the average of several experiments (n = 4) is shown in Fig. 3B, along with controls for membrane-bound (HIV-1 gp120/160env) and cytosolic (β-Gal) proteins. As expected, 100% of the Env protein was found in the P100 fraction, whereas 85% of the β-Gal was in the S100 fraction. Reducing the temperature to 25°C slightly slowed the kinetics; 80% of the Gag was associated with the P100 fraction within 5 to 10 min (not shown). These results are consistent with the synthesis of Gag on cytosolic ribosomes, followed by rapid localization to the P100 fraction. The latter could reflect either membrane binding or the formation of dense cytosolic complexes that copellet with membranes.
FIG. 3.
Time course of Gag localization to the P100 fraction. (A) Cells were transfected with pHXB2ΔBal, pulse-labeled for 2, 5, 10, or 20 min, and subjected to hypotonic lysis and P100 (P) and S100 (S) fractionation. The distribution of radiolabeled Gag at each time point was visualized by immunoprecipitation, SDS-PAGE, and autoradiography. A representative experiment is shown. (B) Graphic depiction of the time course of Gag localization to P100 fractions. Gag was expressed from two constructs: pHXB2ΔBal and pCMVGag, which does not encode other HIV-1 proteins. The quantitations of four experiments were averaged and are shown plotted alongside controls for cytosolic (β-Gal) and membrane-bound (gp120/160env) proteins. The control proteins were detected by Western blotting. (C) The same analysis was performed on two constructs expressing 31 or 69 amino acids of N-terminal Gag sequence fused to GFP.
We next wished to ascertain whether rapid P100 localization requires other HIV-1 proteins. To study this question, we utilized pCMV5-Gag, a Rev-independent Gag expression construct (28). As depicted in Fig. 3B, Pr55gag expressed in pCMV5-Gag-transfected cells exhibited very rapid localization to the P100 fraction, demonstrating that the process does not require other HIV-1 proteins.
Previous work by this laboratory established that the N-terminal 31 amino acids of Gag are required for membrane binding and plasma membrane targeting of Pr55gag (35). In order to determine whether this N-terminal sequence is sufficient to drive rapid membrane binding, we performed the experiment with the Gag31-GFP and Gag69-GFP constructs. As depicted in Fig. 3C, Gag31-GFP and Gag69-GFP both localized rapidly to the P100 fraction, in the same manner as full-length Gag protein. Localization of the Gag-GFP constructs to the P100 fraction likely occurs as a result of bona fide membrane binding and not oligomerization, since the Gag31-GFP and Gag69-GFP constructs lack downstream oligomerization domains. These data indicate that the N-terminal motif of Gag is sufficient to mediate rapid membrane binding. This suggests, in turn, that rapid localization of full-length Gag in the P100 fraction might occur as a result of trafficking of monomers to the membrane. Alternatively, a dominant downstream sequence might direct full-length Gag to dense cytosolic complexes.
Newly synthesized Gag is protease sensitive but gradually progresses to protease resistance due to envelopment in membrane vesicles.
Several recent studies (19, 20, 23, 24) have reported the presence of dense cytosolic Gag complexes that copellet with membranes in the P100 fraction. We therefore employed a trypsin protection assay to distinguish between membrane-bound protein and cytosolic complexes. Gag that is bound to the inner leaflet of a right-side-out vesicle should be protected from proteolysis, whereas cytosolic Gag proteins and Gag present on inside-out vesicles should be protease sensitive.
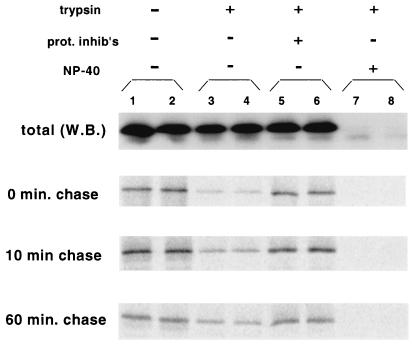
COS-1 cells were transfected with pHXB2ΔBal, pulse-labeled for 5 min, and subjected to hypotonic lysis. The S1 fraction was treated with trypsin, followed by quenching with a protease inhibitor cocktail that included TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone), an irreversible trypsin inhibitor, and immunoprecipitation with anti-p24CA antibody. Alternatively, a denucleated P100 fraction was used with similar results (not shown). As reported previously (16), the majority of the total Gag protein was resistant to proteolysis (Fig. 4, top blot, compare lanes 3 and 4 to lanes 1 and 2). Quantitation of the Western blots revealed that only 35 ± 10% (n = 5) of the Gag was degraded by trypsin. When protease inhibitors were added before the reaction, Gag was not degraded (Fig. 4, lanes 5 and 6), indicating that the proteolysis observed did not occur during the subsequent immunoprecipitation. Addition of 0.2% NP-40, a nonionic detergent that permeabilizes membrane vesicles, rendered >97% of the Gag sensitive to proteolysis (Fig. 4, lanes 7 and 8), implying that the protease resistance is due to the envelopment of Gag in membrane vesicles. In contrast, all the Gag in the S100 fraction was protease sensitive, as expected for a cytosolic protein (not shown).
FIG. 4.
Trypsin protection assay of Gag at various times after synthesis. Denucleated S1 fractions were prepared from transfected cells after pulse labeling and incubation in chase medium for 0, 10, or 60 min. The samples were either untreated (lanes 1 and 2) or treated with trypsin in the absence (−) (lanes 3 and 4) or presence (+) (lanes 5 and 6) of a protease inhibitor cocktail (prot. inhib's.). Samples were treated with trypsin in parallel in the presence of 0.2% NP-40 (lanes 7 and 8). The amounts of total Gag (top panel) and radiolabeled Gag (bottom three panels) in the products were determined by Western blotting (W.B.) and autoradiography, respectively.
The trypsin-sensitive population of Gag observed in the absence of detergent could represent either cytosolic complexes or membrane-bound Gag attached to inside-out plasma membrane vesicles. To confirm that inside-out plasma membrane vesicles were present, cells were cotransfected with a construct expressing c-Raf-1 fused to the C-terminal K-Ras tail. This construct is targeted to the inner leaflet of the plasma membrane (6; Tritel and Resh, unpublished). In contrast to Gag, over 90% of Raf–K-Ras was sensitive to proteolysis (Tritel and Resh, unpublished). Taken together, the protease protection results with Gag and Raf–K-Ras indicate that the majority of the total Gag is bound to the inner leaflet of right-side-out membrane vesicles.
A different protease protection pattern was observed for newly synthesized Gag protein (0-min chase). Most of the newly synthesized protein (85% ± 4%; n = 3) was protease sensitive. A similar result was observed after a 10-min chase, with 79% ± 4% of the Gag being proteolyzed. However, by 60 min, almost half of the Gag (47.7% ± 13%) had become protease resistant. Comparison of the 10- and 60-min chase time points revealed an interesting trend. The total amount of radiolabeled Gag decreased almost threefold from 10 to 60 min (Fig. 4, lanes 1 and 2), while the amount of Gag that was trypsin resistant (lanes 3 and 4) remained constant. These data are consistent with the interpretation that newly synthesized Gag consists primarily of a population of trypsin-sensitive proteins that are either degraded or released from the cell (see below). In addition, a stable population of protease-resistant Gag is present. These molecules are presumably inside membrane vesicles, as they are proteolyzed only when detergent is added to the trypsinization reaction.
The majority of newly synthesized Gag exists in cytosolic complexes.
Because trypsin protection assays cannot distinguish between cytosolic Gag complexes and Gag bound to inside-out membrane vesicles, we employed a sucrose flotation assay to further study Gag's intracellular localization. These assays have been used by other investigators (23, 24, 30) to separate membrane-bound protein, which floats, from dense cytosolic complexes, which remain at the bottom of the centrifuge tube. Denucleated P100 fractions from pulse-labeled, pHXB2ΔBal-transfected COS-1 cells were resuspended in 80% sucrose and overlaid with 65 and 10% sucrose. Following ultracentrifugation, the fractions were analyzed by immunoprecipitation and Western blotting. Most of the pulse-labeled Gag (65.2% ± 7%) remained at the bottom of the centrifuge tube (Fig. 5A). Treatment of the bottom fractions with trypsin resulted in nearly complete proteolysis of the labeled Gag (Tritel and Resh, unpublished), consistent with this fraction representing cytosolic Gag complexes. With longer chase times, the percentage that floated to the 65%-10% interface increased. By 2 h, 92% of the pulse-labeled protein floated, a result similar to the behavior of total Gag protein at steady state (Fig. 5B). Most of the labeled Gag at the interface was resistant to trypsin digestion (Tritel and Resh, unpublished). The membrane marker protein gp120/160env was found at the 65%-10% interface, whereas β-Gal, a cytosolic protein, remained at the bottom of the tube (Fig. 5B). These results indicate that newly synthesized Gag primarily exists in cytosolic complexes. By 30 min after synthesis, nearly all the Gag is membrane bound.
FIG. 5.
Flotation analysis of Gag protein. (A) Transfected cells were subjected to pulse-labeling for 7 min and chase in the presence of excess Met-Cys. Denucleated P100 fractions were prepared, resuspended in 80% sucrose, overlaid with 65 and 10% sucrose, and centrifuged at 200,000 × g for 2 h. The distribution of labeled Gag was analyzed by autoradiography of the gradient fractions. The positions of the bottom of the tube and the 65%/10% interface are indicated. (B) The distribution of labeled Gag in the flotation is shown alongside the distribution of total Gag, as determined by Western blotting. Internal controls for cytosolic (β-Gal) and membrane-bound (gp120/160env) proteins are also shown. Analyses of the newly synthesized and total proteins were performed at least four times. The 10- and 30-min chases were performed once.
Optiprep gradients efficiently reveal detergent sensitivity of intracellular Gag protein.
Several groups (19, 20, 29) have recently reported the presence of Gag in intracellular detergent-resistant complexes. These complexes are presumed to be cytosolic because the mobility of the Gag protein band does not shift on density gradients in the presence of detergent. We therefore used mobility shift on density gradients to determine the percentage of Gag that is membrane bound in the different pulse-labeled samples, as well as the total Gag population. First, the S1 fraction from pHXB2ΔBal-transfected cells was fractionated over a continuous 16 to 60% sucrose gradient (1 to 1.25 g/ml) in the absence or presence of 1% NP-40, a detergent concentration sufficient to dissolve most cellular membranes. A significant fraction of the Gag shifted to the bottom of the density gradient when detergent was present (Fig. 6A), indicating the presence of detergent-sensitive Gag complexes at steady state (see Discussion).
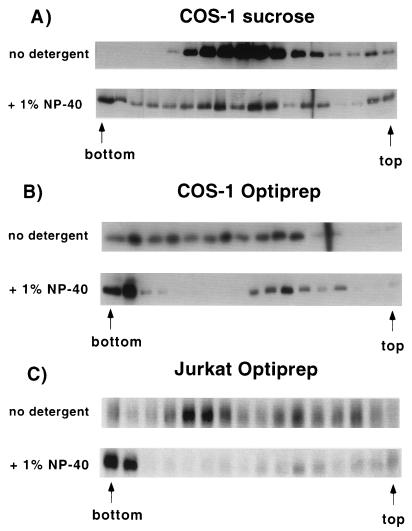
FIG. 6.
Migration of Gag in the absence or presence of detergent in sucrose and Optiprep gradients. (A) Denucleated S1 fractions were prepared from transfected cells, layered over 16 to 60% sucrose gradients after no treatment (top) or treatment with 1% NP-40 (bottom), and centrifuged for 16 h at 100,000 × g. Gag was detected in the gradient fractions by Western blotting. The far left band is the gradient pellet. (B) Denucleated P100 fractions were layered over 0 to 18% Optiprep gradients after no treatment (top) or addition of 1% NP-40 (bottom), centrifuged for 3 h at 100,000 × g, and analyzed for Gag protein. (C) The experiment described for panel B was performed on Jurkat T cells transfected with pHXB2ΔBal.
The experiment was repeated using density gradients of narrower density range to increase the resolution. In addition, Optiprep was used as the centrifugation media instead of sucrose, so that the gradient more closely reflected physiological osmolarity conditions (e.g., an 18% Optiprep solution is 0.35 osM, while 60% sucrose is 1.75 osM). Almost all of the Gag protein shifted to the bottom of the gradient in the presence of 1% NP-40 (Fig. 6B) or CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} (not shown). An identical result was obtained in Jurkat T cells (Fig. 6C), where the level of Gag expression is 15- to 20-fold lower than in COS-1 cells. We conclude that at steady state, Gag protein is primarily membrane bound. The near-iso-osmolar conditions of Optiprep allow these gradients to be more sensitive than sucrose gradients for observation of density shifts.
Newly synthesized Gag is partially detergent resistant but progresses to a detergent-sensitive state.
We next used Optiprep gradients to examine the sizes and localizations of Gag-containing complexes at various stages of assembly. A denucleated P100 fraction was prepared from pulse-labeled, pHXB2ΔBal-transfected cells and loaded over an Optiprep gradient in the absence or presence of 1% NP-40. In addition, the S100 fraction and a P100 fraction from cells harvested after a 4-h chase were analyzed in parallel (Fig. 7A). Gag from the S100 fraction remained mostly near the top of the gradient in both the absence and presence of detergent (Fig. 7A, top). A graph of the distributions in the presence and absence of detergent revealed essentially identical profiles (Tritel and Resh, unpublished), consistent with cytosolic localization in relatively small protein complexes. Gag derived from the P100 fraction after a 4-h chase shifted almost entirely to the bottom of the gradient tube in the presence of detergent, similar to the results obtained for total Gag (Fig. 6) and consistent with its localization in large, membrane-bound complexes. The newly synthesized Gag (P100 pulse-labeled) yielded a biphasic distribution on the gradients in the presence of detergent. The majority of the newly synthesized protein remained in the middle of the gradient, suggestive of localization in cytosolic complexes of intermediate size, between the 4-h-chased Gag and the S100 Gag. However, a small fraction shifted to the bottom of the tube, indicative of localization in a large, membrane-bound protein complex. A similar result was obtained in Jurkat T cells, with 20% of the newly synthesized Gag shifting to the bottom of the tube in the presence of 1% NP-40 (Tritel and Resh, unpublished).
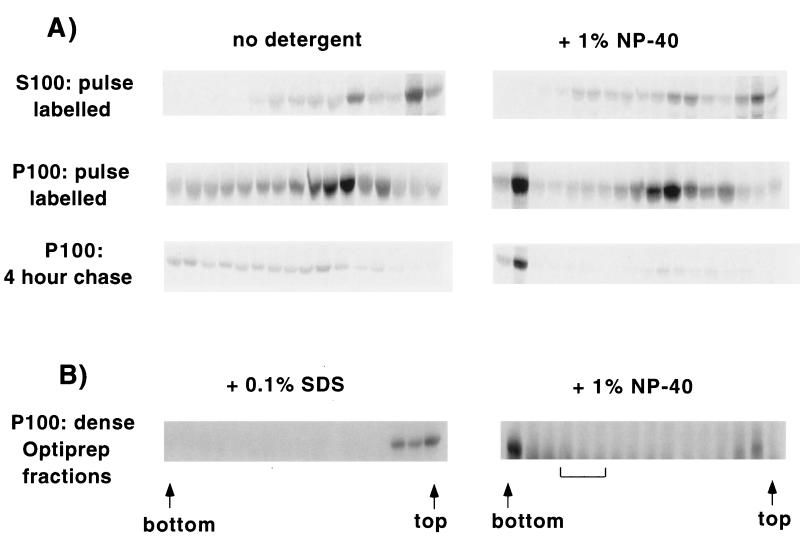
FIG. 7.
Migration of Gag on Optiprep gradients in the absence or presence of detergent. (A) (Top two rows) Transfected cells were pulse-labeled (7 min) and fractionated into S100 and denucleated P100 fractions. Each fraction was fractionated on Optiprep gradients as described in the legend to Fig. 6B in the absence or presence of 1% NP-40. Labeled protein in the gradient fractions was visualized by autoradiography. (Bottom row) The cells were pulse labeled and incubated for 4 h in chase medium. A denucleated P100 fraction was prepared and analyzed on Optiprep gradients. (B) Optiprep gradient analysis was performed on a denucleated P100 fraction from transfected cells as described in the legend to Fig. 1. The fractions corresponding to a density of 1.098 to 1.106 g/ml (brackets) were pooled, treated with 0.1% SDS (left) or 1% NP-40 (right), and fractionated on Optiprep gradients. Gag was detected in the gradient fractions by Western blotting.
Centrifugation in the presence of detergent was also used to verify that the denser fractions of the Optiprep gradient in Fig. 1A contained membrane-bound, assembled Gag complexes. To this end, we pooled the dense fractions (1.098 to 1.106 g/ml), added NP-40, and recentrifuged them through Optiprep gradients. Essentially all the Gag was found at the bottom of the centrifuge tube, indicating that the dense Optiprep fractions contained large, membrane-bound Gag complexes (Fig. 7B, right). To further verify that the Gag found in the bottom fractions of the gradients represented Gag in large protein complexes, samples were pretreated with 0.1% SDS to break protein-protein interactions. All of the Gag shifted to the top of the gradient (Fig. 7B, left).
Comparison of the detergent-containing gradients derived from the pulse-labeled sample, a 2-h chase (not shown), and a 4-h chase (Fig. 7A) revealed an interesting trend. While the intensity of the band in the pellet remained approximately constant, the intensities of bands in the middle of the gradient decreased dramatically by the 2-h chase point, disappearing almost entirely in the 4-h chase. This is suggestive of the presence of two populations of Gag: a stable population in large, membrane-bound complexes (the pellet) and an unstable population in smaller cytosolic complexes (the middle bands). We conclude that Gag assembles into large, membrane-bound complexes in a time-dependent fashion and that Optiprep gradients can be used to resolve different Gag assembly intermediates.
Most newly synthesized Gag is degraded within 2 h, and only the remaining Gag proceeds to assembly and budding.
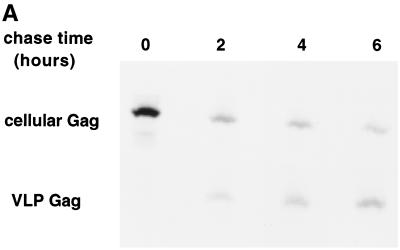
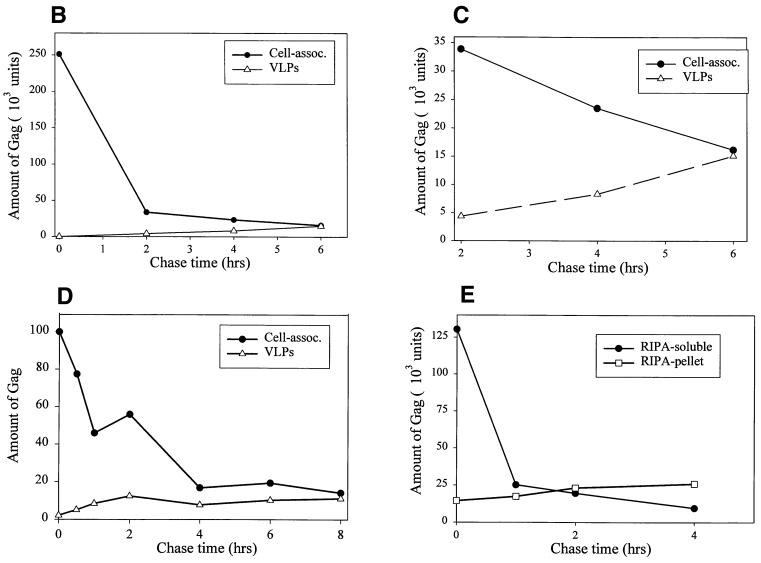
The end point of the assembly process is release of Gag from the cells into virions in the medium. If the Gag populations resolved on the Optiprep gradients represent assembly intermediates, then the kinetics of their disappearance from cellular fractions should parallel the rate of Gag appearance in VLPs. To follow these processes, cells were pulse-labeled and chased for various lengths of time, and the Gag protein remaining in the cell and released into VLPs was visualized (Fig. 8A). The amount of cell-associated, pulse-labeled Gag declined dramatically with increasing chase time; quantitation revealed that by 2 h after its synthesis, only 20% of the original Gag counts per minute was recovered from the cells (Fig. 8B). At least three mechanisms could explain the decline in the radiolabeled Gag counts per minute as a function of time: budding of Gag into VLPs, loss of Gag in the clarification step during sample preparation, and intracellular degradation. Quantitation of extracellular VLP formation in COS-1 cells revealed that very little pulse-labeled Gag was released into VLPs compared to the amount of labeled cellular Gag that had disappeared. However, a slow and steady increase in the Gag counts per minute in VLPs occurred over 2 to 6 h, and this increase closely paralleled the decrease in cellular Gag counts per minute during the same time frame (Fig. 8C). A similar pattern was observed for the loss of cell-associated Gag counts per minute in Jurkat T cells; 80% of the Gag counts per minute was lost from the cells, with only 15 to 20% of the counts per minute recovered as extracellular VLPs (Fig. 8D). The time course of VLP production in Jurkat cells was faster than that in COS-1 cells.
FIG. 8.
Kinetics of disappearance of Gag from cells and its appearance in VLPs. COS-1 cells were pulse-labeled and incubated in chase medium for various lengths of time. The amount of Gag remaining in the cells and in extracellular VLPs was determined by immunoprecipitation and autoradiography. (A) A representative experiment. Medium from twice the number of cells was harvested when producing VLPs. (B) Quantitation of the experiment shown in panel A. (C) Graph of the 2- to 6-h time points from panel B plotted in isolation. (D) The same experiment was repeated with Jurkat T cells transfected with pHXB2ΔBal. (E) The experiment described in panels A to C was repeated, this time retaining the RIPA-insoluble pellet from the clarification step. The pellet was extensively sonicated and immunoprecipitated with anti-CA antibody in parallel with the cell-associated RIPA-soluble fraction.
Since the cellular lysates had been clarified prior to immunoprecipitation, we also examined the clarification pellet for the presence of Gag counts per minute. In both COS-1 cells (Fig. 8E) and Jurkat T cells (not shown), less than 20% of the original Gag counts per minute was recovered from this pellet. In addition, Gag counts per minute in the clarification pellet increased during the time course of the chase, implying that this fraction reflects a time-dependent accumulation of large, insoluble Gag aggregates that do not participate in the assembly process. Since the appearance of Gag counts per minute in neither VLPs nor the clarification pellet can account for the large loss of Gag counts per minute from the pulse-labeled lysate, we conclude that most of the newly synthesized Gag is degraded intracellularly. The Gag that is assembled into VLPs is therefore likely derived from the membrane-bound assembly intermediates characterized in this study.
DISCUSSION
In this study, we have used density gradient centrifugation to resolve distinct intermediates in the process of HIV-1 virion particle assembly. A combination of pulse-chase analysis and biochemical techniques allowed us to temporally order these intermediates and to characterize their sizes and intracellular localizations. Specifically, we have shown a number of differences between the behavior of the newly synthesized and steady-state (total) Gag populations. First, newly synthesized Gag migrates to lighter fractions of an Optiprep velocity gradient than does total Gag (Fig. 1A and B). In addition, a substantial portion of the newly synthesized Gag remains in the middle of the Optiprep gradient after treatment with nonionic detergent, displays protease sensitivity, and remains on the bottom of sucrose flotation gradients. In contrast, nearly all of the total Gag population shifts to the bottom of the Optiprep gradient after detergent treatment, is resistant to protease, and floats up through sucrose gradients (Fig. 4, 5B, and 7). (For the sake of simplicity, the population of newly synthesized Gag that is protease sensitive, etc., will be referred to as “population A”; the population representative of steady-state Gag will be designated “population B”). We have also shown that population A disappears within 2 h of synthesis, with no corresponding increase in population B or in extracellular VLPs (Fig. 8). Population B, by contrast, disappears with kinetics roughly paralleled by the appearance of Gag in VLPs. Finally, we have shown that, as a function of time, intracellular Gag migrates to increasingly dense fractions on Optiprep gradients and that after 6 h, the mobility of these fractions resembles that of VLPs (Fig. 1 and 2). Taken together, these data suggest that a substantial proportion of newly synthesized Gag exists in population A, which does not proceed to true assembly intermediates but instead is presumably targeted for degradation. Moreover, the remaining Gag protein, population B, consists of a number of membrane-bound assembly intermediates. These assembly intermediates can be resolved by velocity centrifugation. Nearly identical results were obtained in Jurkat cells, where the amount of Gag is 20-fold lower than in COS-1 cells. Thus, the level of Gag expression does not appear to influence the overall assembly pathway.
The majority of total Gag at steady state is membrane bound.
We have used three independent biochemical criteria to demonstrate that most of the total Gag population present in the P100 fraction is indeed membrane bound. First, addition of nonionic detergent resulted in a dramatic shift of nearly all the Gag to the bottom of an Optiprep velocity gradient, as expected for a large, membrane-bound protein complex (Fig. 6B). Second, the majority of the total Gag in the P100 fraction displayed protease resistance, as previously reported (16), and became protease sensitive only when the membranes were permeabilized with detergent (Fig. 4). These findings imply that Gag is enveloped in membrane vesicles. It is unlikely that cytosolic complexes of Gag are trapped inside membrane vesicles during homogenization, because β-Gal, a cytosolic protein which forms a tetramer of ∼440 kDa, fractionates primarily in the S100 fraction (Fig. 3B). It is interesting to note that Gag was mostly protease resistant while another protein that is also bound to the inner surface of the plasma membrane, Raf–K-Ras (6), was mostly degraded (not shown). The difference in the protease sensitivities of the two proteins may reflect Gag-induced curvature of the membrane, favoring right-side-out vesicularization of the Gag-containing vesicles. Regions of the plasma membrane containing Gag multimers would exclude other cellular membrane proteins and vesicularize separately, which also explains the ability of assembly domains at different stages to be resolved by centrifugation (see below). Protease resistance provides a conservative estimate of the proportion of Gag that is membrane bound, since some Gag is likely to be associated with inside-out plasma membrane vesicles. The third assay involved flotation through sucrose cushions. Nearly all the steady-state Gag floated to the interface, as expected for a membrane-bound protein (Fig. 5B). Taken together, these data clearly indicate that the majority of the total Gag population is membrane bound.
Several investigators have recently used detergent-resistant migration on sucrose gradients and failure to float on sucrose cushions to argue that a large proportion of Gag is present in intracellular cytosolic complexes (20, 23, 24, 29). Our data, as well as that of Spearman et al. (30), strongly argue that the majority of the total Gag at steady state is membrane bound. The greater resolution afforded by gradients spanning a narrower range of densities increased the sensitivity of our assays. Other investigators used sonication during membrane preparation (23), which may have resulted in the release of membrane-bound proteins. Moreover, some investigators analyzed only the pelleted material from sucrose gradient fractions (20, 29), whereas we utilized the entire sample for analysis. We cannot, however, exclude the possibility that differences in cell type, virus subtype, or membrane preparation methods account for differences in the experimental results. It is also possible that the presence of HIV-1 protease could influence the assembly pathway, although this would likely occur during the late stages of assembly (17).
Newly synthesized Gag rapidly forms cytosolic complexes that copellet with the P100 fraction.
The time course experiments shown in Fig. 3 demonstrate that newly synthesized Gag rapidly localizes to the P100 fraction in a manner that is independent of other viral proteins and downstream Gag oligomerization domains. Several criteria were used to distinguish membrane-bound Gag from cytosolic Gag complexes in the P100 fraction. Approximately 20% of the pulse-labeled Gag floated to the interface of a discontinuous sucrose gradient (Fig. 5) and shifted to the pellet of a continuous Optiprep gradient when detergent was added (Fig. 7). These are characteristics of membrane-bound proteins and likely reflect the presence of a population of plasma membrane-bound protein among the newly synthesized Gag. However, the majority of pulse-labeled Gag exhibited sensitivity to protease digestion and remained at the bottom of the tube in sucrose flotations (Fig. 4 and 5), suggesting that most of the newly synthesized Gag exists in cytosolic complexes that are detergent resistant (Fig. 7).
Most of the newly synthesized Gag is degraded intracellularly.
Quantitation of the levels of Gag at different times after synthesis revealed that ∼80% of the Gag disappeared within 2 h, followed by slower disappearance of the remaining Gag (Fig. 8B and C). The degradation of most of newly synthesized Gag explains the low efficiency of HIV-1 budding that has been observed (34). The rapid disappearance in the first 1 to 2 h occurred selectively in population A. In the sucrose flotation assay, the labeled Gag at the bottom of the tube was depleted within 2 h (Fig. 5A). Similar trends were observed in the Optiprep gradients. In the presence of detergent, the Gag population in the middle fractions disappeared almost entirely within 4 h, while in the absence of detergent, Gag counts per minute in the lighter fractions of the gradient (∼1.06 g/ml) were selectively depleted (Fig. 7A). These data indicate that population A has a relatively short half-life (less than 1 h) while population B has a half-life of several hours. Most of the total Gag population is present in population B.
Optiprep gradients resolve membrane-bound assembly intermediates.
Pulse-chase analysis combined with Optiprep gradients (Fig. 3A) revealed that Gag progressively migrated to denser gradient fractions as a function of time. It should be noted that a subtle density difference between total and newly synthesized Gag was previously reported (32) but was not further characterized. Several lines of evidence indicate that increasing assembly drives the progression to denser gradient fractions. (i) An assembly-deficient Gag mutant fails to display the same behavior (Fig. 3B). (ii) Gag takes 4 to 6 h to migrate through the gradient, similar to the kinetics of its appearance in VLPs (Fig. 3A and 8D). (iii) At later chase points, Gag migrates almost identically to VLPs, suggesting that the Gag-containing cellular domains resemble VLPs (Fig. 3B). (iv) The Gag in the denser fractions exists entirely in large complexes (Fig. 8B). We therefore believe that these gradients separate complexes based on their degrees of assembly. Chemical cross-linking with a homobifunctional cross-linker confirmed that Gag multimers were present in the P100 fraction (Tritel and Resh, unpublished).
Taken together, these data are consistent with the following model for lentivirus assembly. Most of the newly synthesized Gag proceeds to population A, which consists of cytosolic complexes that copellet with the P100 fraction. These complexes are characterized by protease sensitivity, resistance to detergent, and failure to float through a sucrose cushion. Population A is rapidly degraded, and the rate of disappearance is not paralleled by the appearance of Gag in VLPs. The rest of the newly synthesized Gag is present in population B, consisting of large, membrane-bound complexes that also pellet with the P100 fraction. Gag appears in VLPs at approximately the same rate as the loss of population B (Fig. 8B and D). Thus, it is likely that population B represents true assembly intermediates, although the possibility that a minority of the Gag in population A proceeds to population B or VLPs cannot be excluded. At the plasma membrane, Gag complexes undergo increasing multimerization, producing large, dense arrays of Gag protein under the membrane. As Gag multimerization continues, the membrane is deformed outward, host cell proteins are excluded, and other viral components are recruited to the site of assembly. As a result, the protein-to-lipid ratio, and therefore the density of the Gag-containing membrane assembly domains, increases. The sequential assembly complexes isolated on the Optiprep gradients (Fig. 2) therefore likely reflect increasing formation of Gag multimers within assembly domains at the membrane.
In conclusion, we report the novel separation and characterization of sequential in vivo HIV-1 assembly intermediates. These intermediates are large, membrane-bound, Gag-containing complexes. The length of time required for assembly may reflect the transport of Gag to specialized domains in the plasma membrane, Gag multimerization, or recruitment of other viral components into the budding particle. Clearly, the processes involved represent potential targets for therapeutic intervention. Future studies will exploit this system for monitoring HIV-1 assembly to test the role of energy metabolism and identify the cellular and viral components required for assembly.
ACKNOWLEDGMENTS
We thank Luz Hermida-Matsumoto and Wouter van't Hof for helpful discussions and critical reading of the manuscript, Raisa Louft-Nisenbaum for technical assistance, and Debra Alston for secretarial support.
This research was supported by NIH grant CA 72309, as well as NIH Core Grant P30-CA08748.
REFERENCES
- 1.Alland L, Peseckis S M, Atherton R E, Berthaiume L, Resh M D. Dual myristylation and palmitylation of Src family member p59Fyn affects subcellular localization. J Biol Chem. 1994;269:16701–16705. [PubMed] [Google Scholar]
- 2.Andersson S, Davis D N, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P 450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choy E, Chiu V K, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov I E, Philips M R. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 7.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 9.Garnier L, Bradford Bowzard J, Wills J W. Recent advances and remaining problems in HIV assembly. AIDS. 1998;12:S5–S16. [PubMed] [Google Scholar]
- 10.Garoff H, Hewson R, Opstelten D E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 12.Gheysen D, Jabcos E, DeForesta F, Thiriart C, Francotte M, Thines D, DeWilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 13.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson L E, Bowers M A, Sowder R C, Serabyn S A, Johnson D G, Bess J W, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermida-Matsumoto L, Resh M D. Human immunodeficiency virus type I protease triggers a myristoyl switch that modulates membrane binding of Pr55gag and p17MA. J Virol. 1999;73:1902–1908. doi: 10.1128/jvi.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan A H, Swanstrom R. Human immunodeficiency virus type 1 Gag proteins are processed in two cellular compartments. Proc Natl Acad Sci USA. 1991;88:4528–4532. doi: 10.1073/pnas.88.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y M, Liu B, Yu X F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T Cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 21.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morikawa Y, Goto T, Sano K. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J Biol Chem. 1999;274:27997–28002. doi: 10.1074/jbc.274.39.27997. [DOI] [PubMed] [Google Scholar]
- 23.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paillart J C, Gottlinger H G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picard D, Yamamoto K R. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resh M D, Erikson R L. Highly specific antibody to Rous Sarcoma Virus src gene product recognizes a novel population of pp60v-src and pp60c-src molecules. J Cell Biol. 1985;100:409–417. doi: 10.1083/jcb.100.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber B K, Pavlakis G N. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon J H M, Carpenter E A, Fouchier R A M, Malim M H. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J Virol. 1999;73:2667–2674. doi: 10.1128/jvi.73.4.2667-2674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suomalainen M, Hultenby K, Garoff H. Targeting of Moloney murine leukemia virus gag precursor to the site of virus budding. J Cell Biol. 1996;135:1841–1852. doi: 10.1083/jcb.135.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van't Hof W, Resh M D. Rapid plasma membrane anchoring of newly synthesized p59fyn: selective requirement for NH2-terminal myristoylation and palmitoylation at cysteine-3. J Cell Biol. 1997;136:1023–1035. doi: 10.1083/jcb.136.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weclewicz K, Ekstrom M, Kristensson K, Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72:2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]